
Chikungunya Virus Inhibitor Study based on Molecular Docking
Experiments
A. A. Saraiva
2,6 a
, Soares Jeferson
1 b
, Castro Miranda
1 c
, Jose Vigno Moura Sousa
1 d
,
N. M. Fonseca Ferreira
3,4 e
, J. E. S. Batista Neto
6
, Salviano Soares
3 f
and Antonio Valente
2,5 g
1
UESPI - University of State Piaui, Piripiri, Brazil
2
University of Tr
´
as-os-Montes and Alto Douro, Vila Real, Portugal
3
Coimbra Polytechnic - ISEC, Coimbra, Portugal
4
Knowledge Engineering and Decision-Support Research Center (GECAD) of the Institute of Engineering,
Polytechnic Institute of Porto, Portugal
5
INESC-TEC Technology and Science, Porto, Portugal
6
University of S
˜
ao Paulo, S
˜
ao Carlos, Brazil
Keywords:
Chikungunya, Molecular, Docking.
Abstract:
Chikungunya virus disease transmitted by the sting of the mosquito ’Aedes aegypti’ presenting an epidemic in
some regions. In order to have an early diagnosis and the best treatment technique, it establishes the study of
inhibitors for laboratory elaboration of a drug from molecular docking. As a result you have a better chance
of using Suramin followed by Silibin.
1 INTRODUCTION
Numerous factors influence the proliferation of the
Aedes Aegypti mosquito such as standing water and
street litter. This spread of the mosquito is worri-
some because it is the cause of numerous diseases
such as Dengue and Chikungunya (CHIKV). The cur-
rent context is very apprehensive, because according
to (Weaver et al., 2012). So far there is no antiviral
treatment for virus infection and no vaccine licensed
to totally inhibit it.
According to (Monath, 2018). There are sev-
eral clinical and antiviral studies under development
to combat Aedes Aegypti transmitted diseases and
may provide further specifications for the fight against
CHIKV in the future.
The structure used to represent the CHIKV virus
was obtained from the work of (Voss et al., 2010), that
a
https://orcid.org/0000-0002-3960-697X
b
https://orcid.org/0000-0002-0586-4786
c
https://orcid.org/0000-0002-7751-9455
d
https://orcid.org/0000-0002-5164-360X
e
https://orcid.org/0000-0002-2204-6339
f
https://orcid.org/0000-0001-5862-5706
g
https://orcid.org/0000-0002-5798-1298
by X-ray crystallography a model of glycoproteins
E1 and E2 represented in 3D format was obtained as
shown in the figure. 1 .
Figure 1: Chikungunya E1 E2 Envelope Glycoproteins.
Therefore, the present work aims to employ com-
puter simulation in different substances in order to
present the most efficient inhibitor of Chikungunya
virus. The objective of this research is to establish the
relationship of the selected molecules with the virus
and then to hypothesize new drugs through molecular
docking techniques.
Molecular docking is important to predicting the
best instruction aiming to adjust a linker to a protein,
200
Saraiva, A., Jeferson, S., Miranda, C., Sousa, J., Ferreira, N., Neto, J., Soares, S. and Valente, A.
Chikungunya Virus Inhibitor Study based on Molecular Docking Experiments.
DOI: 10.5220/0009118602000205
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 3: BIOINFORMATICS, pages 200-205
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
also characterizing the behavior of small molecules in
connection with the target proteins. Using this virtual
technique, it will be possible to propose structural hy-
potheses of how ligands are connected to their targets
and whether they are possible inhibitors of the virus
in question.
In this context, obtaining Chikungunya protein
was obtained through the work of (Voss et al., 2010)
and ligands through cubichem database.
Research on drugs to inhibit the Chikungunya
virus is relevant to both the scientific and social
spheres. Since most of the population has had Aedes
Aegypti mosquito-related diseases, but so far there is
no vaccination or drug that can inhibit the spread of
the virus in an individual’s body.
The paper is divided into five sections, section 2
deals with possible virus inhibitors, while section 3
emphasizes the computational technique used in the
work, Molecular Docking. In section 4 we describe
the methodology of the work, in section 5 the results
obtained through the simulations and finally the con-
clusion.
2 CHIKV VIRUS INIBITORS
According to (Monath, 2018) The evolution and
spread of the virus is a worldwide concern and the
vaccine is the main aspect that can alleviate epi-
demics. Given this context, there are several options
for virus inhibitors to analyze in different ways. In
this work Andrographolide, Epigallocatechin gallate,
Harringtonine, Silibinin and Suramin were used.Such
inhibitors will be classified below.
Andrographolide is a labdane diterpene produced
by the plant Andrographis paniculata, which has a
wide range of therapeutic applications such as anti-
inflammatory and platelet aggregation activities and
potential antineoplastic properties. (Gupta et al.,
2018) employs Andrographolide as an important
bioactive with anti-inflammatory properties, in its
work the compounds found in this substance reduce
inflammation in various diseases. In research con-
ducted (Gupta et al., 2018),showed the effect of in
vitro research of andrographolide in action on the
treatment of CHIKV virus, these experiments yielded
encouraging results.
Being a compound obtained from green tea, epi-
gallocatechon gallate (EGCG) has inhibitory effects
on various viruses. (Weber et al., 2015) described
it as an antiviral compound for a variety of viruses,
although the exact mechanism of the inhibitory ef-
fects are not yet understood, it was included in this
research as a candidate for a future drug capable of
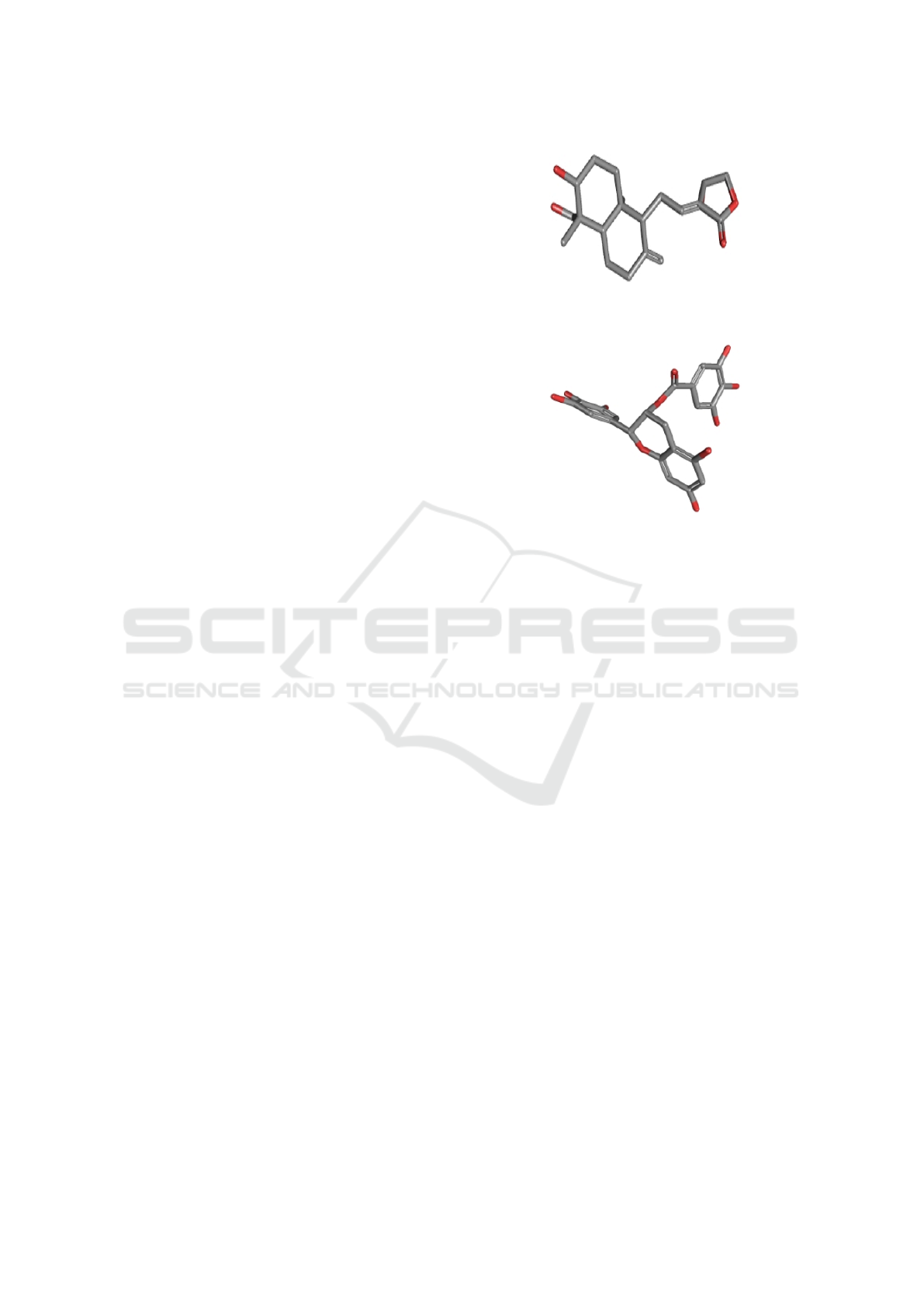
Figure 2: Andrographolide 3D Structure. Source: Pub-
Chem. (PubChem, 2005).
Figure 3: Epigallocatechin gallate 3D structure. Source:
PubChem. (PubChem, 2005).
fighting the Chikugunya virus shown in the figure.
3. (Raekiansyah et al., 2018) reported how difficult
it is to develop an effective and safe vaccine to com-
bat dengue mosquito-borne diseases. In this work, we
investigated the combination of EGCG treatment with
suramin drug, this context increased chikungunya in-
hibition.
Harrigtonine is a substance of the alkaloid family,
where they are derived mainly from plants, but may
also be derived from fungi, bacteria and even animals.
(Kaur et al., 2013) conducted a study on Harringto-
nine’s action in inhibiting CHIK cell replication and
subsequently confirmed its effectiveness against the
virus. In this study the results indicated that harring-
tonine acts in the post-initial stage of CHIKV replica-
tion and strongly interferes with the viral protein syn-
thesis process. Given this, Harrigtonine is a strong
candidate for research on efficient inhibitors of the
CHIKV virus. In the figure 6 Harrigtonine molecule
in 3D format obtained from the Pubchem database is
presented.
A compound of the flavonoid family, silibinin
is used to treat a variety of diseases such as hep-
atitis, liver cirrhosis, and chemical fig-leaf injury.
(Lani et al., 2015) carried out research with different
flavonoid types among them silibinin, where it pre-
sented promising results for CHIVK virus inhibition.
Suramin is a polyanionic compound with an un-
known mechanism of action. It is used parenterally in
the treatment of African trypanosomiasis and clinical
hypotheses have been created to be used as a CHIVK
Chikungunya Virus Inhibitor Study based on Molecular Docking Experiments
201
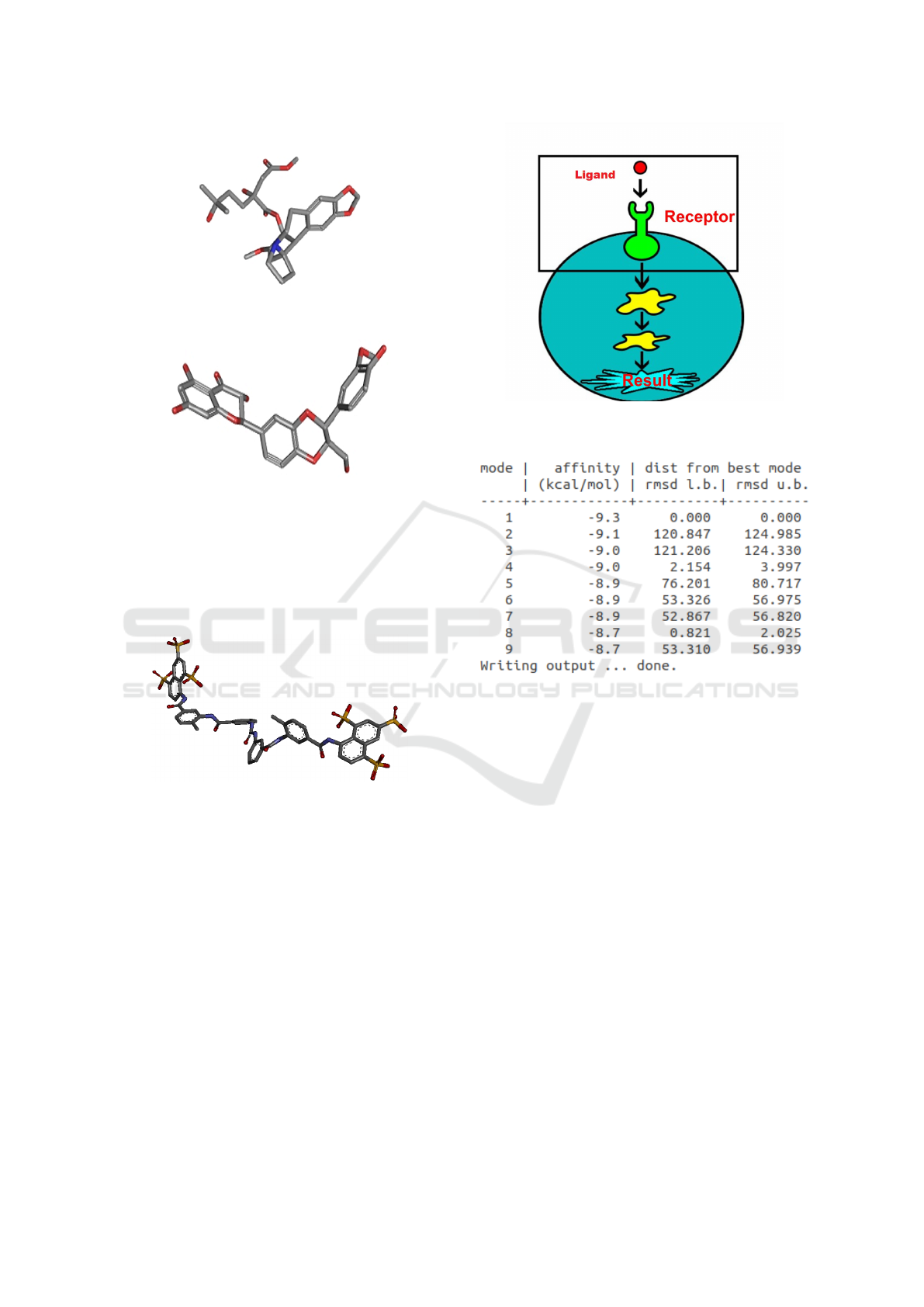
Figure 4: Harringtonine 3D Structure. Source:PubChem.
(PubChem, 2005).
Figure 5: Estrutura 3D Silibinin. Source: PubChem. (Pub-
Chem, 2005).
inhibitor. According to (Albulescu et al., 2015) In in
vitro experiments suramin proved to be an antipara-
sitic drug, which obtained satisfactory results in the
inhibition and replication of CHIKV and other al-
phaviruses.
Figure 6: Suramin 3D structure. Source: RCSB PDB.
(Berman et al., 2000).
3 MOLECULAR DOCKING
(Ferreira et al., 2015),says molecular docking is a ver-
satile computational technique for the study of biolog-
ical macromolecules, this technique studies the pro-
duction of drugs based on molecular structures where
they are simulated through numerical interactions by
algorithms, where the objective is to predict the bound
conformations and binding affinity. between receptor
and ligand.
Docking can be defined as a ”key-lock” problem,
which has the purpose of predicting the modes of in-
teraction between two molecules knowing only their
isolated three-dimensional structures.
Figure 7: Illustration of the ligand with its receptor
molecule.
Figure 8: Ligand-Receiver Affinity Result Example.
Ligands are molecules produced by cells that in-
teract like a puzzle with its receptor as shown in Fig-
ure 7. The receptor, on the other hand, is the target
protein in which the interaction between the parts to
verify compatibility information is desired.
The practice of this coupling method is for identi-
fication and characterization of the binding sites in the
target proteins, generating evaluation values of the in-
teraction potential between the target and the ligand.
In the figure 8 There is an example of a table show-
ing the results of a simulation performed in autodock
vina, which is showing affinity values.
Also according to (Ferreira et al., 2015), The soft-
ware combines two main components: search algo-
rithm and the score function, in which the algorithm
is responsible for searching for possible combinations
in the links and the score demonstrates the best bind-
ing results obtained during the procedure. The algo-
rithms allow the exploration of various angles, both
rotational, translational and conformational of the lig-
and in the target protein.
In the image 8, a result table is exemplified af-
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
202
ter the molecular dock on the software autodock
vina.This table shows the nine best ligand-to-receptor
nesting results, where the first column shows the se-
quence of the numbered results and the second col-
umn shows the binding affinity in kcal / mol, repre-
senting the highest energy. In the next columns, two
variants of RMSD metrics are provided: the rmsd/lb
(lower limit of RMSD) an the rmsd/ub (upper limit
of RMSD). The rmsd/ub combines each atom in one
conformation with itself in the other conformation,
ignoring any symmetry, whereas rmsd/lb is defined
as follows: rmsd/lb (c1, c2) = max(rmsd’(c1, c2),
rmsd’(c2, c1)).
4 METHODOLOGY OF WORK
The methodology has two main objectives: prediction
of conformation, and binding affinity.
In the simulations developed in this work the fol-
lowing configurations were used: core i7 fourth gen-
eration, 12 GB ram, nvidia p6000 video card, HD Sdd
240 gb.
The molecules presented as ligands in this work
were extracted from PubChem, a highly diverse
database of molecules maintained by the National
Center for Biotechnology Information. The receivers
were obtained through the work of (Voss et al., 2010),
where the chikungunya virus glycoproteic structure is
available in the RCSB PDB Prontein data bank.
The experiment consists in performing the molec-
ular docking simulations using the molecules pre-
sented in the section 3, together with the target pro-
tein (CHIKV virus). From the results of these exper-
iments, we seek to investigate which ligand had the
best protein affinity to stipulate a better candidate for
chikungunya inhibitor.
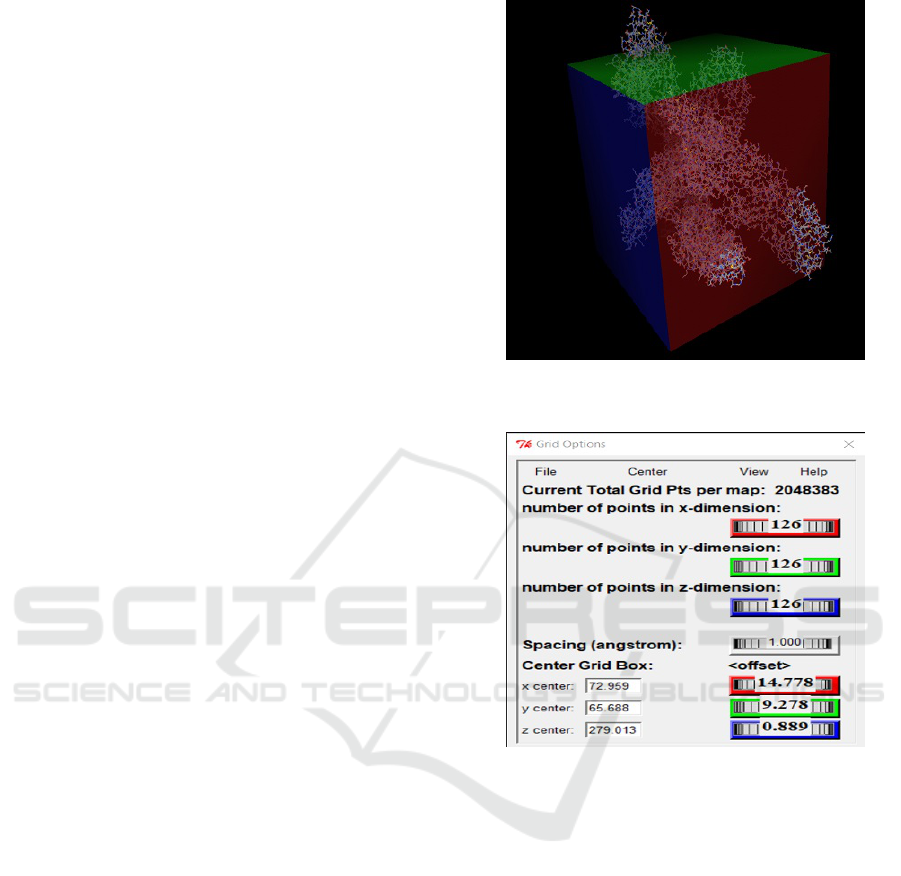
Importantly, before the molecular docking pro-
cess, the protein goes through a mapping phase of the
region where the software must perform the ligand-
protein docking simulations. (Vina, 2010) Called this
function AutoGrid where you can manually choose
the best ligand coupling location, this process will
avoid unnecessary processing effort as the region size
to try to couple is smaller. As shown 9The entire pro-
tein area was used, in which the parameters shown
in the image were selected for precise ligand fitting
at the best protein site to obtain the best quadrant in
common with all ligands.
The coordinates, that is, the parts of the proteins
where the ligand will dock, are presented numerically
in the image. 10.
To perform the molecular docking process was
used autodock vina (performing simulations with re-
Figure 9: Grid Box Example on Chikungunya Virus
Molecule.
Figure 10: Grid Box Coordinates at Chikungunya Virus
Molecule.
ceptor and ligands), Mgltools (format conversion of
molecules) and PyMol (for visualization of results).
5 RESULTS
On the table 1, The best results of molecular docking
among ligands are shown. The results that released
the most energy (represented by the lowest value re-
sults) are the best ligand-receptor fittings. (Shityakov
and F
¨
orster, 2014) explains that the lower the value
presented more significance it will present to the bind-
ing found, in which the affinity values in the molecu-
lar docking process are favorable only when they are
negatively represented. That is, the more negative the
value obtained, the better the interaction.
According to table has suramin as a result of lower
value. And as a demonstration of your connection you
Chikungunya Virus Inhibitor Study based on Molecular Docking Experiments
203
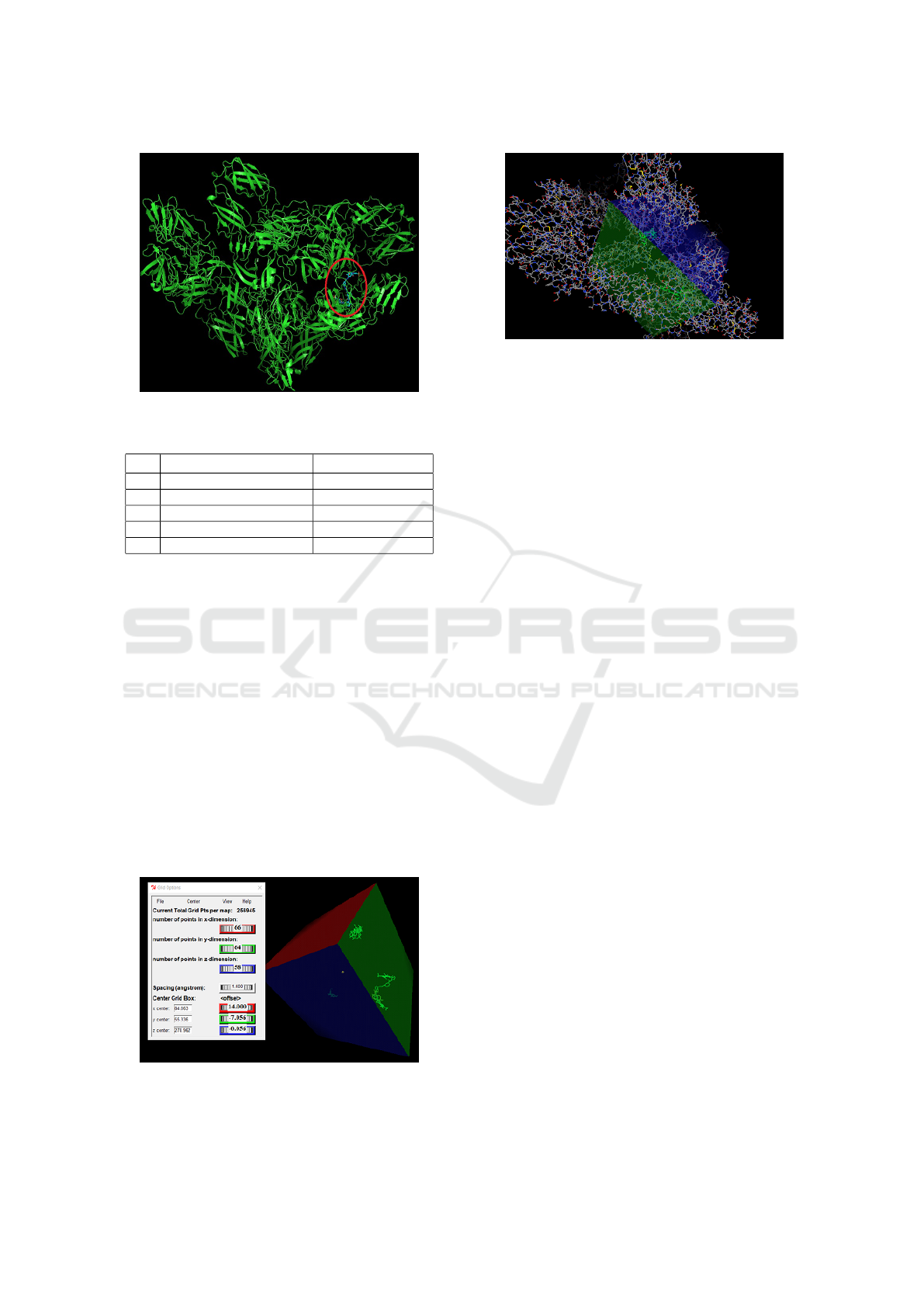
Figure 11: Suramin + Chivk.
Table 1: Affinity Table Of CHUCK Ligands.
Ligante Affinity(kcal/mol)
(a) Andrographolide -7.9
(b) Epigallocatechin gallate -9.1
(c) Harringtonine -8.3
(d) Silibinin -9.4
(e) Suramin -12.7
have the image 11 which represents the Suramin lig-
and on the protein. In green the main target, the chiku-
gunya virus and in blue the suramin ligand circled in
red.
Another interesting factor to note is the quad-
rant where the active molecules (Andrographolide,
Epigallocatechin gallate, Harringtonine, Silibinin and
Suramin) had the highest binding value. In the fig-
ure 13, the best connecting regions are presented,
in which the molecules are coupled, and from these
strands can test other possible inhibitors in the quad-
rant obtained. Since it was observed that in this quad-
rant the melecules obtained higher binding value. Af-
ter this phase, one can propose the place where the
protein will release the most energy and thus define
the guidelines for future inhibitors.
Figure 12: Best Results Coordinates.
Figure 13: Best ligand Results.
6 CONCLUSION
Computational receptor-ligand docking methodolo-
gies are very important tools in the intelligent plan-
ning of new drugs. The applied methodology al-
lowed comparing the inhibitors for chikungunya virus
through molecular docking techniques, where the in-
hibitor that obtained the best prediction results of con-
formation and binding affinity in this simulation was
Suramin followed by Silibin.
The conclusion is observed in the values presented
in the table 1 que The best result of all ligands was
suramin, where it presented -12.7 protein affinity, the
binding models obtained are divided into files for
multimodal visualization in three-dimensional format
to observe where it best fit the protein.
As a continuation of this work consists in in vitro
simulations of the combination of these inhibitors
with other substances. It can also, as a suggestion of
future work, consider the use of other ligands in the
coordinates presented in this work, to verify an opti-
mization.
ACKNOWLEDGMENTS
The elaboration of this work would not have been
possible without the collaboration of the Engineering
and Decision Support Research Center (GECAD) of
the Institute of Engineering, Polytechnic Institute of
Porto, Portugal and FAPEMA.
REFERENCES
Albulescu, I. C., Van Hoolwerff, M., Wolters, L. A., Bot-
taro, E., Nastruzzi, C., Yang, S. C., Tsay, S.-C.,
Hwu, J. R., Snijder, E. J., and Van Hemert, M. J.
(2015). Suramin inhibits chikungunya virus replica-
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
204

tion through multiple mechanisms. Antiviral research,
121:39–46.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat,
T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E.
(2000). The protein data bank. Nucleic acids research,
28(1):235–242.
Ferreira, L., dos Santos, R., Oliva, G., and Andricopulo, A.
(2015). Molecular docking and structure-based drug
design strategies. Molecules, 20(7):13384–13421.
Gupta, S., Mishra, K. P., Dash, P. K., Parida, M., Ganju, L.,
Singh, S. B., et al. (2018). Andrographolide inhibits
chikungunya virus infection by up-regulating host in-
nate immune pathways. Asian Pacific Journal of Trop-
ical Medicine, 11(3):214.
Kaur, P., Thiruchelvan, M., Lee, R. C. H., Chen, H., Chen,
K. C., Ng, M. L., and Chu, J. J. H. (2013). Inhi-
bition of chikungunya virus replication by harring-
tonine, a novel antiviral that suppresses viral protein
expression. Antimicrobial agents and chemotherapy,
57(1):155–167.
Lani, R., Hassandarvish, P., Chiam, C. W., Moghaddam, E.,
Chu, J. J. H., Rausalu, K., Merits, A., Higgs, S., Van-
landingham, D., Bakar, S. A., et al. (2015). Antiviral
activity of silymarin against chikungunya virus. Sci-
entific reports, 5:11421.
Monath, T. P. (2018). Chikungunya and zika: The future.
In Chikungunya and Zika Viruses, pages 367–377. El-
sevier.
PubChem (2005). ata deposited in or computed by pub-
chem. [Online; acesses November 27, 2018].
Raekiansyah, M., Buerano, C. C., Luz, M. A. D., and
Morita, K. (2018). Inhibitory effect of the green
tea molecule egcg against dengue virus infection.
Archives of virology, 163(6):1649–1655.
Shityakov, S. and F
¨
orster, C. (2014). In silico predic-
tive model to determine vector-mediated transport
properties for the blood–brain barrier choline trans-
porter. Advances and applications in bioinformatics
and chemistry: AABC, 7:23.
Vina, A. (2010). Improving the speed and accuracy of dock-
ing with a new scoring function, efficient optimiza-
tion, and multithreading trott, oleg; olson, arthur j.
Journal of Computational Chemistry, 31(2):455–461.
Voss, J. E., Vaney, M.-C., Duquerroy, S., Vonrhein,
C., Girard-Blanc, C., Crublet, E., Thompson, A.,
Bricogne, G., and Rey, F. A. (2010). Glycoprotein
organization of chikungunya virus particles revealed
by x-ray crystallography. Nature, 468(7324):709.
Weaver, S. C., Osorio, J. E., Livengood, J. A., Chen, R.,
and Stinchcomb, D. T. (2012). Chikungunya virus and
prospects for a vaccine. Expert review of vaccines,
11(9):1087–1101.
Weber, C., Sliva, K., von Rhein, C., K
¨
ummerer, B. M., and
Schnierle, B. S. (2015). The green tea catechin, epi-
gallocatechin gallate inhibits chikungunya virus infec-
tion. Antiviral research, 113:1–3.
Chikungunya Virus Inhibitor Study based on Molecular Docking Experiments
205
