
PySpot: A Python based Framework for the Assessment of
Laser-modified 3D Microstructures for Windows and Raspbian
Hannah Janout
1 a
, Bianca Buchegger
2,3 b
, Andreas Haghofer
1 c
, Dominic Hoeglinger
2
,
Jaroslaw Jacak
2 d
, Stephan Winkler
1 e
and Armin Hochreiner
2 f
1
University of Applied Sciences Upper Austria, School of Informatics, Communications and Media,
Softwarepark 11, 4232 Hagenberg, Austria
2
University of Applied Sciences Upper Austria, School of Medical Engineering and Applied Social Sciences,
Garnisionstraße 21, 4240 Linz, Austria
3
Johannes Kepler University Linz, Institute of Applied Physics, Altenberger Straße 69, 4040 Linz, Austria
Keywords:
Image Processing Methods, Fluorescence Microscopy, 3D Microstructures, Image Analysis, Software
Development, Bioinformatics.
Abstract:
Biocompatible 3D microstructures created with laser lithography and modified for optimal cell growth with
laser grafting, can imitate the 3D structure cells naturally grow in. For the evaluation of the quality and success
of those 3D microstructures, specialized software is required. Our software PySpot can load 2D and 3D images
and analyze those through image segmentation, edge detection, and surface plots. Additionally, the creation
and modification of regions of interest (ROI) allow for the quality evaluation of specific areas in an image by
intensity analysis. 3D rendering allows for identifying complex geometrical properties. Furthermore, PySpot
is executable on Windows as well as on Raspbian, which makes it flexible to use.
1 INTRODUCTION
1.1 Cell Research using Laser
Generated and Manipulated
Structures
2D and 3D writing of biocompatible microstructures,
as well as surface modification of such structures,
have gained importance in the fields of cell research,
material science, and biophysics. One way to create
such a microstructure is the technique of laser lithog-
raphy, where micrometer- and nanometer-sized struc-
tures can be fabricated (Maruo S., 1997)(Kawata S.,
2001). Functionalized structures, e.g., improving
the imitation of natural cell growth, can be achieved
either by using different materials (Wollhofen R.,
2017) (Buchegger B., 2019) or by surface modifi-
cation of the written structures, called laser grafting
a
https://orcid.org/0000-0002-0294-3585
b
https://orcid.org/0000-0003-4346-8415
c
https://orcid.org/0000-0001-6649-5374
d
https://orcid.org/0000-0002-4989-1276
e
https://orcid.org/0000-0002-5196-4294
f
https://orcid.org/0000-0002-0027-1535
(Ovsianikov A., 2012). Laser grafting provides a se-
lective and precise way to apply coating and function-
alization even to the tiniest forms of objects. Often,
the functionality and success of these microstructures
can be analyzed by using a fluorescence microscope.
Images obtained with fluorescence microscopes have
to be interpreted with the appropriate software. In this
work, we introduce PySpot – an easy-to-use image
analysis tool for the use on Windows as well as Rasp-
bian.
1.2 Problem Definition and Goals
Images taken using a fluorescence microscope can
come, amongst others, in the form of SPE or Tiff
files, which are 2D image file formats. SPE files come
from the Princeton Instruments CCD Cameras, while
TIFF files are a file format type, which stores raster
graphics images. Additionally, since this technique is
used for 3D areas, some images come in the form of
a python source file. Those images were created by a
C++ program implemented by the researchers at the
University of Applied Sciences in Linz. They contain
a 3D list of values, which are further processed as a
3D Numpy array and then loaded into the program.
Janout, H., Buchegger, B., Haghofer, A., Hoeglinger, D., Jacak, J., Winkler, S. and Hochreiner, A.
PySpot: A Python based Framework for the Assessment of Laser-modified 3D Microstructures for Windows and Raspbian.
DOI: 10.5220/0008948001350142
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 2: BIOIMAGING, pages 135-142
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
135
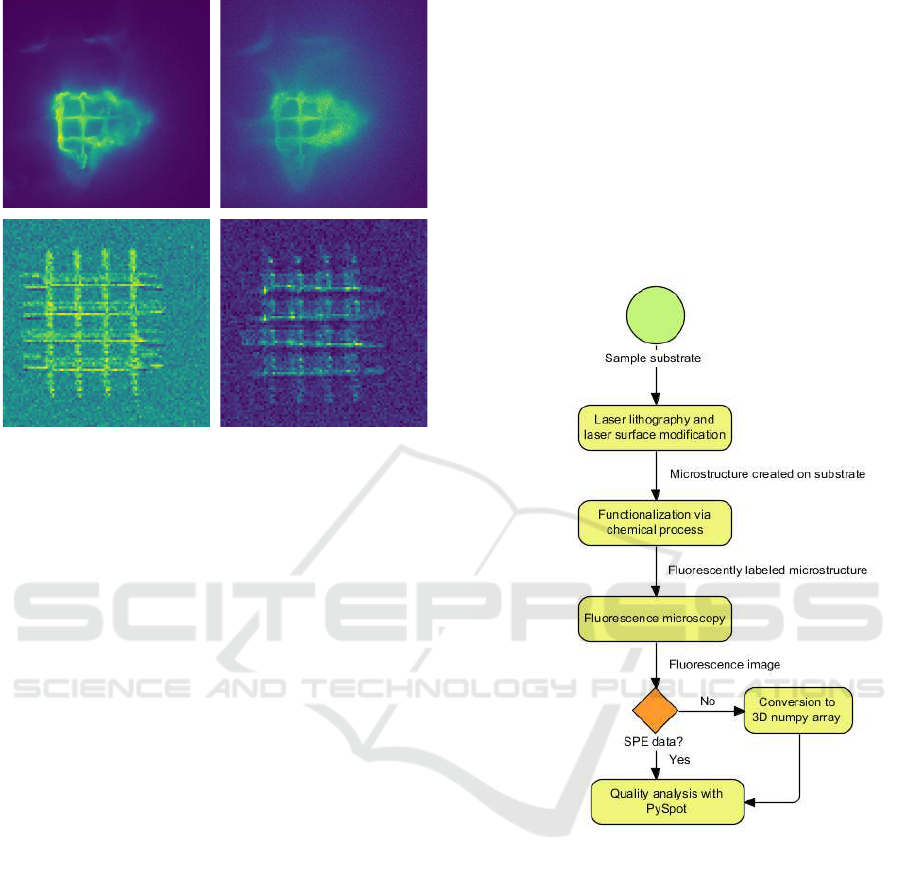
a b
c d
Figure 1: Different images resulting from the workflow de-
scribed in the chapter above. (a) and (b) show the result of
2D data, while (c) and (d) show individual layers of a 3D
image.
Figure 1 (a) and (b) show examples of 2D data
received through SPE files. Those images contain a
grid drawn through the application of laser lithogra-
phy and laser grafting. The images (c) and (d) show
different layers of a 3D microstructure originating
from a 3D Numpy array. (c) shows the third layer
of a structure, and (d) shows the fourth layer. In both
of those images, the intensities are rather low, mak-
ing it difficult to analyze the structure quality without
proper software. Thus, we require an image analy-
sis software to assess the reliability and quality of the
newly created technique. One of the most famous and
widely used image analysis tools is the open-source
processing software ImageJ. Opening and modifying
an image of the SPE format is not possible in the base
version of ImageJ and requires the installation of a
plugin. The 3D Numpy arrays cannot be opened or
modified with the base version but would require the
implementation of a custom plugin. ImageJ includes
a vast number of functionalities, making it sometimes
too complicated and unsuitable for unfamiliar users.
Another frequently used tool for image analysis is
provided by the computing environment and language
MATLAB and its image processing toolbox. While
MATLAB and its toolbox offer most functionalities
needed for the review, it requires the implementation
of a GUI from scratch and the integration of the spe-
cific functions. The variety of image formats and the
lack of easy-to-use software for image analysis raised
the need for specialized software. Our software en-
ables loading images in the form of SPE and TIFF
files, as well as Numpy arrays. Detailed analysis re-
quires image segmentation, edge detection, and the
creation and manipulation of ROIs. To evaluate the
quality of the created image, the information con-
tained inside an ROI’s borders need specific statisti-
cal evaluation by intensity analysis. PySpot is a user-
friendly graphical interface implemented in Python
for the analysis and evaluation of the 2D and 3D im-
ages. A further advantage is that PySpot is executable
on Windows as well as on Raspbian. PySpots func-
tionality, as well as its methods, are described in detail
in this paper.
Figure 2: Analysis of microstructures using imaging and
image analysis with PySpot.
2 IMPLEMENTATION
PySpot is an image analysis tool implemented in
Python and was created to easily analyze and evaluate
images of 2D and 3D laser-created microstructures,
see Figure 2. PySpot accepts data from SPE files
created by the Princeton Instruments CCD Cameras,
TIFF files, as well as in the form of 3D Numpy arrays
for analysis, and all available functionalities work for
all three formats. PySpot provides features for the
creation of regions of interest (ROI) and their analysis
through statistical evaluation. Furthermore, it allows
for the evaluation of the image through thresholding,
BIOIMAGING 2020 - 7th International Conference on Bioimaging
136

edge detection, and surface plots on Windows as well
as on Raspbian.
2.1 Cross-sections
A cross-section is defined as ,,something that has been
cut in half so that you can see the inside or a model
or picture of this.” (Rationality, 2019). In PySpot, a
line represents a cross-section. It provides informa-
tion regarding the intensities of pixels located on the
line representing the cross-section, as well as its coor-
dinates and length. Additionally, a histogram allows
for a simple visualization of the intensity difference
of the respective pixels.
2.2 Region of Interest
A region of interest (ROI) is a subset of an image, that
has been identified for a particular reason. In connec-
tion with image analysis, an ROI is a specific section
of an image that marks the region to be analyzed by
the program. In PySpot, an ROI can either be formed
like a rectangle or polygon and give information on
the ROI’s area, dimension, coordinates, and the sta-
tistical evaluation of its containing pixels.
2.3 Automatic ROI Detection
PySpot includes a feature for the automatic detection
of ROIs on the currently displayed image. This fea-
ture uses the principle of thresholding and contour
finding. Since both algorithms work with grayscale
images, the first step consists of converting the dis-
played image from RGBA to grayscale. The resulting
image will be given to a thresholding function to cre-
ate a binary image with the foreground in white and
the background in black, which will then be used to
single out the contours inside the image. The received
contours vary greatly in size and can be as small as
just one pixel. Therefore, each contour below a cer-
tain height and width is singled out. The user chooses
those limits. If a contour meets the requirements, its
points will be saved into an array, added as a new ROI,
and automatically drawn onto the image. In the case
of a 3D image, the ROIs will be searched for in the
currently displayed layer and not for every individual
one. The resulting ROIs created by this step will be
inserted in every layer of the 3D image, resulting in
a way to supervise the change in intensity through-
out the layers.Thresholding is an image segmentation
technique, which isolates certain values on an image
by converting it to a binary image. The image given
to a thresholding function needs to be in a greyscale
format, and its components will then be partitioned
into foreground and background based on their inten-
sity values. Every pixel with a value above a spe-
cific limit will be assigned a new value. Pixel with a
value equal or below the limit will be assigned a dif-
ferent value (Sahoo P., 1988). A contour is a curved
line, which connects all consecutive points with the
same value along a boundary. In image analysis, con-
tours provide a proper way to get important informa-
tion regarding object representation and image recog-
nition (Satoshi Suzuki, 1985) (Seo J., 2016). Contour
finding algorithms use binary images with white as
the color for the foreground and black for the back-
ground. Due to this requirement, it is best to convert
the desired image to greyscale and then apply thresh-
olding. Contour finding algorithms find a start point
near a white-black border and from there on track ob-
jects alongside the boundary between the white fore-
ground and black background. The contours coordi-
nates are saved in memory in the respective tracing
order (Satoshi Suzuki, 1985) (Seo J., 2016).
2.4 Definition of ROI Boundaries
Finding the right borders for ROIs can be quite diffi-
cult in certain images. Thus, PySpot includes an edge
detection feature, which will highlight the image’s ob-
ject edges. Edge detection allows for an easy way to
segment an image and visually extract data from it.
This can help in the image analysis and localization
of objects and therefore, the boundaries of potential
ROIs. The edge detection feature uses well-known al-
gorithms like the Gaussian-blur and Canny edge de-
tection. Both of those algorithms require grayscale
images. Hence, the first step consists of the image’s
conversion from RGBA to grayscale. Once it has been
converted, it is given to the Canny edge detection al-
gorithm, which uses the discontinuities of brightness
to detect the edges. Due to a great variety in the im-
ages that have to be analyzed, PySpot will automat-
ically display a second edge detection image, which
got smoothed by an additional Gaussian-blur before-
hand. This results in a smoother version of the im-
age and will make small, insignificant edges and un-
wanted noise disappear from the end-result.
The Gaussian-blur, also called Gaussian-smoothing,
is an image processing technique used to reduce noise
and the amount of detail in an image by smoothing
the intensity differences with the Gaussian function
(Gedraite E., 2011) (Deng G., 1993).
g(x, y) =
1
2πσ
2
e
−
x
2
+y
2
2σ
2
(1)
PySpot: A Python based Framework for the Assessment of Laser-modified 3D Microstructures for Windows and Raspbian
137

Due to the property of an image to display a pixel
value as one single value, the equation shown in equa-
tion 1 has to be approximated. The Gaussian-blurs
kernel decides the radius around a pixel in which the
approximation should be realized (Gedraite E., 2011)
(Deng G., 1993). Canny edge detection was devel-
oped by John F. Canny in 1986 and is a widely used
edge detection operation, which works by detecting
an image’s discontinuities in brightness. Edge de-
tection algorithms are used for the segmentation and
data extraction by tracing the boundaries of objects in-
side an image (J., 1986). This algorithm is composed
of five different steps, which can only be applied to
greyscale images. In the first step, the Canny edge
detection algorithm smooths the given image with the
Gaussian-blur function. This results in a smoother
image with less noise, which could be interpreted as
the wrong edges. Afterward, in the second step, four
different filters are being used to allow the detection
of horizontal, vertical, and diagonal edges. This is
done by applying an edge detection operator on the
image, calculating the first derivative in the horizon-
tal or vertical direction. In the next step, the edges of
the image will be thinned out. Therefore, smaller, in-
significant edges disappear. This is done by compar-
ing the intensity of each pixel with the next pixel at its
positive and negative gradient direction. If the current
pixels intensity is higher than its neighbor’s, it is be-
ing kept as an edge; otherwise, it will be suppressed.
The edges remaining after this step will be catego-
rized as either ,,strong” or ,,weak” edges based on the
value of their derivatives. To filter out the remaining
noise and the actual edges, in the last step of the algo-
rithm, pixels marked as weak will be transformed into
strong ones, as long as they are neighbors to at least
one other strong pixel in the image (J., 1986).
2.5 Using Thresholding for Improved
ROI Detection
Even through the use of the automatic ROI detection
feature or the visual boundaries achieved by edge de-
tection, finding the perfect form and size for an ROI
can be quite difficult. The automatic detection might
not find the exact ROIs desired by the user due to a
wrong set of parameters or too much noise in the im-
age. The edge detection functionality cannot always
provide a full outline of the objects, because of in-
tensity fluctuations alongside the border. One way
to improve the detection step and narrow down the
amount of potential ROIs is thresholding. A thresh-
olding functionality sets all pixels of an image to ei-
ther fore- or background based on a limit given by the
user. This creates a sharp edge between the contained
elements and their background, facilitating the pro-
cess of finding an ROI’s boundary when used as the
base image used for automatic detection.
2.6 Image Rescaling
Images acquired by the workflow can vary in size, de-
pending on the use of camera settings and the data to
be analyzed. Thus, using a fixed dimension for the
image displayed by PySpot can be quite tricky, due
to loss in quality. Therefore, images in PySpot are
displayed in their original size. While this works per-
fectly for most images, some images’ original size is
too small to see the elements clearly or to select ROIs
with the utmost precision. Therefore, PySpot includes
a feature to change the size of the images by a specific
factor between one and ten. A user can freely change
the scaling of the image and zoom in and out as they
wish to. Resizing an image can always be done during
the analysis process. Hence, all elements contained
are adapted to the new size of the image as well.
2.7 Axes Visualization
A 3D image in PySpot is displayed layer by layer,
using a scrollbar to scroll up and down the individ-
ual sections of an image alongside one axis. This al-
lows for an easy way to modify and analyze the cur-
rently displayed axis. As a side-effect, it is not possi-
ble to look at the axes besides the one currently dis-
played, requiring the need for a particular feature to
view all axes simultaneously. The axes visualized are
the XY-axis (which most analysis processes use), the
XZ-axis, and YZ-axis. A new window opens upon
starting the feature to prevent an overfilling of the
main window. This requires the implementation of an
entirely new application. This new application con-
tains three individual graphical views for each axis
to be displayed, scrollbars to allow scrolling and a
checkbox to allow switching between a colored and
grayscaled image. Upon opening the application, for
each image alongside an axis, its pixel values are ex-
tracted from the Numpy array. The extracted values
are converted into an image and appended to an ar-
ray of images. This array contains all images of an
axis. This structure allows for an easy way to scroll
through the layers of an axis. Once every image was
converted and saved into its respective array, the ap-
plication opens. Visualizing the axes in a single win-
dow.
BIOIMAGING 2020 - 7th International Conference on Bioimaging
138
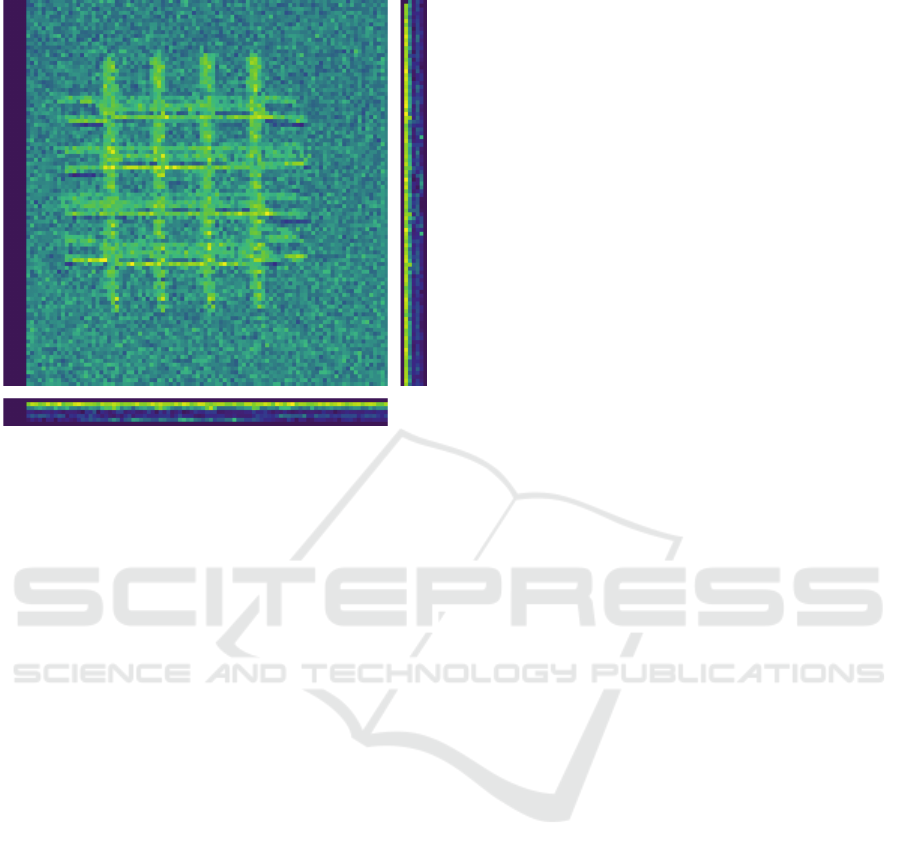
XY YZ
XZ
Figure 3: Axes visualization of 3D data. The individual
axes (XY, XZ, YZ) are displayed next to each other, giving
a great overview.
2.8 3D Rendering
Often, it is important to get a 3D understanding of the
construct of a 3D image. This cannot be visualized
simply by looking at the individual layers of the axes.
3D rendering provides an easy way to get a better 3D
understanding of the objects in the image. The 3D
rendering feature in PySpot is based on PyQtGraph,
which relies on PyOpenGL for the 2D and 3D visu-
alization of objects. For this feature, the image firstly
loaded into PySpot is given to another script as a pa-
rameter. This script creates a new window, which will
contain the 3D render. Information about the image’s
individual pixels is taken and modified. Through this
modification, negative values are removed, and the
overall values smoothed down. This array of mod-
ified pixels is added to a GLViewWidget as an item
and shown as a 3D Render. The initial intensity deter-
mines the representation of the individual pixels. The
user can set a lower and upper limit before creating
the image, which decides the coloration of the pixel
in the 3D rendering. Pixels below the lower limit are
omitted, while green represents those in between the
lower and upper limits. Those above the upper limit
are blue.
2.9 Grayscaling for Improved Intensity
Differentiation
Each color has a different influence on the way a per-
son perceives an image and its containing objects.
Therefore, PySpot offers functionality to switch be-
tween an image’s colored and grayscaled version.
Colored images provide a better way to visualize the
degree of intensity of the objects of an image, while
grayscaled images offer a better way to analyze and
locate the transition between intensities. To provide
this feature, the loading step of an image creates an ar-
ray for each color scheme. During the next step, each
layer of the loaded image is converted into a colored
RGBA and grayscaled image and saved into the re-
spective array. Through a checkbox in the GUI, a user
can switch between those two color schemes. Allow-
ing the adaptation of the image’s presentation to their
current need.
3 RESULTS
We evaluated the accuracy of the laser manipula-
tion methods used to create 3D microstructures using
PySpot. In the first step, an image is loaded into the
program and displayed as an RGBA image. Example
images are shown in Figure 1 (a) and (b) and came
from an SPE file. Image (a) contains a grid as a car-
rier structure where a small part was modified using
laser manipulation to add a thin layer that enables the
binding of fluorescence-labeled proteins. Whereas
(b) shows another grid, which has not been modi-
fied. (c) and (d) show the third and fourth layer of
a 3D Numpy array, containing another carrier struc-
ture. The automatic ROI detection feature is used to
mark the important regions for the analysis. It is based
on the principle of thresholding and contour finding.
Therefore, applying a threshold beforehand provides
an easy way to segment the regions of an image se-
lectively. Also, narrowing down the area significant
for ROI detection saves computing time, and unnec-
essary calculations. In Figure 4, a thresholding step
was taken before applying the ROI detection, narrow-
ing down the regions marked. As an alternative to
the thresholding feature, another method used to get a
better understanding of an area’s outline and the bor-
der of potential ROIs is the edge detection feature.
This feature uses a Gaussian-blur and Canny edge de-
tection to filter out the outlines of an image’s elements
and displays them in two different result windows.
The first of those windows contains an edge detection
solely with the Gaussian filter already implemented
in the Canny algorithm. Therefore, also containing
PySpot: A Python based Framework for the Assessment of Laser-modified 3D Microstructures for Windows and Raspbian
139
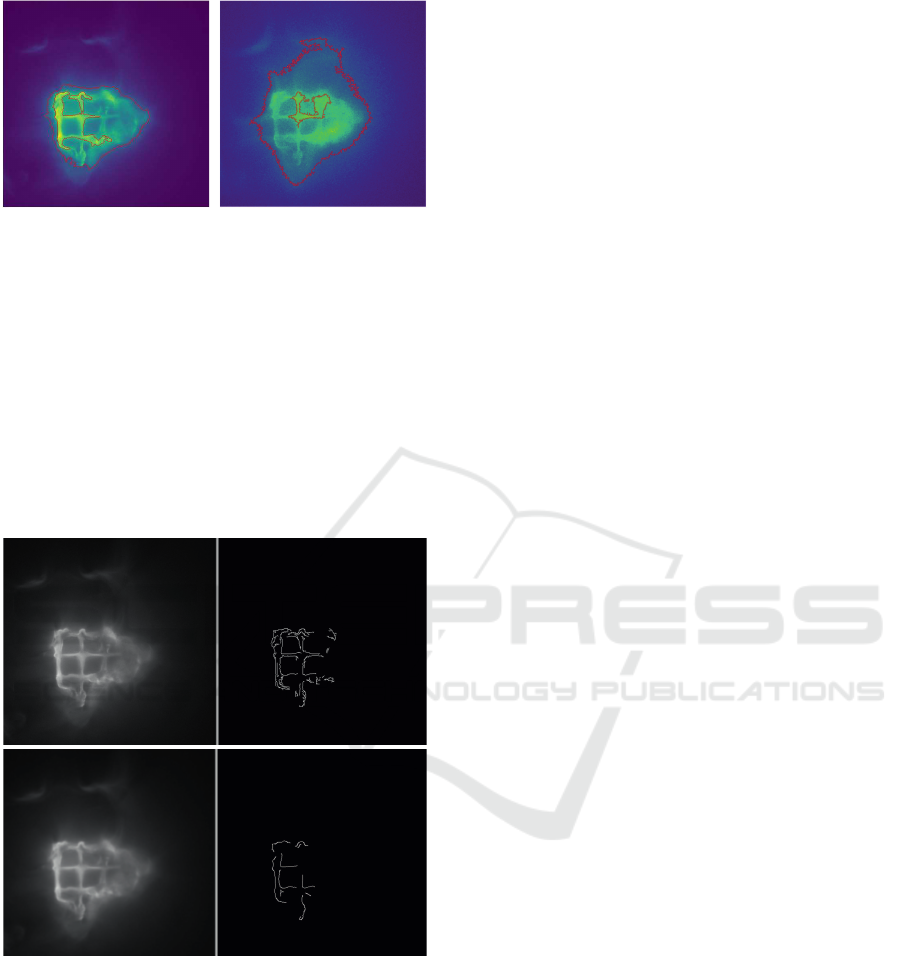
a b
Figure 4: Automatically detected ROIs on the correspond-
ing images from Figure 1. The ROIs in (a) are rather small
and outline the brighter structure almost perfectly. Contrary,
the ROIs in (b) cover a much greater area due to the brighter
background.
the edges of smaller, potentially insignificant borders.
Thus, the second window displays an image, which
had an additional Gaussian-blur applied to it before
detecting the edges. This results in a smoother in-
put image and fewer detected borders. The user can
determine the parameters used for the Gaussian-blur
and the limits required for the Canny edge detection
algorithm.
a
b
Figure 5: Image (a) shows the original image in grayscale
and the result of the Canny edge detection without and extra
blur. In the image (b) a Gaussian-blur was applied before
the edge detection.
As seen in Figure 5, a detection without additional
Gaussian-blur results in more detected edges. Since
most ROIs only need the main outline of an element
and not the small edges in between, this image of-
ten contains too much information. As an alternative,
the image from Figure 5 (b) has had a Gaussian-blur
with a dimension of 5, and a sigma of 3 applied to
it, significantly reducing the detected edges. This re-
duces the contained information in the image and will
display an element’s outlines clearer in some cases
than the version with all edges. Since this feature is
solely to get a better understanding of the edges of
the elements, the result is not taken into account dur-
ing an automatic ROI detection. Regardless of the
way, an ROI is drawn, be it automatically or manu-
ally, the evaluation of their pixels is always the same.
First of all, every ROI displays its dimensions and
area, as well as coordinates, in a table as basic in-
formation. Another second table displays informa-
tion about the intensity of all pixels inside an ROI.
This includes the minimum, average, and maximum
intensity of all contained pixels, giving a general view
of the brightness of the marked region. The brighter
an ROI’s intensity, the more particles have accumu-
lated inside its borders. Naturally, the overall inten-
sity of pixels is not enough to evaluate the success
of the cultivation. Therefore, a third table contains
information about each pixel inside of the currently
selected ROI. Each pixels’ intensity is compared to
its background, creating a direct relationship between
its quality and intensity. The first comparison made
is on a percentage basis and calculated by dividing
the background’s intensity by the intensity of the par-
ticular pixel, and saved as the contrast of the pixel
to its background. The second comparison is named
Delta and gives the absolute difference between pixel
intensity and background. Histograms visualize the
individual intensities and their frequency in an ROI,
offering a great addition to the statistical evaluation,
due to the qualities connected to the intensities of the
pixels, as seen in Figure 6. The visualization with a
histogram helps in the detection of outliners, which
could have a significant influence on the quality and
evaluation. Through the knowledge of the intensity
distribution, a thresholds limit, and ultimately the out-
lines of an ROI can be set with more precision. As
seen in Figure 6, the results can vary greatly from one
ROI to another. While the ROI in (a) mostly has inten-
sity values around 4.000 counts per pixel, the ROI in
(b) shows much higher overall intensity values, with
the majority ranging between 5.000 and 6.000 counts
per pixel. This shows that segmenting the image with
different thresholds has a great impact on the selected
region’s overall quality. Since the first ROI spans
over a greater area than the second one, its intensities
are far more diverse than those concentrated in (b),
taking thinly cultivated regions into account as well.
Through a comparison of (a), (b) with (c), and (d), the
difference between the two analyzed images becomes
prominent. When judging the images with the naked
eye, the difference between the images does not seem
too severe. Through comparison of the images with
BIOIMAGING 2020 - 7th International Conference on Bioimaging
140
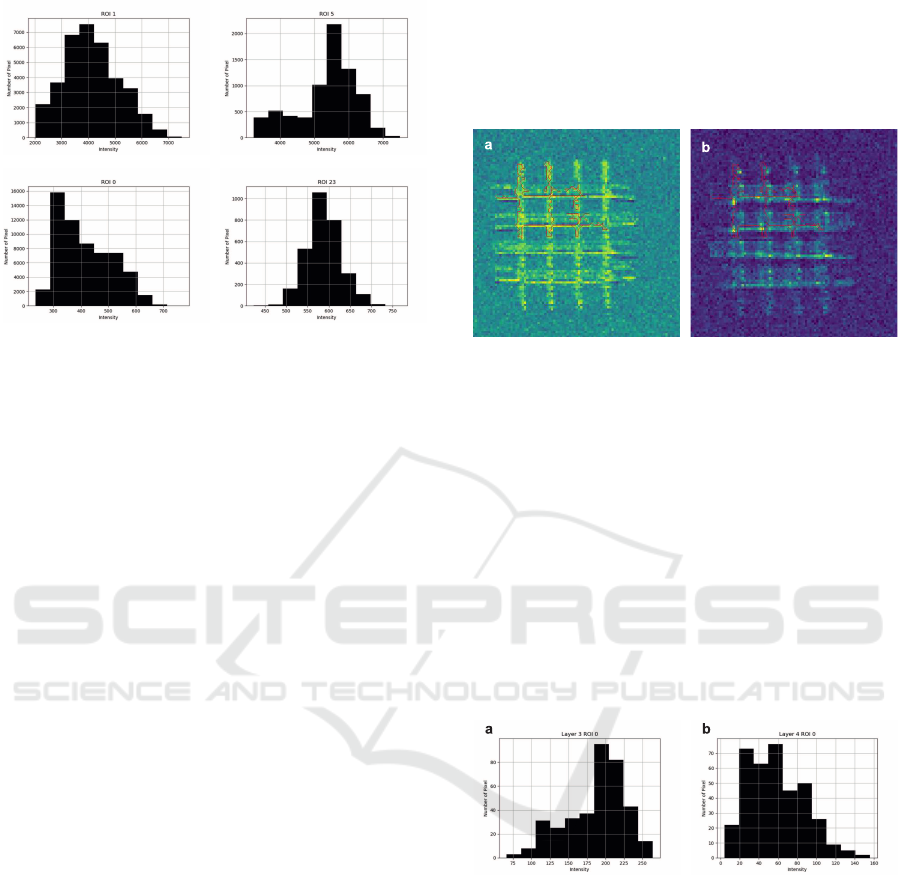
a b
c d
Figure 6: Histograms of the biggest ROIs in Figure 4, y-axis
displays the number of pixels and the x-axis the different
intensities. (a) and (b) show the histograms for the ROIs
outlined in Figure 4 (a). (c) and (b) those outlined in Figure
4 (b). (a) and (c) contain the intensities of the big ROIs
created with a threshold of 100, and (b) and (d) those of the
smaller ROIs.
the program, the major difference becomes clear. The
ROI on Figure 4 (a) is around ten times higher than
those in (b).
All those steps can be taken with 3D images, as
well. Visualizing the difference between the individ-
ual layers of a 3D area created through later modi-
fication. The first step consisted of a grayscale con-
version to get a better understanding of the individual
transitions between fore- and background. Afterward,
thresholding is applied to the image to segment the
previously found structures. The result of this step
can vary greatly among layers. This variation can be
due to the quality of the modification on the different
layers, causing particles to more prominently accu-
mulate on one layer than another. While this might
be the case for several layers in some images, oth-
ers only contain one layer with such a great differ-
ence and show certain outliners. This makes the se-
lection of a matching ROI quite difficult since there
is no complete consistency in the present data. Due
to this inconsistency among some layers, the layer
chosen for an automatic ROI detection has a great in-
fluence on the resulting quality evaluation of the im-
age. Figure 7 shows the biggest ROI resulting from
an automatic detection on layer three with a threshold
limit of 150. As seen in the figure, the shown ROI
includes many pixels with an intensity above 150 on
layer three, while it contains almost no pixels above
this value on layer four. Visualizing the great differ-
ence in intensity and density of fluorescent particles
on those two layers. Choosing another layer as the
base for the detection, the resulting ROI would have
marked a different area. Therefore, the layer chosen
for the detection determines the main feature of the
quality evaluation of the whole 3D image. The great
difference is even more evident in the statistical eval-
uation of the ROI’s pixel values, as seen in the his-
tograms of Figure 8.
Figure 7: Automatically detected ROIs with threshold limit
150 on layer 3 in (a) and layer 4 in (b).
The intensity of the contained pixels varies greatly
between the layers. On layer three, most of the con-
tained pixels show an intensity value of 180 counts
per pixel, with the majority collecting around 200
and the highest going up to just above 250 counts
per pixel. Contrary to this, the pixels on layer four
show a way lower intensity, with values ranging be-
tween 5 and 160. The largest share shows an intensity
of around 40 counts per pixel, with an average of 58
counts per pixel. While the histograms show the dis-
tribution of counts per pixel, the statistical evaluation
is provided by different tables in the same way as for
the 2D data.
Figure 8: Histograms of the ROI shown in Figure 7. The
y-axis displays pixel quantitiy and the x-axis intensities. (a)
displays the values of layer three, while layer four is repre-
sented in (b).
Retrieving all necessary information out of a 2D rep-
resentation is often difficult and also does not provide
the visualization needed to get a proper understanding
of the current 3D structure. Therefore, the 3D render-
ing can be used to get a more natural visualization of
the data, as seen in Figure 9. The visualized data is
placed on top of a grid, which functions as a base and
helps in judging the datas dimension.
PySpot: A Python based Framework for the Assessment of Laser-modified 3D Microstructures for Windows and Raspbian
141

Figure 9: 3D rendering of a hair knot with a lower limit of
65 and an upper limit of 80. (a) shows the rendered image
from the side, while (b) shows the top view.
4 CONCLUSION
In this paper, we show an easy-to-use software for
analyzing and evaluating SPE, TIFF, and Numpy ar-
ray data. The features of the software were illustrated
using 2D and 3D imaged microstructures. PySpot
provides standard functionalities, e.g., histograms and
thresholding and even more specialized features like
automated detection of ROIs and visualization of
three orthogonal layers (XY, XZ, and YZ) in one fig-
ure. For improved analysis of 3D data, functionalities
for the tracking of individual pixels’ intensities or an
improved 3D rendering with the option of data ex-
traction can be implemented in the program. Since
Python is an interpreter language, PySpot is not as
fast during calculations and operations as other image
analysis software packages. Instead, its benefits lie in
its simplicity, compatibility with different operating
systems, and easy adaptability. There is the possi-
bility to use it on Windows as well as on Raspbian.
Therefore, it can be used on a Raspberry Pi, which
are cheap, single-board computers. Additionally, this
basis allows for an easy way to add self-implemented
features into the program, adapting it to individual re-
quirements and different types of data. PySpot can
be downloaded free of charge at the homepage of the
Bioinformatics Research Group Hagenberg (http:
//bioinformatics.fh-hagenberg.at). Due to its
simplicity, yet vast array of functionalities, PySpot is
suited for inexperienced as well as experienced users.
ACKNOWLEDGEMENTS
This work was funded within the TIMED Center
for Technical Innovations in Medicine of the Univer-
sity of Applied Sciences Upper Austria (TC-LOEM)
and by the Basic Research Funding Program of the
University of Applied Sciences Upper Austria (AB-
SAOS).
REFERENCES
Buchegger B., Vidal C., N. J. B. B. K. A. H. A. K. T. A.
J. J. (2019). Gold nanoislands grown on multiphoton
polymerized structures as substrate for enzymatic re-
actions. ACS Materials Letters, pages 399–403.
Deng G., C. L. (1993). An adaptive gaussian filter for noise
reduction and edge detection. volume 3, pages 1615 –
1619.
Gedraite E., H. M. (2011). Investigation on the effect of
a gaussian blur in image filtering and segmentation.
pages 393–396.
J., C. F. (1986). A computational approach to edge detec-
tion. IEEE Transactions on Pattern Analysis and Ma-
chine Intelligence, PAMI-8:679–698.
Kawata S., Sun H.-B., T. T. T. K. (2001). Finer features for
functional microdevices. Nature, 412(6848):697–698.
Maruo S., Nakamura O., K. S. (1997). Three-dimensional
microfabrication with two-photon-absorbed pho-
topolymerization. Opt. Lett., 22(2):132–134.
Ovsianikov A., Li Z., T. J. S. J. L. R. (2012). Selective func-
tionalization of 3d matrices via multiphoton grafting
and subsequent click chemistry. Advanced Functional
Materials, 22(16):3429–3433.
Rationality (2019). The Cambridge Dictionary.
Sahoo P., Soltani S. and, W. A. (1988). A survey of thresh-
olding techniques. Computer Vision, Graphics, and
Image Processing, 41:233–260.
Satoshi Suzuki, K. A. (1985). Topological structural anal-
ysis of digitized binary images by border following.
Computer Vision, Graphics, and Image Processing,
30(1):32 – 46.
Seo J., Chae S., S. J. K. D. C. C. H. T. (2016).
Fast Contour-Tracing Algorithm Based on a Pixel-
Following Method for Image Sensors.
Wollhofen R., Buchegger B., E. C. J. J. K. J. K. T. A. (2017).
Functional photoresists for sub-diffraction stimulated
emission depletion lithography. Opt. Mater. Express,
7(7):2538–2559.
BIOIMAGING 2020 - 7th International Conference on Bioimaging
142
