
Combining Image and Caption Analysis for Classifying Charts in
Biodiversity Texts
Pawandeep Kaur
1 a
and Dora Kiesel
2 b
1
Heinz-Nixdorf Chair for Distributed Information Systems, Friedrich Schiller University, Jena, Germany
2
Bauhaus University, Faculty of Media, VR Group, Weimar, Germany
Keywords:
Caption Analysis, Chart Classification, Natural Language Processing, NLP, Caption Classification, Visualiza-
tion Recommendation, Chart Recognition
Abstract:
Chart type classification through caption analysis is a new area of study. Distinct keywords in the captions
that relate to the visualization vocabulary (e.g., for scatterplot: dot, y-axis, x-axis, bubble) and keywords from
the specific domain (e.g., species richness, species abundance, phylogenetic associations in the case of biodi-
versity research), serve as parameters to train a text classifier. For better chart comprehensibility, along with
the visual characteristics of the chart, a classifier should also understand these parameters well. Such concep-
tual/semantic chart classifiers then will not only be useful for chart classification purposes but also for other
visualization studies. One of the applications of such a classifier is in the creation of the domain knowledge-
assisted visualization recommendation system, where these text classifiers can provide the recommendation
of visualization types based on the classification of the text provided along with the dataset. Motivated by this
use case, in this paper, we have explored our idea of semantic chart classifiers. We have taken the assistance
of state-of-the-art natural language processing (NLP) and computer vision algorithms to create a biodiversity
domain-based visualization classifier. With an average test accuracy (F1-score) of 92.2% over all 15 classes,
we can prove that our classifiers can differentiate between different chart types conceptually and visually.
1 INTRODUCTION
Automatic chart type classification is an increasingly
common pursuit in visualization. In the majority of
cases, the overall goal of the chart classification is au-
tomatic chart comprehension. Nonetheless, previous
studies (Liu et al., 2013),(Abhijit Balaji, 2018),(Savva
et al., 2011),(Jobin et al., 2019) have paid little at-
tention to the role of the free text information pro-
vided along with the charts in the form of captions.
Processing only the visual chart elements in the im-
age can recognize the type of chart well, however,
this recognition alone is not enough to understand
the chart semantics. Captions on the other hand pro-
vide clear goals of the represented charts and resolve
the ambiguity that arises due to visually similar chart
types. For example, column charts look similar to
histograms but their representative goals are differ-
ent. Column charts are used to represent the com-
parison among various sizes of the data series while
histograms show the frequency with which specific
a
https://orcid.org/0000-0002-3073-326X
b
https://orcid.org/0000-0002-6283-2633
values occur in the dataset (Harris, 2000). Due to the
different chart semantics, an image classifier alone,
which might decode a histogram as a column chart
would not be able to provide an accurate and efficient
automatic interpretation.
After manually surveying a vast number of cap-
tion samples during the corpus creation process, we
know that chart captions provide information about
a) representative goals of the author which are not
directly visible from the image itself, b) domain-
specific tasks, depicted through the chart and c) chart
layout and pictorial elements (e.g. text, colors, lines,
shapes). Consider, for instance, Figure 1 and the orig-
inal caption to the visualization depicted in Figure 1
(Moody and Jones, 2000).
”Fig. 5. Boxplots comparing the distribution of
the measured soil variables at the different canopy
positions at trunk, midcanopy, the canopy edge, and
outside the canopy, respectively. The upper and lower
boundaries of each box represent the interquartile
distance (IQD). The horizontal midline is the median
value. The whiskers extend to 1.5x IQD. Outliers are
displayed as horizontal lines beyond the range of the
Kaur, P. and Kiesel, D.
Combining Image and Caption Analysis for Classifying Charts in Biodiversity Texts.
DOI: 10.5220/0008946701570168
In Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2020) - Volume 3: IVAPP, pages
157-168
ISBN: 978-989-758-402-2; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
157
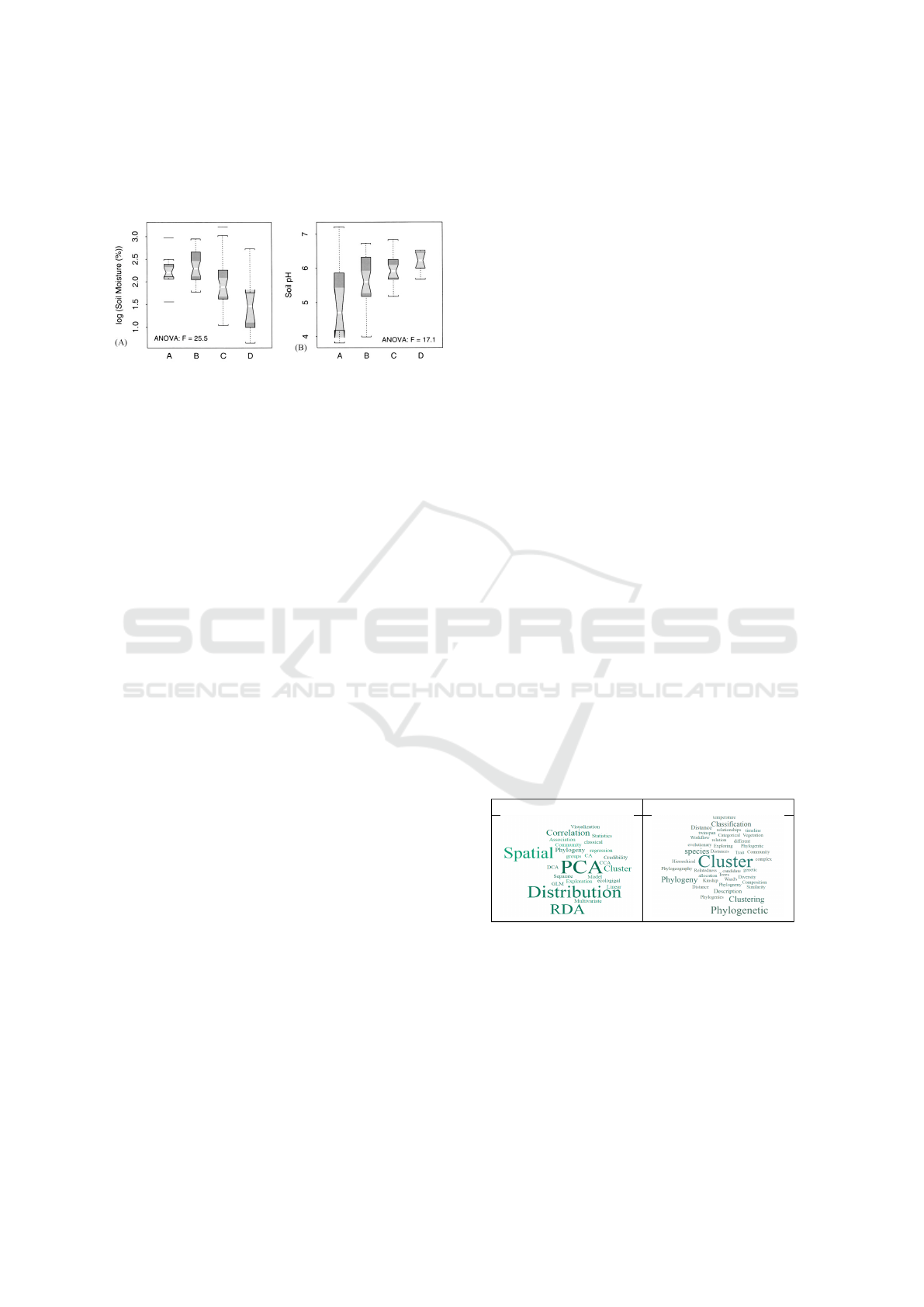
whiskers. If the notches of any two boxes do not over-
lap vertically, this suggests a significant difference at
a rough 5% confidence interval.”
Figure 1: Example image adapted from a biodiversity pub-
lication (Moody and Jones, 2000).
The caption of this figure provides clues about the
following information:
Chart Type: Boxplots, box, horizontal midline,
whiskers, horizontal lines, notches.
Representative Goals: comparing, distribution.
Domain Specific Variables: soil variables, canopy
positions, trunk, midcanopy, canopy edge.
Statistical or Analytic Information: interquartile
distance (IQD), median value, confidence
interval.
These identifiers can serve as important features
for a text classifier.
In this work, we have taken the assistance of both
computer vision and NLP technology in the produc-
tion of a very first text classifier that identifies the best
fitting chart from a set of fifteen different chart types
given a biodiversity caption or data description. The
classifier was incrementally trained, starting with a
dataset of 4073 manually annotated biodiversity vi-
sualization captions and achieves an average test ac-
curacy (F1-score) of 92.2% over all 15 classes. The
classifier understands a specific chart vocabulary and
related biodiversity vocabulary used for each partic-
ular chart type and will, therefore, only work on the
biodiversity text. However, the described workflow
can be used to train classifiers for other different do-
mains as well.
Our primary contribution is the chart classification
workflow that classifies a chart not only on the basis
of chart elements in the image but also on the basis of
captions. Following this workflow creates a semantic
chart classifier that understands the visualization type
along with the high level visualization goals. We also
contribute by providing a novice approach in the field
of data visualization recommendation system, to rec-
ommend visualization schema via visualization clas-
sifier based on the knowledge gathered at the train-
ing process. In our knowledge, we are the first who
conceptualize and implement a visualization classifier
that understands the semantics of the chart and can
infer representative goals from the biodiversity text
based on the different chart types.
In Section 2 we present the motivation for this
work, state-of-the-art in Section 3, classification pro-
cess is presented in Section 4, result and discussion in
Section 5, challenges and research directions in Sec-
tion 6 and conclusion in Section 7.
2 MOTIVATION
The chart type classification we present in this pa-
per will constitute one important step in a biodiver-
sity knowledge-assisted visualization recommenda-
tion system that will suggest suitable visualization
types based on biodiversity research data and goals.
To build such a system, a visualization designer
needs to know the domain-based and the visual goals
of the user (Munzer, 2009). In the earlier stages of our
study, we tried to gather this information via a survey.
Due to the limited responses, we could not get good
representatives of the dataset. An excerpt of the re-
sult is shown in Table 1. It shows that scientists use
scatterplot prominently to convey the result of differ-
ent ordination analysis techniques (PCA, RDA, DA)
and Dendrogram is more prominently used to rep-
resent Phylogeny, Classification and Clustering. To
gather this knowledge in bulk and to use this knowl-
edge to classify future biodiversity texts serve as the
main motivation in the creation of biodiversity visual-
ization classifier.
Table 1: Visualization types and the purposes they are used
for in biodiversity domain (Kaur et al., 2018).
Scatterplot Dendrogram
The proposed system, depicted in Figure 2, will
work as follows: After the user provides a data table
and a description of its contents, along with how it
was created, its intended research goals, and so forth,
a biodiversity visualization text classifier is applied to
suggest the most suitable visualization types to rep-
resent the data based on its vast knowledge of similar
data and research goals. In a second step, based on the
visualization type, our context-aware algorithm will
select the suitable data variables that can be mapped
IVAPP 2020 - 11th International Conference on Information Visualization Theory and Applications
158

to the visualization.
The goal of the chart type classification task de-
scribed in this paper is the creation of a knowledge
base for and the training of the biodiversity text clas-
sifier that will be responsible for inferring the best
suited chart types from given metadata text which
provides information about the what and why of the
dataset. The biodiversity text classifier will output a
’predicted visualization list’, which is a ranked list of
suitable chart types for this dataset and related vocab-
ulary. This information can then be used by scientists
to create suitable visualizations and gain new insights
into the selected dataset.
In this paper, we will focus on the creation of the
biodiversity visualization text classifier.
3 STATE OF THE ART
In this section, we present the state of the art of chart
image classification and image caption analysis.
3.1 Chart Image Classification
Many studies have used different computer vision al-
gorithms for the classification of chart types from dig-
ital images. Typical representative of them are Ta-
ble 2. We have found our study different from them
in the following aspects:
• Data Source: We have found that some studies
have used automatically generated synthetic data
(Abhijit Balaji, 2018) and others have used well
curated data from previous studies (Jung et al.,
2017). In their work, (Savva et al., 2011) have
carefully downloaded their dataset from online
search. The resulting dataset does not really re-
flect reality, where intra-class variation is a lot
higher. In our work, we tried to reflect this by
creating our corpus from scientific publications
which have huge intra-class variations (see Ta-
ble 3). Moreover, we have considered various
forms of different charts by grouping them not
only based on their visual similarities but also on
their chart semantics.
• Visualization Classes: The range of the classes
that we have considered (15) is much larger than
most of the previous studies, except (Jobin et al.,
2019), who have also considered other document
figure types as their classes. Moreover, their train-
ing dataset was far more balanced than what we
had. In our case, we were not aware of the types
of visualizations available in the downloaded cor-
pus in advance. Therefore, it was important for
us to include as many different visualization types
as possible in our training set. This leads to the
assignment of unequal proportion of examples for
many classes.
Table 2: A summary of different chart image classification
studies.
Name Year Data source Classes Accuracy
(Savva et al., 2011) 2011 Online 10 96%
(Jung et al., 2017) 2017 Online 10 76,7%
(Abhijit Balaji, 2018) 2018 Synthetic 5 99,72%
(Jobin et al., 2019) 2019 Conferences 28 92,86%
Table 3: Intra-class visual similarity for different variation
of Column or Bar Chart.
(A) (B)
(C) (E)
We consider our work to be closed to (Jobin et al.,
2019), who has also done scientific document figure
classification on publications from different confer-
ences. They had also created their categories based
on what was available to them in their downloaded
corpus. Like us, they pre-selected the categories,
grouped them into super classes and then had used it-
erative learning to gather more trained data. However,
for their classification process, they had considered all
different types of figures available in scientific docu-
ments, whereas our work is limited to visualization
images only. They have only considered image clas-
sification, whereas we have taken the benefits of both
text and image classifiers for semantically classifying
the visualization types.
3.2 Use of Captions for Classification
Purposes
Image captions are an important source of informa-
tion and have been a long-studied subject in differ-
ent domains. Earlier studies have used captions with
computer vision algorithms to identify human faces
in the newspapers (Srihari, 1991). Caption analysis
has been predominantly used in biomedical domain
for its various research goals. In their study, (Murphy
Combining Image and Caption Analysis for Classifying Charts in Biodiversity Texts
159
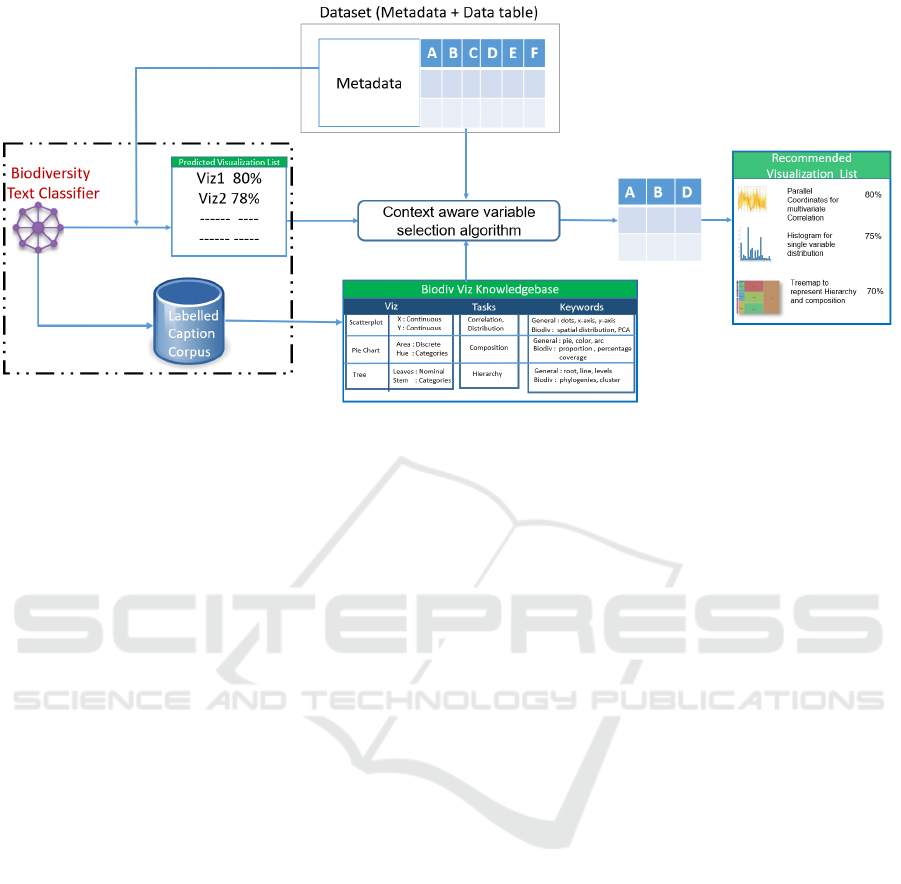
Figure 2: A conceptual diagram of biodiversity knowledge-assisted visualization recommendation system.
et al., 2004) used joint caption image features in clas-
sifying protein sub-cellular location in images. The
study by (Rafkind et al., 2006) was the first to use
text identifiers from biomedical image captions (”di-
ameter”, ”gene-expression”, ”histogram” etc) to iden-
tify graph chart images as a separate category from
other biomedical image categories. More recent stud-
ies have utilized the information from the captions in
biological documents for the purpose of image index-
ing and search (Charbonnier et al., 2018; Xu et al.,
2008; Lee et al., 2017). So far, caption data has been
used along with the computer vision algorithms to
distinguish graph images from others, however, we
could not find any study that has used caption data
alone or caption data in conjunction with other me-
dia, to classify different chart types. Classifying chart
types from caption data is still in a novice state. We
have observed different use cases in which research in
caption analysis could be beneficial for visualization
as well as for linguistics research:
• Visualization Research: The foremost is in the
creation of domain specific visualization knowl-
edgebases or corpus, 2) a machine learning
model trained on the visualization captions can be
evolved and reused by other users for different do-
main knowledge-assisted visualization products,
3) a text classifier trained on different chart types
can be used for tagging, indexing and searching
visualization documents, 4) it could be a valuable
source for future theoretical visualization research
problems (Chen et al., 2017) like the creation of
visualization ontologies on the basis of classified
visualization concepts.
• Computer Linguistics Research: Research on
caption classification will help 1) to better under-
stand the requirements of classifications on very
short and convoluted texts, 2) to study the influ-
ence of domain specific language on classifica-
tion and possibly exploit domain specific regulari-
ties to improve classification results and 3) to find
effective ways to integrate domain expert knowl-
edge into the classification process.
4 CLASSIFICATION PROCESS
The process of creating the biodiversity text classifier
consists of a sequence of complex steps, visualized
in Figure 3. In order to reach the highest possible
quality in recognizing the best suited chart types from
biodiversity text – e.g. biodiversity image captions
– the underlying training corpus needs to have a suf-
ficient size (we estimated at least 15 000 samples to
be enough, ideally 1 000 for each of the 15 classes)
and quality. So, the first step was to manually create
a starting dataset, that associates caption texts with
their respective chart types. This set is then incremen-
tally extended using a combination of image and cap-
tion classification in order to gain the highest possible
quality on the automatic labeling of unlabeled data.
The resulting dataset is then used as training set for
the biodiversity text classifier, that can be integrated
into the biodiversity knowledge assisted visualization
recommendation system described in Section 2.
IVAPP 2020 - 11th International Conference on Information Visualization Theory and Applications
160
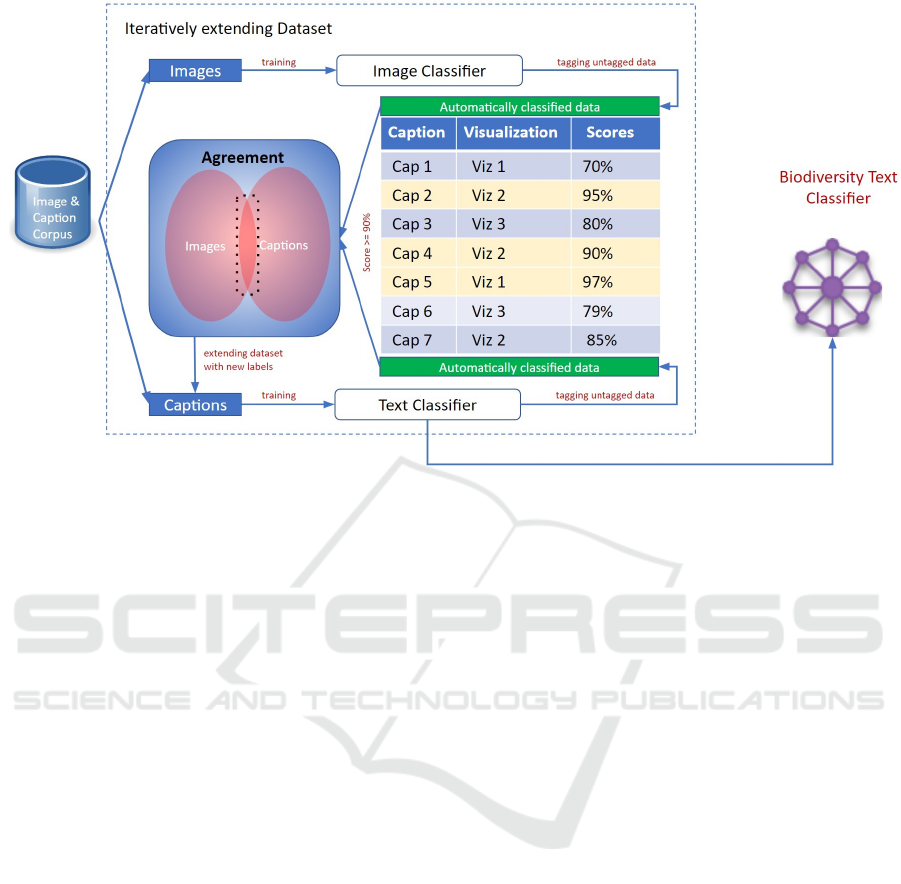
Figure 3: Workflow for the creation of the biodiversity visualization text classifier.
4.1 Data Preparation
In our data collection process, first we selected the
reputed biodiversity journals which represents differ-
ent biodiversity sub-domains. For details please re-
fer to Section 2 of appendix. We had downloaded
all available volumes and issues of these journals till
2016 which is the year when this download was done.
For creating the initial dataset, we downloaded 26 588
biodiversity publications through Elsevier ScienceDi-
rect article retrieval API (api, 2018), which allows
the download of a complete publication in an XML
format. From these 26 588 downloaded publications,
96 837 images and their captions were extracted using
a python script.
4.1.1 Class Formation and Annotation Process
Out of 96 837 image and caption samples, we cre-
ated our training data by randomly selecting a sub-
set of 4 073 visualization image captions and labeling
them with their respective visualization types manu-
ally. The labelling task was done by one of the author
of this paper who is a Ph.D. student in data visual-
ization with four years of experience in this domain.
In case of disambiguation regarding the selection of
the visualization label for a particular caption, ade-
quate guidance from visualization publications and
reference books (Harris, 2000) was taken. Due to
the sheer richness of different visualization types –
a closer study revealed the sample to contain 59 dif-
ferent visualizations (see Section 1 of appendix) –
we continued our annotation process in the following
stages:
Class Grouping: In order to gain adequate sample
sizes for each of the visualization types or classes, we
split / merged the original 59 classes into super / sub
classes:
> 50 Samples in Class: Since we considered 50 ex-
amples to be sufficient for classification, all
classes with same or more examples were kept as
a super classes.
< 10 Samples in Class: These classes had very
small set of examples and were not suitable
match for our super classes. Therefore, they were
rejected from the further annotation process.
All Other Classes: All classes, that do not figure fre-
quently enough to suffice for the classification
task have been merged into super classes either
based on their visual similarity or their represen-
tational goals. For example, chart types that use
the same coordinate space (e.g. xy plot) and same
visual marks (e.g. bars) were considered visually
similar and then were merged. This way, all the
chart types which are visually similar to Column
Chart e.g. Bar Chart, Stacked Bar Chart, Multiset
Bar Chart etc. were all merged into the super class
Column Chart.
On the other hand, Chord Diagrams, Alluvial Dia-
grams and Network diagrams are visually dissim-
ilar but have the common representational goal of
connecting entities. Same with the Pie Chart and
Stacked Area Chart which are visually dissimilar
Combining Image and Caption Analysis for Classifying Charts in Biodiversity Texts
161

but represent a common visual goal of represent-
ing proportion or composition among data enti-
ties.
Thus, they all were grouped in the class ’Net-
work’. All non-visualization images (e.g.,
camera-clicked pictures, conceptual diagrams
etc.) were grouped into the ’NoViz’ class. Due to
the variant structure of non-visualization images,
NoViz class was also excluded from the image
classification process. An overview of retained
classes are provided in Section 3 of appendix.
Doing so, we ended up with 15 different super
classes.
Assignment of Classes for Caption Classification:
Once, we had formed the classes, we did another
round of annotation. We have now labelled our se-
lected corpus of 4 073 captions with these 15 classes.
For detailed information about these classes, refer to
Section 4 of appendix.
Assignment of Classes for Image Classification:
All classes were considered for image classification
except for those which are visually similar to other
classes. Histogram is visually similar to the column
chart and timeseries is visually similar to the line
chart (see appendix 3). Thus, histogram and time-
series were ignored from the image classification pro-
cess. Alongside, due to the variant structure of non-
visualization images, ’NoViz’ class was also excluded
from the image classification process.
We have provided the frequency distribution of
classes for image and caption classification in Table 4.
In appendix Section 4, we provide examples for each
class, that consist of the replicated original image and
caption from open-access publications. Due to the
copyright issues, we are unable to provide original ex-
amples from our dataset.
4.2 Image Classification
4.2.1 Image Classification Model
For image classification, we have used Convolutional
Neural Networks (CNNs). CNNs are a specialized
kind of neural networks for processing data that has a
known grid-like topology. Since images can also be
thought as 2D-grid of pictures, thus CNNs have been
tremendously successful in application to image data
(Goodfellow et al., 2016).
For training, we have used reusable pre-trained
neural network modules provided by Tensorflow Hub
(TFH, 2019). TensorFlow Hub is a library for the pub-
lication, discovery, and consumption of reusable parts
of machine learning models.
Table 4: Frequency distribution of our manually annotated
training dataset for Caption and Image classification.
Classes Caption Classes Image Classes
Ordination Plot 503 278
Map 529 277
Scatterplot 399 272
Line Chart 320 283
Dendrogram 282 243
Column Chart 427 302
Heatmap 147 124
Boxplot 210 104
Area Chart 159 95
Network 58 32
Histogram 57 -
Timeseries 319 -
Noviz 511 -
Pie Chart - 134
Proportion 157 -
Total 4073 2144
Out of the available CNN modules in TFHub, we
chose MobileNet V2 (Sandler et al., 2018) as our
CNN architecture. MobileNet V2 is a family of neu-
ral network architectures for efficient on-device im-
age classification and related tasks.
Mobilenet
v2 module of Tensorflow Hub contains
a trained instance of the network, packaged to do
the image classification. This Tensorflow Hub mod-
ule uses the Tendorflow Slim implementation of ’Mo-
bilenet v2’ with a depth multiplier of 1.0 and an in-
put size of 224x224 pixels. For training the classifier
we used Keras (Chollet et al., 2015) with Tensorflow
(Abadi et al., 2016) backend. To train the classifica-
tion network on our data, we resized the images to
a fixed size of 224 x 224 x 3 and normalized them
before feeding into the network. We used Adam op-
timizing function (Kingma and Ba, 2014) with the
learning rate of 0.001.
4.2.2 Results from Image Classification
For evaluation, we have used Keras’ in-build evalu-
ation function. When provided with the suitable pa-
rameters, Keras seperates and retains a portion from
the training data and then uses that unseen retained
data for evaluating the model. For evaluating our
image model, 20% of the examples from the image
dataset were retained from training. Our model has
achieved a classification accuracy of 75% on automat-
ically selected batch of 100 images. Then this classi-
fier was used to classify the original corpus of 96837
IDs. Our image classifier was able to annotate 54%,
i.e., 52921 IDs out of 96837 IDs, with the confidence
interval of 95% and more.
IVAPP 2020 - 11th International Conference on Information Visualization Theory and Applications
162
4.3 Caption Classification
The 4 073 manually labeled image captions served as
training set for the initial supervised classifier. In or-
der to be able to optimize the classifier for each of the
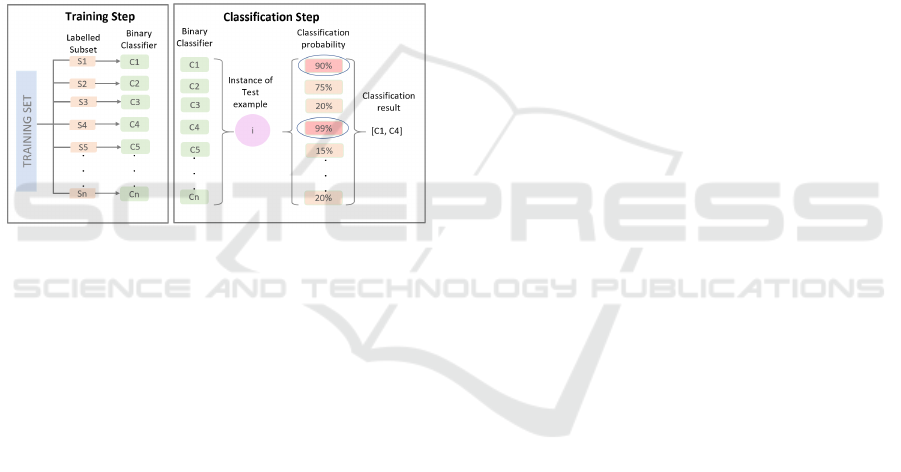
identified classes separately, we decided to build bi-
nary classifiers, that can distinguish one specific class
from all others. From these specialized binary clas-
sifiers an assembly classifier is constructed (see Fig-
ure 4, Training Step). Given an input, the assembly
asks each classifier to process the input separately (as
detailed in Figure 4, Classification Step), and receives
a probability score that states how likely it is, that the
given sample is of the respective class. The classes of
all classifiers, that give a positive response with a cer-
tain preset confidence (in our case usually 90%), will
then be returned as result vector.
Figure 4: Workflow for the creation of the biodiversity vi-
sualization text classifier.
To find out which binary classifiers to incorpo-
rate into the assembly, we implemented and opti-
mized three standard classifiers in text classification
(Joachims, 1998; Sebastiani, 2002): Support Vector
Machines (SVM), CNN and Random Forests. SVMs
(Cortes and Vapnik, 1995) are inherently binary su-
pervised learners. In their linear form, they find the
maximum-margin hyperplane in data space that best
separates the data points of one class from the data
points of the other class. Kernels (Boser et al., 1992)
have been introduced to generalize the principle to
polynomial, radial or sigmoid functions. Random
Forests (Ho, 1995) are a assemblies of a – usually
rather large – number of Decision Trees that con-
tribute to the main decision in form of a majority
vote. Additionally, in resemblance to the image clas-
sifier, a neural network solution – specifically a mul-
tilayer perceptron classifier with the same stochastic
gradient-based optimizer as the image classifier and
the same constant learning rate of 0.001 – was used.
4.3.1 Preprocessing
As is standard in natural language processing (Aggar-
wal and Zhai, 2012), the labels have been broken into
tokens, stemmed and stop words have been removed
before processing them. Additionally, some standard
phrases that have been identified during manual n-
gram evaluation of the data and are unrelated to the
contents of the image, like phrases to make people
aware of the modalities of the online version of the
paper – e. g. ”For interpretation of the references
to color in this figure legend, the reader is referred to
the web version of this article”)– , have been automat-
ically removed. In order to keep the training data as
pure as possible, captions referring to multiple visual-
izations were filtered out, leaving a dataset of 4 066.
The resulting word vectors contain the term fre-
quency - inverse document frequency (tf-idf) scores
(Ramos et al., 2003) per word.
4.3.2 Model Optimization or Parameterization
Each binary classifier has been trained separately on
a data set consisting of all samples of the target class
and an equal number of samples uniformly distributed
over all other classes. Classifiers have been evaluated
using a 5-fold-cross-validation, that splits the data set
into 5 equal parts training on 4 parts and testing on
the last. The final evaluation result constitutes as the
average of all five runs.
In order to reach the best results, we optimized
both the pre-processing of the data as well as the pa-
rameters of the classifier itself. On data level, we op-
timized the maximum size of the vocabulary, the min-
imum number of documents each word figures in and
which n-grams should be included into the analysis.
Applying an exhaustive grid search over the range of
sensible parameters for each feature (vocabulary size:
[250 to 1250 (steps of 250)], minimum document fre-
quency: [0 to 4], n-grams: [1 to 6]), we achieved the
best results using a base vocabulary that consists of
the 750 most important words and 2-grams, that oc-
cur in at least 3 documents in the whole corpus.
We also optimized SVM for its kernel function
(linear, polygonal, sigmoid, and radial basis func-
tion), finding a linear kernel to give the best results,
and the Random Forest classifier for the number of
Decision Trees in the assembly (100 to 2000 in steps
of 100), finding that the impact on the classification
accuracy is rather small. The neural network has been
tested with different node sizes in its hidden layers (2,
10, 15, 50). The best result has been achieved with 15
nodes.
Figure 5 shows the best results on each class for
each of the classifiers. The results show that Random
Combining Image and Caption Analysis for Classifying Charts in Biodiversity Texts
163
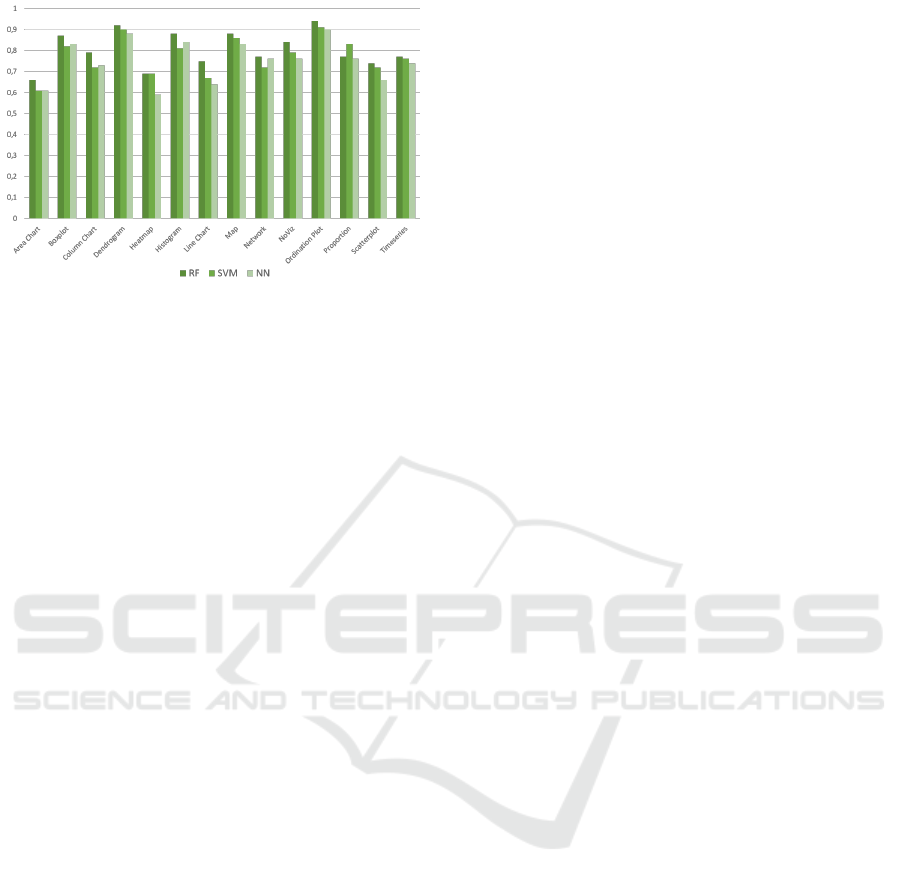
Figure 5: Classification results (F1 score) of the Random
Forest (RF), Support Vecor Machine (SVM) and Neural
Network (NN) classifiers for each class in our corpus.
Forests outperform the results of SVM and the neural
network in all classes with up to 7% increase in the
F1 score. One possible conclusion to draw from these
results is that, the linguistic properties of caption data
can be modelled more precisely through a series of
parallel boolean operations than through a maximum-
margin method. Following this finding, we will use
Random Forests as binary classifiers for the assembly
classifier.
4.3.3 Incremental Learning and Caption
Dataset Extension
The purpose of the caption classification is twofold.
First, we want to extend the existing dataset to re-
duce the risk of overfitting the single binary classi-
fiers. Second, to train the binary classifiers to under-
stand the words and phrases describing the underlying
data of the chart in order to finally recommend chart
types based on data set descriptions.
Incremental learning and an additional agreement
step with the classification result of the image classi-
fier (see Figure 3) was used to increase the size of the
original 4 066 captions to a dataset of 22 881 captions.
The one iteration of the incremental learning algo-
rithm includes the following steps:
Learning: Conduct 5-fold cross-validation on the
current dataset to evaluate the quality of the set
(results see Figure 6). Train all binary classifiers
on the whole dataset.
Annotation: Use the assembly to label as many cap-
tions of the remainder of the untagged data as pos-
sible with at least 90 % confidence. Include new
labels and captions into the extended dataset.
The two steps are repeated until a finishing cri-
terion is met. Since we were focusing on extending
the dataset in this phase, we stopped the algorithm
when the number of newly included tags fell under-
neath a preset threshold (0.01% of the whole corpus
in our case). This way, the caption classifier was able
to annotate 44% of the total corpus which amounts to
43 256 IDs with a confidence interval of 90%.
4.3.4 Refining the Knowledgebase
In order to ensure highest quality in the creation of our
knowledge base, we refined the resultant data from
image and caption classification in a multi-step pro-
cess:
• We start with 52 921 labeled images in Image Cor-
pus (IC), and 43 256 labeled captions in Caption
Corpus (CC).
• To get only the visualization image IDs, first we
removed the ’NoViz’ labelled IDs from the cap-
tion corpus. Leaving behind 43 256-451= 42 805
to be merged. After the merging process, these
IDs were put back to the corpus for iterative learn-
ing.
• We merged the two corpora by only keeping those
IDs that have been tagged by both classifiers. This
set contains a total of 22 817 common IDs.
• This set is, then, reduced to only contain the most
reliable ID/label pairs:
1. ID/label pairs with full or partial agreement in
classified labels from image and caption clas-
sifier (11 108 samples). Partial agreement is
reached if the label given by the image clas-
sifier is contained in the class list provided by
the caption classifier; full agreement is reached
if the caption classifier only provides one label
and this label matches the class of the image
classifier.
2. ID/label pairs with more than 98% confidence
from image Classifier (10 728 samples)
3. ID/label pairs whose classes were absent in im-
age classifiers (Area Chart, Time Series, His-
togram, Proportion), if the source of disagree-
ment between image and caption classifiers
stems from these classes, like ’Timeseries’ in
CC and ’Line Chart’ in IC or ’Histogram’ in CC
and ’Column Chart’ in IC. All other conflicts
have been resolved manually. In our manual
verification, ’Proportion’ has performed bad
due to its similar vocabulary with ’Pie Chart’
and all other classes that represent some ’Pro-
portion’ or ’Composition’ representation goals
for example different stacked chart: Stack Area
or Stack Column. To avoid such confusions for
incremental learning round, we had to merge
some of these example to ’Pie Chart’, ’Stack
IVAPP 2020 - 11th International Conference on Information Visualization Theory and Applications
164
Area Chart’ and ’Column Chart’. Rest of the
examples were ignored.
4. ID/label pairs that have been manually checked
upon due to the multi-assignment of the cap-
tion classifier and assigned the single true class
if possible. Captions representing multiple vi-
sualizations have been rejected.
This leaves us with 22 248 high-quality ID/label
pairs.
• Finally, the automatically created dataset has
been merged with the manually annotated dataset
to further increase the quality and size of our
knowledge-base (22 866 samples in total, 1 468
Ordination Plots, 4 989 Maps, 1 669 Scatterplots,
6 173 Line Charts, 452 Dendrograms, 5 459 Col-
umn Charts, 603 Heatmaps, 303 Boxplots, 99
Area Charts, 187 Network Diagrams, 69 His-
tograms, 330 Timeseries, 448 Noviz, 304 Pie
Charts and 313 Stack Area Charts).
5 RESULTS AND DISCUSSION
5.1 Results
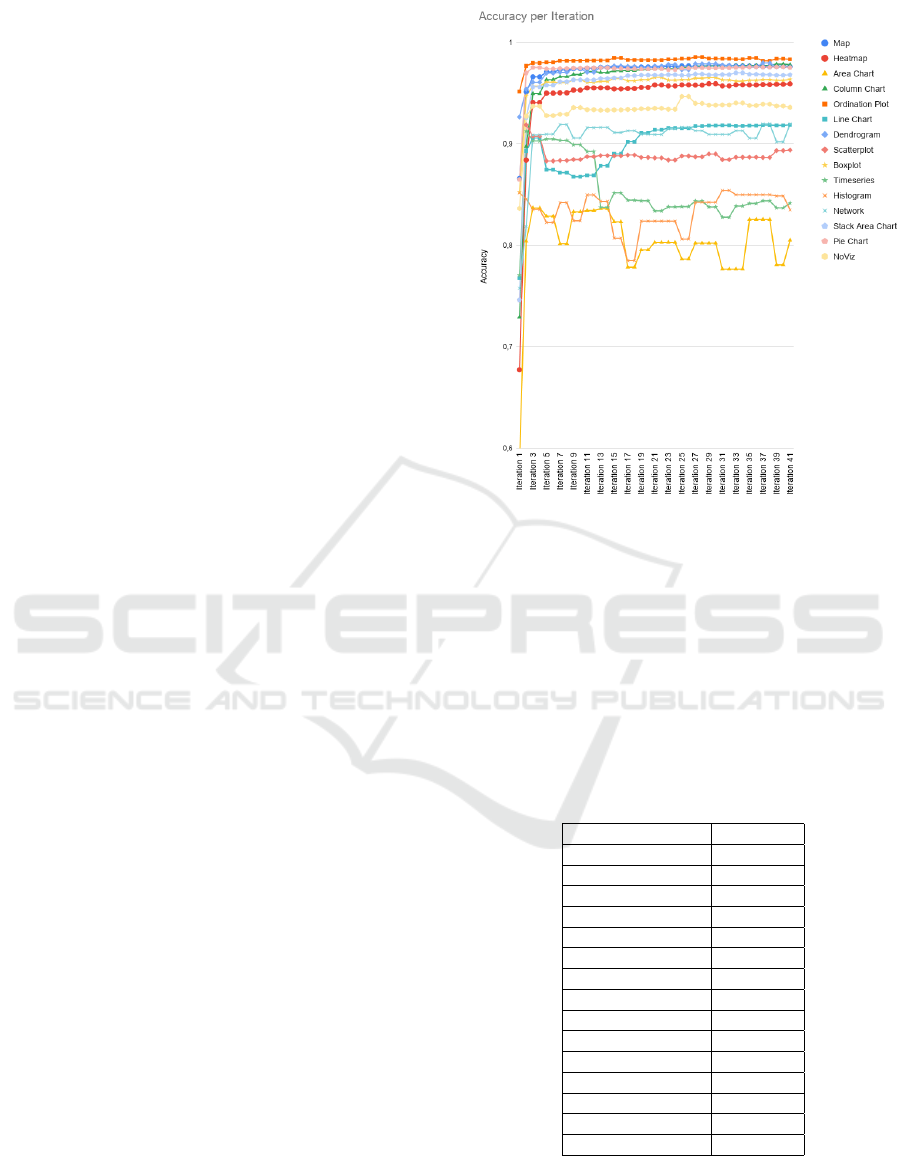
Figures 6 and 7 show the development of the quality
of the classifiers as well as the number of samples for
each label over the course of the 41 iterations neces-
sary to reach the ending criterion (a tag rate of less
then 0.01 % of the unlabeled samples of the corpus).
In most cases, the quality of the classifiers rises the
most within the first 3 iterations. After that phase,
most classifiers do not change in quality any more.
Exceptions are the line chart, with a drop after the
steep rise in the beginning, the time series, with a drop
at the eleventh iteration, and the histogram and area
charts that fluctuate around 80% accuracy. The drops
in the performances of both line chart and time series
classifiers coincide with steep rises in the numbers of
examples for the respective classes, suggesting that
the classifier needed some iterations to adapt to the
new dataset. The fluctuations in the quality of his-
togram and area chart classifiers stem from the small
sample sizes for the respective classes. For the confu-
sion matrix, please refer to Section 5 of appendix.
We have found the most consistent confusions be-
tween boxplots and column charts and maps and pie
charts. The confusion between boxplots and column
charts could stem from the presence of error bars in
boxplots and a special type of column charts. The
confusion between maps and pie charts could be ex-
plained with the presence of certain images where pie
charts were overlaid on the maps. Another similar
Figure 6: Line graph of the development of the accuracy
of each binary classifier during the iterative learning phase.
Notably, even though most classifiers began with classifica-
tion accuracy of less than 80%, almost all of them increase
their accuracy drastically after the first five iterations.
case is between pie charts and stacked area charts.
The reason could be because both visualizations share
a similar representation goal as ’Proportion’ and the
division of some examples from ’Proportion’ into
these two charts at the previous stage (see subsubsec-
tion 4.3.4).
Table 5: Scores from Incremental Learning.
Classes Accuracy
Ordination Plot 0.98
Map 0.97
Scatterplot 0.89
Line Chart 0.91
Dendrogram 0.97
Column Chart 0.97
Heatmap 0.95
Boxplot 0.96
Area Chart 0.80
Network 0.91
Histogram 0.83
Timeseries 0.84
Noviz 0.93
Pie Chart 0.97
Stack Area Chart 0.96
Combining Image and Caption Analysis for Classifying Charts in Biodiversity Texts
165
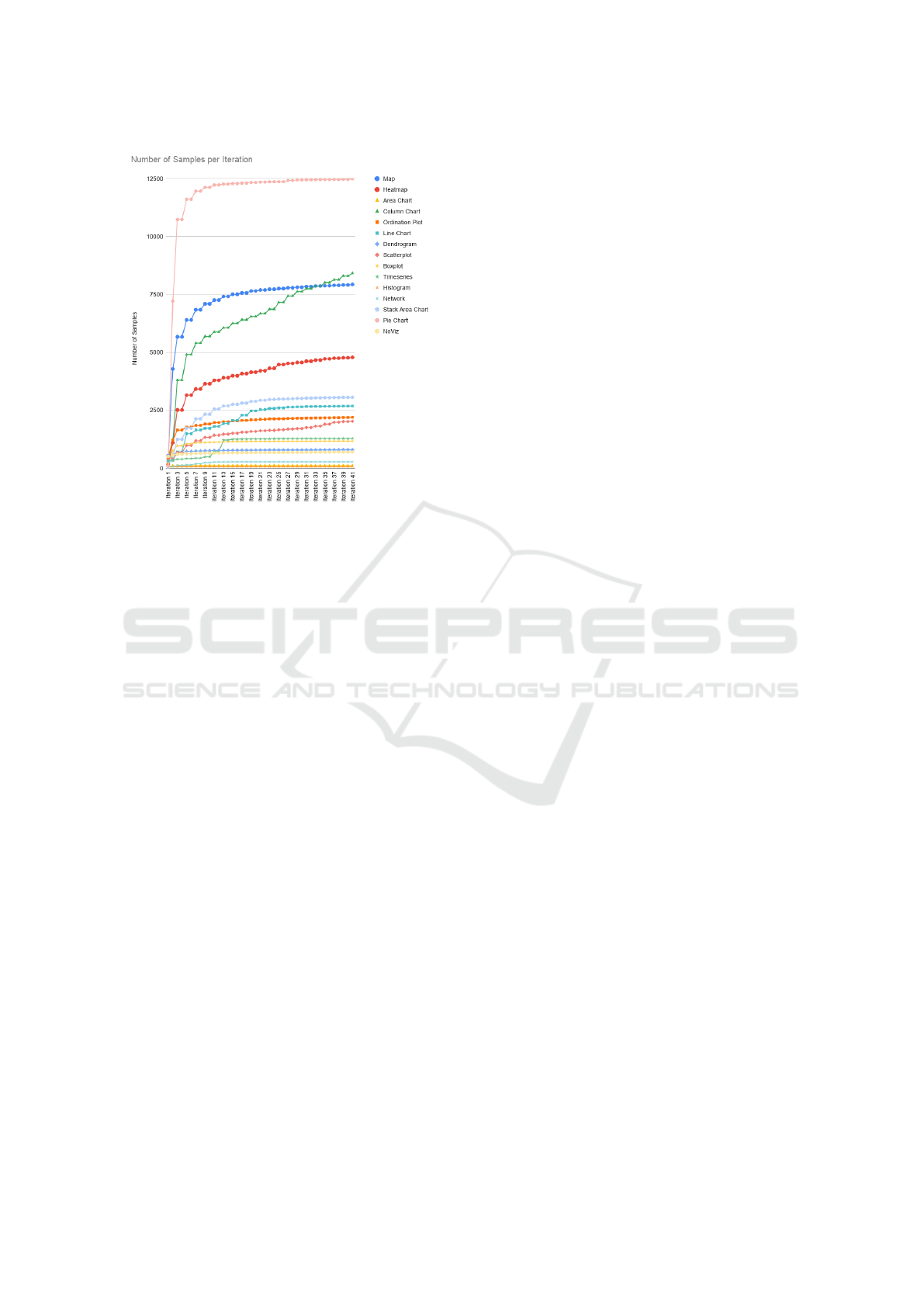
Figure 7: The development of the sample sizes for each
class during the iterative annotation. Similar to the increase
in accuracy of the binary classifiers in Figure 6, the numbers
of sample sizes increase very quickly within the first few
iterations.
5.2 Reasons for Misclassifications
In our extensive study of the misclassification cases,
we were able to extract several categories of reasons,
why these misclassifications happened.
• Mixed Vocabulary from Different Chart
Types: The main reasons for this problem was
a) often, multiple different visualizations are
used in conjunction in one image, showing, for
example, pie charts on different locations on a
map. We have observed that the classifier could
not perform well on those image captions, as
the information about multiple chart types in the
same text seemed to offer conflicting clues. b)
In one image, multiple different visualizations
are used to represent multidimensionality of
the results. For example, the use of scatterplot
for showing the distribution of some species
and in the same image use of column chart for
illustrating the comparison with other species.
Although all efforts were made to remove such
instances from our training set, however, we
can’t deny the existence of them in the rest of the
corpus.
• Similar Representational Goal: Histograms,
boxplot and scatterplot share same goal of
showing distribution among continuous variables.
Where histogram shows the frequency distribu-
tion of a variable, boxplot provides detail informa-
tion about this distribution among different quar-
tiles. Then, scatterplot shows relationship and
causation of this distribution with other variable/s.
Unfortunately, although the visual representation
is different, the language describing both visual-
izations tends to use similar wording, likely caus-
ing misclassifications.
• Mixture of Definition/Description and Inter-
pretation Vocabulary: A caption can be used
to fulfill different tasks: define/describe the con-
tents and/or interpret them. As the language dif-
fers very heavily from one task to the other and
the ratio between definitions and interpretations
varies from sample to sample even within a given
class, a classifier might be drawn to either spe-
cialize in the definition/interpretation parts of the
samples (high precision, low recall) or generalize
to a point that it cannot exclude other classes (low
precision, high recall).
• Level of Abstraction of Some Classes: Due
to limited examples for some of the classes, we
had to form superclasses of visualization types.
For example, ’Column Chart’ class is created
by merging examples from 14 related visual-
izations. This also leads confusion with other
classes. For example boxplots are confused with
column charts due to the presence of error bars in
certain types of column chart.
• Wrongly Mentioned Visualization Types: In
addition to the regular vocabulary, the binary clas-
sifiers also look for specific visualization name in
the caption texts. Unfortunately, in some captions,
wrong visualization names are referred, mistaking
for example a column chart for a histogram.
5.3 Comparison
Table 5 provides individual scores for different
classes. Due to the special goal and characteristics
of our study, currently we do not have any base study
to compare our results with. None of the previous
studies have considered both aspects of charts (visu-
als from images and chart semantics from captions)
for chart classification. In Figure 8, we have provided
the comparison among scores from common classes
in 3 different studies.
Figure 8 shows that in comparison to other stud-
ies, we are only lacking in two classes i.e Scatterplot
and Area Chart. In our work, ’Ordination Plot’ and
’Stack Area Chart’ which is similar to ’Scatterplot’
IVAPP 2020 - 11th International Conference on Information Visualization Theory and Applications
166
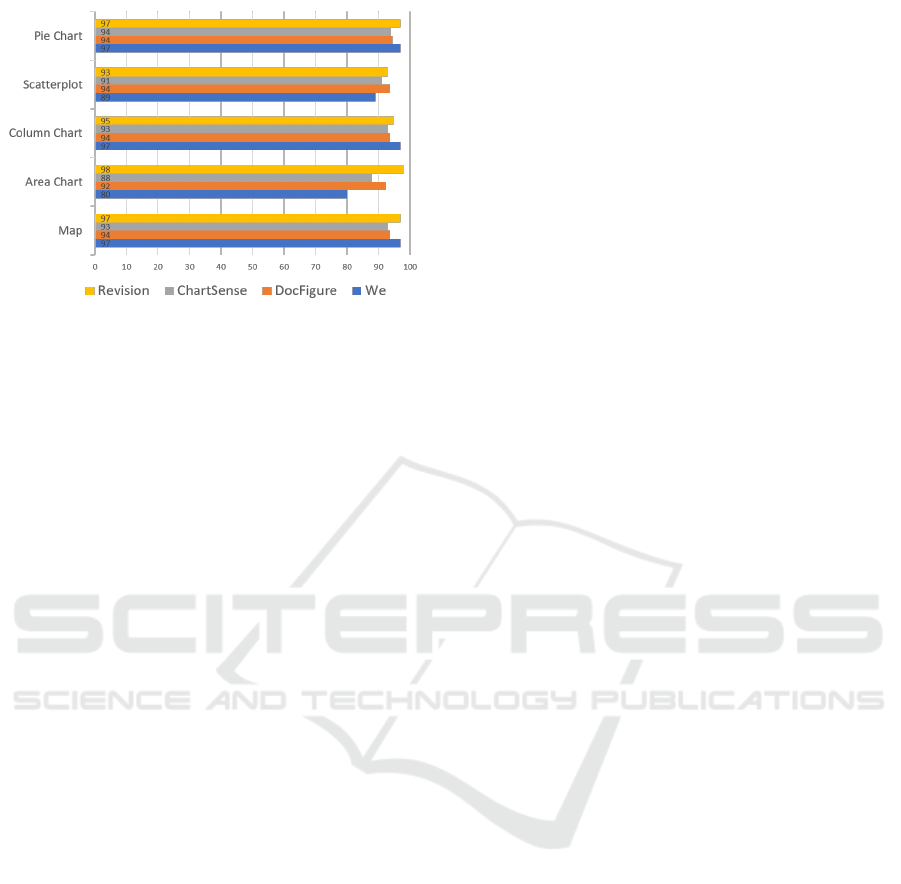
Figure 8: Comparison with other studies: Revision (Savva
et al., 2011), ChartSense (Jung et al., 2017) and DocFigure
(Jobin et al., 2019).
and ’Area Chart’ were considered as a separate class
based on their representative goal and visual dissim-
ilarities. No such fine differences were made in the
other studies. Scores of Ordination plot is 98% and
Stack area chart score 96% and if we compare them
with the other studies, then our performance is bet-
ter. With an average accuracy (F1-score) of 92.2%
we have proved that our approach of chart classifi-
cation is better than other chart image classification
technique.
6 CHALLENGES AND
RESEARCH DIRECTION
The result from our study lays a strong foundation that
for better chart classification apart from visual chart
elements, caption and other chart related text could
be a major source of information.It is just a prelim-
inary step. For an enhanced semantic chart recogni-
tion systems, there are many problems that needs to
be answered-
• As our classifiers were only trained on the biodi-
versity text, we are not sure how well they will
perform on general text or text from other do-
mains. More studies are needed in this direction
wherein a classifier trained in one domain can be
generalized to other domain. This work is out of
scope for our research therefore, we leave it on
future studies.
• Apart from only using the captions in the train-
ing process, text in the publication refers to the
chart images could yield better results. Moreover,
if the data is enriched with more semantic knowl-
edge like synonyms, concurrent words, ontolo-
gies etc, then better classification accuracy can be
achieved.
• The real population is not always as clean as the
training data that is fed to the classifiers. There-
fore, more studies are required that can under-
stand the common variations found in the visual-
ization images and captions. For example, hybrid
visualizations, multi-embed visualizations etc.
• Semantic chart classification classify the charts
not only based on the chart elements but also their
representation goals. We have found that those vi-
sualizations that tend to share the same goals are
the one with most false positives. The way out for
this is to create the classes solely based on the vi-
sualization goals. Then such work will be more
helpful for task-based visualization recommenda-
tion systems.
7 CONCLUSION AND FUTURE
WORK
In this work on chart classification, along with the vi-
sual similarity of different chart types, we have also
considered the conceptual similarities of the charts.
We have manually labelled the chart images and cap-
tions from biodiversity publications. We have trained
both the image and chart classifiers on this data. From
the best results of these two classifiers, we have in-
crementally trained our text classifier. Doing so,
we have achieved an average (F1-score) of 92.2%
from ensembles of binary caption classifiers. Our re-
sult proves that conceptual/semantic chart classifiers
can efficiently differentiate between those chart types
which are visually similar and are as efficient as im-
age classifiers. Along with that due to the conceptual
understanding of such classifiers, they can be used
for different purposes. One of these is the creation
of knowledge-assisted visualization recommendation
systems. We will be using these classifiers to infer
different visualizations/chart types from biodiversity
text.
ACKNOWLEDGEMENTS
The work has been funded by the DFG Priority Pro-
gram 1374 Infrastructure-Biodiversity-Exploratories
(KO 2209 / 12-2). We would like to thanks our
project principal investigator Birgitta K
¨
onig-Ries for
her valuable feedback.
Combining Image and Caption Analysis for Classifying Charts in Biodiversity Texts
167

REFERENCES
(2018). Elsevier sciencedirect apis. https://dev.elsevier.
com/sciencedirect.html#/Article Retrieval.
(2019). https://tfhub.dev/google/imagenet/mobilenet v2
050 96/feature vector/2.
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z.,
Citro, C., Corrado, G. S., Davis, A., Dean, J., Devin,
M., et al. (2016). Tensorflow: Large-scale machine
learning on heterogeneous distributed systems. arXiv
preprint arXiv:1603.04467.
Abhijit Balaji, Thuvaarakkesh Ramanathan, V. S. (2018).
Chart-text: A fully automated chart image descriptor.
CoRR, abs/1812.10636.
Aggarwal, C. C. and Zhai, C. (2012). Mining text data.
Springer Science & Business Media.
Boser, B. E., Guyon, I. M., and Vapnik, V. N. (1992).
A training algorithm for optimal margin classifiers.
In Proceedings of the 5th Annual ACM Workshop
on Computational Learning Theory, pages 144–152.
ACM Press.
Charbonnier, J., Sohmen, L., Rothman, J., Rohden, B., and
Wartena, C. (2018). Noa: A search engine for reusable
scientific images beyond the life sciences. In Pasi, G.,
Piwowarski, B., Azzopardi, L., and Hanbury, A., ed-
itors, Advances in Information Retrieval, pages 797–
800, Cham. Springer International Publishing.
Chen, M., Grinstein, G., Johnson, C. R., Kennedy, J., and
Tory, M. (2017). Pathways for theoretical advances in
visualization. IEEE computer graphics and applica-
tions, 37(4):103–112.
Chollet, F. et al. (2015). Keras. https://keras.io.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
In Machine Learning, pages 273–297.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep
Learning. MIT Press. http://www.deeplearningbook.
org.
Harris, R. L. (2000). Information graphics: A comprehen-
sive illustrated reference. Oxford University Press.
Ho, T. K. (1995). Random decision forests. In Proceedings
of 3rd international conference on document analysis
and recognition, volume 1, pages 278–282. IEEE.
Joachims, T. (1998). Text categorization with support vec-
tor machines: Learning with many relevant features.
In European conference on machine learning, pages
137–142. Springer.
Jobin, K. V., Mondal, A., and Jawahar, C. V. (2019).
Docfigure: A dataset for scientific document figure
classification. In 2019 International Conference on
Document Analysis and Recognition Workshops (IC-
DARW), volume 1, pages 74–79.
Jung, D., Kim, W., Song, H., Hwang, J.-i., Lee, B., Kim,
B., and Seo, J. (2017). Chartsense: Interactive data
extraction from chart images. In Proceedings of the
2017 CHI Conference on Human Factors in Comput-
ing Systems, pages 6706–6717. ACM.
Kaur, P., Klan, F., and K
¨
onig-Ries, B. (2018). Issues and
suggestions for the development of a biodiversity data
visualization support tool. In Proceedings of the Eu-
rographics/IEEE VGTC Conference on Visualization:
Short Papers, EuroVis ’18, pages 73–77.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. arXiv preprint
arXiv:1412.6980.
Lee, P.-s., West, J. D., and Howe, B. (2017). Viziometrics:
Analyzing visual information in the scientific litera-
ture. IEEE Transactions on Big Data, 4(1):117–129.
Liu, Y., Lu, X., Qin, Y., Tang, Z., and Xu, J. (2013). Review
of chart recognition in document images. In Visualiza-
tion and Data Analysis 2013, volume 8654, pages 384
– 391. International Society for Optics and Photonics,
SPIE.
Moody, A. and Jones, J. A. (2000). Soil response to canopy
position and feral pig disturbance beneath quercus
agrifolia on santa cruz island, california. Applied Soil
Ecology, 14(3):269 – 281.
Munzer, T. (2009). A nested model for visualization design
and validation. IEEE Transactions on Visualization
and Computer Graphics, 15(6):921–928.
Murphy, R. F., Kou, Z., Hua, J., Joffe, M., and Cohen,
W. W. (2004). Extracting and structuring subcellu-
lar location information from on-line journal articles:
The subcellular location image finder. In Proceedings
of the IASTED International Conference on Knowl-
edge Sharing and Collaborative Engineering, pages
109–114.
Rafkind, B., Lee, M., Chang, S.-F., and Yu, H. (2006).
Exploring text and image features to classify im-
ages in bioscience literature. In Proceedings of the
HLT-NAACL BioNLP Workshop on Linking Natural
Language and Biology, LNLBioNLP ’06, pages 73–
80, Stroudsburg, PA, USA. Association for Computa-
tional Linguistics.
Ramos, J. et al. (2003). Using tf-idf to determine word rele-
vance in document queries. In Proceedings of the first
instructional conference on machine learning, volume
242, pages 133–142. Piscataway, NJ.
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., and
Chen, L. (2018). Inverted residuals and linear bottle-
necks: Mobile networks for classification, detection
and segmentation. arXiv preprint arXiv:1801.04381.
Savva, M., Kong, N., Chhajta, A., Fei-Fei, L., Agrawala,
M., and Heer, J. (2011). Revision: Automated clas-
sification, analysis and redesign of chart images. In
Proceedings of the 24th annual ACM symposium on
User interface software and technology, pages 393–
402. ACM.
Sebastiani, F. (2002). Machine learning in automated
text categorization. ACM computing surveys (CSUR),
34(1):1–47.
Srihari, R. K. (1991). Piction: A system that uses captions
to label human faces in newspaper photographs. In
AAAI, pages 80–85.
Xu, S., McCusker, J., and Krauthammer, M. (2008). Yale
image finder (yif): a new search engine for retriev-
ing biomedical images. Bioinformatics, 24(17):1968–
1970.
APPENDIX
Appendix and scripts are available online at:
https://github.com/fusion-jena/Biodiv-Visualization-
Classifier
IVAPP 2020 - 11th International Conference on Information Visualization Theory and Applications
168
