
Evaluation of Phenotyping Errors on Polygenic Risk Score
Predictions
Ruowang Li, Jiayi Tong, Rui Duan, Yong Chen and Jason H. Moore
Department of Biostatistics, Epidemiology & Informatics, University of Pennsylvania Philadelphia, PA, 19104, U.S.A.
Keywords: Polygenic Risk Score, PRS, Electronic Health Record, Phenotyping Error, Risk Prediction.
Abstract: Accurate disease risk prediction is essential in healthcare to provide personalized disease prevention and
treatment strategies not only to the patients, but also to the general population. In addition to demographic
and environmental factors, advancements in genomic research have revealed that genetics play an important
role in determining the susceptibility of diseases. However, for most complex diseases, individual genetic
variants are only weakly to moderately associated with the diseases. Thus, they are not clinically informative
in determining disease risks. Nevertheless, recent findings suggest that the combined effects from multiple
disease-associated variants, or polygenic risk score (PRS), can stratify disease risk similar to that of rare
monogenic mutations. The development of polygenic risk score provides a promising tool to evaluate the
genetic contribution of disease risk; however, the quality of the risk prediction depends on many contributing
factors including the precision of the target phenotypes. In this study, we evaluated the impact of phenotyping
errors on the accuracies of PRS risk prediction. We utilized electronic Medical Records and Genomics
Network (eMERGE) data to simulate various types of disease phenotypes. For each phenotype, we quantified
the impact of phenotyping errors generated from the differential and non-differential mechanism by
comparing the prediction accuracies of PRS on the independent testing data. In addition, our results showed
that the rate of accuracy degradation depended on both the phenotype and the mechanism of phenotyping
error.
1 INTRODUCTION
Understanding the risk factors underlying diseases
has long been pursued in healthcare in order to screen
and prevent disease onset for high-risk individuals.
Proper quantification of the risk factors could help
stratify patients based on their risk profiles, which in
turn can be beneficial for developing personalized
disease prevention and treatment strategies
(Torkamani, Wineinger, & Topol, 2018). With the
development of high-throughput sequencing
technologies, it is now a reality to systematically
evaluate the genotypes’ contribution to disease risks.
Genetic twin studies have shown that many human
phenotypes and diseases are highly heritable;
however, early genome-wide association studies have
identified many single nucleotide polymorphisms
(SNPs) that are only weakly to moderately associated
with the diseases. In addition, for the associated
SNPs, they only explain a small amount of the disease
risks (Lo, Chernoff, Zheng, & Lo, 2015; Manolio et
al., 2009; Visscher et al., 2017). Recent studies have
demonstrated that many phenotypes are polygenic in
nature, meaning a phenotype is associated with more
than one gene (Purcell et al., 2009; Yang et al., 2010).
Thus, the polygenic risk score (PRS) method was
developed to capture the small effects from many
genetic factors in order to combine their effects into a
single predictive variable (Euesden, Lewis, &
O’Reilly, 2015; Purcell et al., 2009). The PRS has
been evaluated for its role in determining disease risk
in many complex diseases including coronary artery
disease, atrial fibrillation, type 2 diabetes,
inflammatory bowel disease, breast cancer (Khera et
al., 2018), obesity (Khera et al., 2019), schizophrenia
(Schizophrenia Working Group of the Psychiatric
Genomics Consortium, 2014), and antipsychotic drug
treatment (J.-P. Zhang et al., 2019). For some of the
diseases, the predictive power of PRS has reached
clinical significance similar to that of monogenic
mutations (Khera et al., 2018).
For the past decade, electronic health record
(EHR) linked genetic data has proven to be a valuable
data source for identifying genetic associations for
diseases. EHR with linked genetic data has the
Li, R., Tong, J., Duan, R., Chen, Y. and Moore, J.
Evaluation of Phenotyping Errors on Polygenic Risk Score Predictions.
DOI: 10.5220/0008935301230130
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 3: BIOINFORMATICS, pages 123-130
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
123

advantages of having a large sample of the patient
population as well as a rich source of matching
clinical phenotypes to conduct genomics research. In
addition, several EHR data have already been used to
conduct PRS research, including the UK Biobank
(Khera et al., 2018) and eMERGE (Li, Chen, &
Moore, 2019). While the genetic data is an integral
part of PRS prediction, the phenotype used to
construct PRS is equally as important. A crucial step
in constructing a PRS is to determine the marginal
association of each SNP with the phenotype. Thus,
the quality of the associations determines the utility
of the constructed PRS. However, there are
unavoidable biases and measurement errors
associated with the EHR derived phenotypes.
Existing studies have evaluated the impact of
phenotyping errors on statistical inference and
showed that the errors could increase false negatives
(Duan et al., 2016) as well as inflate the number of
false positives (Chen, Wang, Chubak, & Hubbard,
2019) of the associations. Nevertheless, so far, there
has been no investigation on the impact phenotyping
error on the predictive ability of PRS.
In this study, we used real EHR data from
eMERGE to simulate three types of phenotype under
two phenotyping error mechanisms, differential
(error differs across covariates’ levels) and non-
differential (error is consistent across covariates’
levels). We systematically quantified the PRS
predictive ability in different phenotypes under
different severities of phenotyping error and error
mechanisms. Our results showed that as more errors
were added to the phenotypes, non-differential
phenotyping errors lowered the PRS prediction
accuracies similarly among different phenotypes. In
contrast, differential phenotyping errors affected the
PRS prediction differently depending on the
underlying phenotype model. We believe that our
results could better inform researchers and clinicians
of the robustness of PRS when assessing disease risk.
2 METHOD
To evaluate the impact of phenotyping error on PRS
prediction, we used simulated datasets where we
knew the ground truth to quantify the change in
prediction accuracy. The evaluation was carried out
in five stages. 1) Use real patients’ genetic data from
eMERGE EHR as input to construct PRS. 2) Simulate
known phenotypes under various underlying true
models. The phenotypes were constructed to have
true associations with demographic, environmental,
clinical, and genetic factors (PRS). 3) Inject errors
into the known phenotypes under two different error
generating mechanisms: differential and non-
differential 4) Adjust the strength of the phenotyping
error 5) Quantitatively evaluate the predictive ability
of PRS on the testing data under each simulation
scenario.
2.1 eMERGE EHR Genetic Data
In order to simulate realistic PRS, we utilized the
patients’ genetic data from the electronic medical
records and genomics network (eMERGE, dbGaP
accession: phs000888.v1.p1) (McCarty et al., 2011).
Recent studies suggested that PRS does not perform
well across multiple ethnic groups; thus we restricted
our study samples to only one ethnicity (Martin et al.,
2017, 2019). To maximize the sample size, we
extracted white patients from nine different hospitals
under eMERGE: Children's Hospital of
Pennsylvania, Cincinnati Children's Hospital Medical
Center/Boston's Children's Hospital, Geisinger
Health System, Group Health/University of
Washington, Essentia Institute of Rural Health,
Marshfield Clinic, Pennsylvania State University
(Marshfield), Mayo Clinic, Icahn School of Medicine
at Mount Sinai School, Northwestern University, and
Vanderbilt University. The SNP genotyping was
performed using the Illumina 660W-Quad BeadChip
at the Center for Genotyping and Analysis at the
Broad Institute, Cambridge, MA. Genome imputation
was performed by eMERGE according to the
standard pipeline (Verma et al., 2014). Overall,
31,183 patients’ 38,040,165 autosomal SNP
genotypes were extracted.
2.2 Phenotype Simulation
We simulated three types of phenotype under
different underlying true models (Figure 1, solid
arrows on top). First, a phenotype was simulated to
be associated with the demographic variables, a set of
causal SNPs, and an environmental factor. All
variables were independently associated with the
phenotype; thus, it was named the independent model.
Second, a phenotype was simulated to be associated
with the demographic variables, a set of casual SNPs
and a related diagnosis. In this case, the related
diagnosis was also associated with a subset of the
causal SNPs, though the associations were different
from that of the phenotype. For example, a subset of
causal SNPs may have pleiotropic effects between
hypertension and heart failure, but the pleiotropic
associations with the two diseases are distinct. In
addition, diagnosis in hypertension is also one of the
factors in determining heart failure status. Because
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
124
the related diagnosis (hypertension) shared a subset
of causal SNPs with the phenotype (heart failure) and
the associations were distinct, the model was called
the weakly correlated model. Finally, a phenotype
was similarly simulated to be associated with
demographic variables, a set of causal SNPs, and a
related diagnosis as in the weakly correlated model.
However, the set of pleiotropic SNPs had the same
effects on the related diagnosis as on the phenotype.
An example would be that a subset causal SNPs are
similarly associated with cardiac arrest (related
diagnosis) as well as heart failure (phenotype).
Figure 1: Phenotypes generating mechanism. The
phenotypes were generated using patients’ age, gender,
SNP genotypes, and an environmental factor or a related
diagnosis status. The top solid arrows represent the true
phenotype generating mechanism. In the independent
model, all factors were independently associated with the
phenotype. In the weakly correlated model, the related
diagnosis and the phenotype shared a subset of causal
SNPs, but the associations
and
were independent. In
the strongly correlated model, the subset of shared casual
SNPs had the same associations, as in
is a subset of
.
The bottom dotted arrows indicate the phenotype error
generating mechanism. The biased phenotypes were
generated based on the values of the true phenotype and the
environmental factor or the related diagnosis.
Furthermore, cardiac arrest is also associated with
heart failure diagnosis. In this study, this model was
named strongly correlated model. The SNPs in all
models were randomly selected from the common
SNPs (minor allele frequency > 5%) in the eMERGE
EHR genetic data. The mathematical models for the
phenotype simulation are presented in the following
sections.
2.2.1 Independent Model
In this model, the phenotype Y was generated through
the logistic model.
Phenotype:
Logit
PY 1
~30.3∗Age0.1∗Gender
∑
β
∗
2∗_
The coefficients for the intercept, age, gender, and
environmental factors (Env_factor) were selected so
that the disease prevalence was around 30%. The
same coefficients were also used for the weakly
correlated and the strongly correlated model so that
the models were comparable. The distributions of the
random variables in all equations were listed in Table
1.
2.2.2 Weakly Correlated Model
In the weakly correlated model, a related diagnosis
was first generated using q SNPs, where q was a
subset of p SNPs that were used to generate the
phenotype. In addition, the coefficients for
generating the related diagnosis were independent of
β that were used to generate the phenotype.
Table 1: Parameter values for phenotype simulation.
Variable Value
Total randomly selected SNPs 500
Phenotype associated SNPs p = 100
Diagnosis associated SNPs q = 50
Age Normal (40, 10)
Gender Bernoulli (p = 0.5)
Environmental factor (Env_factor) Bernoulli (p = 0.5)
Phenotype ~ SNP associations
~ Normal (0, 0.3)
Related diagnosis ~ SNP associations
~ Normal (0, 0.3)
Related diagnosis:
Logit
PRelateddiagnosis 1
~
∗
;q⊆
Phenotype:
Logit
PY 1
~ 3 0.3∗Age0.1∗Gender
∑
β
∗
2∗
Evaluation of Phenotyping Errors on Polygenic Risk Score Predictions
125
2.2.3 Strongly Correlated Model
The strongly correlated model was the same as the
weakly correlated model except that the related
diagnosis and the phenotype shared a subset of q
SNPs as well as their coefficients.
Related diagnosis:
LogitPRelateddiagnosis 1~
∑
β
∗
;q ⊆
;
⊆
Phenotype:
Logit
Y1
~30.3∗Age0.1∗Gender
∑
β
∗
2∗
2.3 Biased Phenotype Due to Errors
As shown in Figure 1, the biased phenotypes were
generated based on the value of the true phenotypes
as well as the environmental factor or the related
diagnosis (Figure 1, dotted arrows at the bottom). The
intuition was that, first, the biased phenotype would
be expected to be a deviation from the true phenotype.
Second, many of the phenotyping algorithms utilized
by EHR systems used environmental and diagnosis
variables to determine the phenotype or disease
status, thus, the precision of the phenotype was also
associated with these factors (Kirby et al., 2016;
Robinson, Wei, Roden, & Denny, 2018; Wei &
Denny, 2015). Mathematically, the phenotyping
errors were determined by the sensitivity and
specificity:
Sensitivity and specificity measures how close the
biased phenotype is to the true phenotype. As an
example,
in the independent model, the biased
phenotype was generated using the following 2x2
tables. The 2x2 table shows that the biasness of the
phenotype depends on both the phenotype as well as
an environmental factor. Thus, there were two pairs
of sensitivity and specificity values for the two
variables.
Y=1 Y=0
ENV_FACTOR=1 Sensitivity
Exposure
Specificity
Exposure
ENV_FACTOR=0 Sensitivity
non_Exposure
Specificity
non_Exposure
Mathmatically, the Sensitivity
Exposure
controlled
the sensitivity of the biased Y when the true
phenotype Y = 1 and Env_factor = 1. The new
phenotype value under this combination was
generated using the Bernoulli distribution with the
probability equaled to
Sensitivity
Exposure
. In contrast,
the
Specificity
Exposure
determined the probably of the
biased Y = 0, when true Y = 0 and the Env_factor =
0. The value was generated by Bernoulli (1-
Specificity
Exposure
). Thus, the degree of phenotyping
errors was controlled by the values of the sensitivity
of specificity. As a special case, a phenotype was the
gold standard when sensitivity = specificity = 100%.
For biased phenotypes, the phenotyping error was
non-differential when the sensitivities (e.g. a) and
specificities (e.g. b) were the same across the two
Env_factor levels; otherwise, the error was
differential (e.g. a, b, c, and d). For instance, a
phenotype that is more error-prone for patients with
lower levels of environmental exposure would be
differentially biased.
NON‐DIFFERENTIALPHENOTYPINGERROR
Y=1 Y=0
ENV_FACTOR=1 a% b%
ENV_FACTOR=0 a% b%
DIFFERENTIALPHENOTYPINGERROR
Y=1 Y=0
ENV_FACTOR=1 a% c%
ENV_FACTOR=0 b% d%
2.4 Biased Phenotype Generation
For all phenotypes (independent, weakly correlated,
and strongly correlated), a range of phenotyping
errors were introduced using different levels of
sensitivity and specificity. In addition, differential
and non-differential error generating mechanisms
were applied at each sensitivity and specificity level.
To simplify the presentation of the results, the same
value of sensitivity and specificity for the non-
differential phenotyping error was used (Table 2). For
differential phenotyping error, one sensitivity and one
specificity were kept at 99%, while the others varied
(Table3). Overall, 60 biased phenotypes were
generated.
2.5 Evaluation of PRS Prediction
The effect of the phenotyping errors on PRS
prediction was evaluated in the following steps.
1. Split the data into training and testing
2. Sample sizes of the data split
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
126

3. Obtain coefficients for the SNPs using the
training data
4. Construct PRSs in both training and testing data
5. Build a predictive model using the PRS in the
training data
6. Apply the model to the test data
7. Compare the predicted phenotype value to the
true phenotype value
The data was split into the training and testing
datasets, with the testing dataset being held out for
evaluation. Using the training data, all SNPs’
marginal association, β
,
, with the biased
phenotypes were obtained. The marginal associations
from the training data were then used to construct
PRSs in both the training and testing data. Next, a
predictive model was built using the PRS in the
training data; the model was subsequently applied to
the testing PRS to obtain the predicted phenotype.
The predicted phenotype was compared with the true
phenotype in the testing data to obtain the testing
area-under-the-curve (AUC) value. The entire
process, from phenotype simulation to PRS
prediction, was repeated 100 times using different
random seeds to obtain 100 replications of the results.
3 RESULT
In all simulations, gold standard results were included
to serve as the baselines for comparison. The gold
standards demonstrated the maximum obtainable
prediction accuracies from PRSs that were generated
using the true phenotype. Figure 1 showed a change
in PRS prediction accuracy as more non-differential
errors were added into the phenotype. The accuracies
gradually decreased from gold standard to 50%
sensitivity and specificity. At 50% sensitivity and
specificity, the biased phenotype was generated the
same way as coin-flipping. Thus, the prediction
accuracies of PRS at this error level was also around
50%. Notably, the gold standard accuracies were also
different even when the simulation parameter values
were the same for all three phenotypes.
4 DISCUSSION
Disease risk prediction utilizing genetic information
via PRS has shown great promise in many complex
human diseases. With the increasing availability of
linked genetic data in EHR systems, PRS prediction
can be widely applied to many phenotypes and
diseases to identify high-risk patients for better
disease prevention and treatment care. Nevertheless,
patients’ true disease statuses are often unknown.
Thus, the observed disease status is only a proxy for
the true disease status, and the observed status will be
biased due to phenotyping errors. In this study, we
quantified the degradation of PRS prediction using
three different types of phenotype under the
differential and non-differential phenotyping errors.
Table 2: Sensitivity and specificity for the non-differential phenotyping error.
Error model name Error mechanism
Gold standard No error
X = (95, 90, 85, 80, 75, 70,
65, 60, 55, 50)
Y=1 Y=0
Env_factororDiagnosis=1
Sensitivity=X% Specificity=X%
Env_factororDiagnosis=0
Sensitivity=X% Specificity=X%
Table 3: Sensitivity and specificity for the non-differential phenotyping error.
Error model name Error mechanism
Gold standard No error
X = (95, 90, 85, 80, 75, 70,
65, 60, 55, 50)
Y=1 Y=0
Env_factororDiagnosis=1
Sensitivity=X% Specificity=99%
Env_factororDiagnosis=0
Sensitivity=99% Specificity=X%
Evaluation of Phenotyping Errors on Polygenic Risk Score Predictions
127
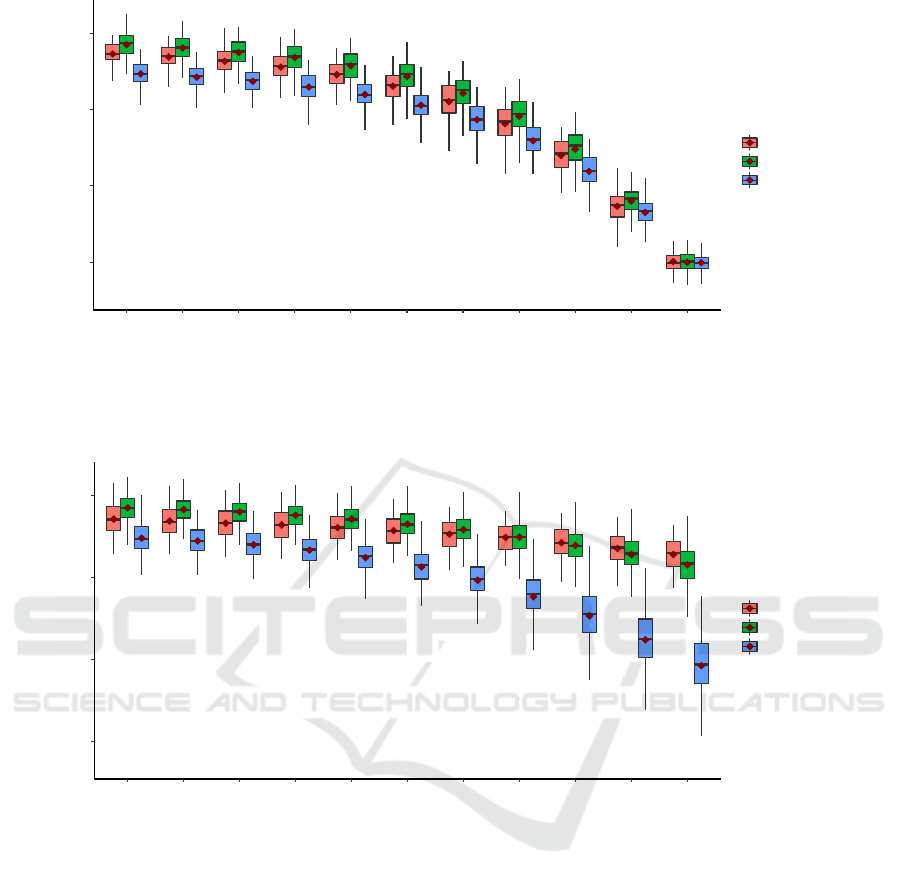
Figure 2: Performance of PRS prediction under non-differential phenotyping error. Each boxplot represents 100 replications
of the same experiment using different datasets. The x-axis indicates the sensitivity and specificity level set by variable X in
table 2. The y-axis shows the prediction AUC on the testing data.
Figure 3: Performance of PRS prediction under differential phenotyping error. Each boxplot represents 100 replications of
the same experiment using different datasets. The x-axis indicates the sensitivity and specificity level set by variable X in
table 3. The y-axis shows the prediction AUC on the testing data.
We utilized the eMERGE EHR genetic data so
that the SNPs had the minor allele frequency
distribution and correlation structure that are
observed in the real patients’ data. Using the SNPs
data along with other demographic and clinical
variables, we simulated three different phenotypes
with increasing levels of complexity (Figure 1). For
the phenotype generated under the independent
model, all variables independently related to the
phenotype. Here, we assumed that an individual’s
genetic factors do not affect one’s environmental
exposure. Under the weakly correlated model, we
used a related diagnosis status to determine the
phenotype status, and the two were associated with a
common subset of SNPs through pleiotropic effects.
In this case, we assumed that the associated effects
were different between the related diagnosis and the
phenotype. This is likely when the phenotypes are
regulated through different biological mechanisms,
such as between heart diseases and mental disorders
(Andreassen et al., 2013; Li, Duan, et al., 2019; X.
Zhang et al., 2019). Finally, in the strongly correlated
model, the diagnosis and the phenotype were assumed
to be more similar due to the shared underlying SNPs
as well as their coefficients. This reflects a possible
scenario when a subtype of disease is used to
diagnose the main disease.
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
0.747
0.786
0.773
0.743
0.781
0.769
0.737
0.775
0.764
0.73
0.769
0.756
0.72
0.758
0.746
0.706
0.744
0.731
0.687
0.722
0.711
0.659
0.691
0.682
0.619
0.648
0.64
0.565
0.58
0.573
0.499
0.5
0.501
0.5
0.6
0.7
0.8
G
o
l
d
s
t
a
n
d
a
r
d
9
5
%
9
0
%
8
5
%
8
0
%
7
5
%
7
0
%
6
5
%
6
0
%
5
5
%
5
0
%
Testing AUC
Simulation model
Independent
Weakly correlated
Strongly correlated
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
0.748
0.785
0.772
0.745
0.783
0.769
0.74
0.78
0.767
0.734
0.776
0.764
0.725
0.772
0.761
0.713
0.765
0.758
0.697
0.759
0.753
0.677
0.75
0.749
0.653
0.74
0.743
0.624
0.728
0.736
0.593
0.716
0.728
0.5
0.6
0.7
0.8
G
o
l
d
s
t
a
n
d
a
r
d
9
5
%
9
0
%
8
5
%
8
0
%
7
5
%
7
0
%
6
5
%
6
0
%
5
5
%
5
0
%
Te s t i n g AU C
Simulation model
Independent
Weakly correlated
Strongly correlated
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
128

As expected, as more phenotyping errors were
added to the three phenotypes, the prediction
accuracy of PRS decreased. However, the rates of the
decrease depended on the type of phenotyping errors.
First, the gold standards’ accuracy in Figure 2 and
Figure 3 were similar because they both represented
PRS predictive power without any phenotyping
errors. Interestingly, the PRS achieved the best
performance in the phenotype generated from the
weakly correlated model, followed by the
independent and strongly correlated model. This can
be explained by the different amount of genetic
contribution to the phenotype. In the weakly
correlated model, SNPs contributed to the phenotype
through two mechanisms: 1. direct associations with
the phenotype. 2. Indirect associations through the
related diagnosis. Because the indirect associations
were independent of the direct associations, the SNPs
contributed “twice” to the phenotype. In contrast, in
the independent model, the SNPs were only
associated with the phenotype through their direct
associations. And in the strongly correlated model,
the SNPs’ associations were diminished because part
of the associations was mediated by the related
diagnosis. Second, non-differential phenotyping
errors similarly affected all phenotypes. The relative
order of PRS prediction accuracies did not change as
more non-differential phenotyping errors were added.
Finally, differential phenotyping errors, which are
more likely to be observed in real data, exhibited
different accuracy trajectories for the phenotypes.
The independent model was affected the least, likely
because the SNPs and the environmental factor were
independent. Thus, differential phenotyping errors
induced by the environmental factor did not have a
major impact on the PRS prediction accuracy.
However, in the weakly correlated and strongly
correlated model, both the phenotype and the related
diagnosis were associated with the SNPs. Thus,
differential errors based on these variables had a
severe impact on the PRS, with the strongest impact
in the strongly correlated model. In summary, non-
differential phenotyping errors affected PRS
prediction equally among the phenotypes.
Differential phenotyping errors had an increased
impact on PRS prediction if the target phenotype and
the variables used to determine the phenotype have a
shared genetic component.
While it is useful to understand the impact of
phenotyping errors on PRS prediction, it is also
important to identify approaches that can minimize
the error. One effective approach to reducing error is
through manual chart review of patients’
comprehensive clinical histories by doctors or
domain experts. However, manual review is both
time-consuming and expensive. A potential
alternative approach is to chart review a subset of
patients to determine the amount of phenotyping error
as well as the error mechanism. Then, the results
presented in this study could serve as a guideline to
determine whether the errors are within the
acceptable range. If not, the phenotype quality needs
to be improved. For future studies, the impact of
phenotyping errors on the continuous outcome can be
explored. Furthermore, phenotype differences across
individuals or populations depend on both genetic and
environmental factors(Rosenberg, Edge, Pritchard, &
Feldman, 2019). To evaluate the relative importance
of these factors, it is essential to verify the accuracies
of the measurements. Thus, more complex error
patterns that depend on multiple environmental or
clinical variables are likely to be more realistic and
should be investigated. Finally, AUC is the current
standard measurement for PRS performance.
However, some studies suggested that AUC may not
be the best metric for evaluating classification
accuracy. Thus, other accuracy metrics, such as net
reclassification improvement or integrated
discrimination improvement can be used (Pencina,
D’Agostino, D’Agostino, & Vasan, 2008).
REFERENCES
Andreassen, O. A., Djurovic, S., Thompson, W. K., Schork,
A. J., Kendler, K. S., O’Donovan, M. C., … Dale, A.
M. (2013). Improved detection of common variants
associated with schizophrenia by leveraging pleiotropy
with cardiovascular-disease risk factors. American
Journal of Human Genetics, 92(2), 197–209.
https://doi.org/10.1016/j.ajhg.2013.01.001
Chen, Y., Wang, J., Chubak, J., & Hubbard, R. A. (2019).
Inflation of type I error rates due to differential
misclassification in EHR-derived outcomes: Empirical
illustration using breast cancer recurrence.
Pharmacoepidemiology and Drug Safety, 28(2), 264–
268. https://doi.org/10.1002/pds.4680
Duan, R., Cao, M., Wu, Y., Huang, J., Denny, J. C., Xu, H.,
& Chen, Y. (2016). An Empirical Study for Impacts of
Measurement Errors on EHR based Association
Studies. AMIA ... Annual Symposium Proceedings.
AMIA Symposium, 2016, 1764–1773. Retrieved from
http://www.ncbi.nlm.nih.gov/pubmed/28269935
Euesden, J., Lewis, C. M., & O’Reilly, P. F. (2015).
PRSice: Polygenic Risk Score software.
Bioinformatics, 31(9), 1466–1468. https://doi.org/
10.1093/bioinformatics/btu848
Khera, A. V., Chaffin, M., Wade, K. H., Zahid, S.,
Brancale, J., Xia, R., … Kathiresan, S. (2019).
Polygenic Prediction of Weight and Obesity
Evaluation of Phenotyping Errors on Polygenic Risk Score Predictions
129

Trajectories from Birth to Adulthood. Cell, 177(3),
587-596.e9. https://doi.org/10.1016/j.cell.2019.03.028
Khera, A. V, Chaffin, M., Aragam, K. G., Haas, M. E.,
Roselli, C., Choi, S. H., … Kathiresan, S. (2018).
Genome-wide polygenic scores for common diseases
identify individuals with risk equivalent to monogenic
mutations. Nature Genetics, 50(9), 1219–1224.
https://doi.org/10.1038/s41588-018-0183-z
Kirby, J. C., Speltz, P., Rasmussen, L. V, Basford, M.,
Gottesman, O., Peissig, P. L., … Denny, J. C. (2016).
PheKB: a catalog and workflow for creating electronic
phenotype algorithms for transportability. Journal of
the American Medical Informatics Association, 23(6),
1046–1052. https://doi.org/10.1093/jamia/ocv202
Li, R., Chen, Y., & Moore, J. H. (2019). Integration of
genetic and clinical information to improve imputation
of data missing from electronic health records. Journal
of the American Medical Informatics Association.
https://doi.org/10.1093/jamia/ocz041
Li, R., Duan, R., Kember, R. L., Rader, D. J., Damrauer, S.
M., Moore, J. H., & Chen, Y. (2019). A regression
framework to uncover pleiotropy in large-scale
electronic health record data. Journal of the American
Medical Informatics Association. https://doi.org/
10.1093/jamia/ocz084
Lo, A., Chernoff, H., Zheng, T., & Lo, S.-H. (2015). Why
significant variables aren’t automatically good
predictors. Proceedings of the National Academy of
Sciences of the United States of America, 112(45),
13892–13897. https://doi.org/10.1073/pnas.151828
5112.
Manolio, T. A., Collins, F. S., Cox, N. J., Goldstein, D. B.,
Hindorff, L. A., Hunter, D. J., … Visscher, P. M.
(2009). Finding the missing heritability of complex
diseases. Nature, 461(7265), 747–753. https://doi.org/
10.1038/nature08494
Martin, A. R., Gignoux, C. R., Walters, R. K., Wojcik, G.
L., Neale, B. M., Gravel, S., … Kenny, E. E. (2017).
Human Demographic History Impacts Genetic Risk
Prediction across Diverse Populations. The American
Journal of Human Genetics, 100(4), 635–649.
https://doi.org/10.1016/j.ajhg.2017.03.004
Martin, A. R., Kanai, M., Kamatani, Y., Okada, Y., Neale,
B. M., & Daly, M. J. (2019). Current clinical use of
polygenic scores will risk exacerbating health
disparities. BioRxiv, 441261. https://doi.org/10.1101/
441261
McCarty, C. A., Chisholm, R. L., Chute, C. G., Kullo, I. J.,
Jarvik, G. P., Larson, E. B., … eMERGE Team. (2011).
The eMERGE Network: a consortium of
biorepositories linked to electronic medical records
data for conducting genomic studies. BMC Medical
Genomics, 4, 13. https://doi.org/10.1186/1755-8794-4-
13
Pencina, M. J., D’Agostino, R. B., D’Agostino, R. B., &
Vasan, R. S. (2008). Evaluating the added predictive
ability of a new marker: From area under the ROC
curve to reclassification and beyond. Statistics in
Medicine, 27(2), 157–172. https://doi.org/10.1002/
sim.2929
Purcell, S. M., Wray, N. R., Stone, J. L., Visscher, P. M.,
O’Donovan, M. C., Sullivan, P. F., … Sklar, P. (2009).
Common polygenic variation contributes to risk of
schizophrenia and bipolar disorder. Nature, 460(7256),
748. https://doi.org/10.1038/nature08185
Robinson, J. R., Wei, W.-Q., Roden, D. M., & Denny, J. C.
(2018). Defining Phenotypes from Clinical Data to
Drive Genomic Research.
https://doi.org/10.1146/annurev-biodatasci
Rosenberg, N. A., Edge, M. D., Pritchard, J. K., & Feldman,
M. W. (2019). Interpreting polygenic scores, polygenic
adaptation, and human phenotypic differences.
Evolution, Medicine, and Public Health, 2019(1), 26–
34. https://doi.org/10.1093/emph/eoy036
Schizophrenia Working Group of the Psychiatric Genomics
Consortium. (2014). Biological insights from 108
schizophrenia-associated genetic loci. Nature,
511(7510), 421–427. https://doi.org/10.1038/
nature13595
Torkamani, A., Wineinger, N. E., & Topol, E. J. (2018).
The personal and clinical utility of polygenic risk
scores. Nature Reviews Genetics, 19(9), 581–590.
https://doi.org/10.1038/s41576-018-0018-x
Verma, S. S., de Andrade, M., Tromp, G., Kuivaniemi, H.,
Pugh, E., Namjou-Khales, B., … Ritchie, M. D. (2014).
Imputation and quality control steps for combining
multiple genome-wide datasets. Frontiers in Genetics,
5, 370. https://doi.org/10.3389/fgene.2014.00370
Visscher, P. M., Wray, N. R., Zhang, Q., Sklar, P.,
McCarthy, M. I., Brown, M. A., & Yang, J. (2017). 10
Years of GWAS Discovery: Biology, Function, and
Translation. The American Journal of Human Genetics,
101(1), 5–22. https://doi.org/10.1016/J.AJHG.2017.
06.005
Wei, W.-Q., & Denny, J. C. (2015). Extracting research-
quality phenotypes from electronic health records to
support precision medicine. Genome Medicine, 7(1),
41. https://doi.org/10.1186/s13073-015-0166-y
Yang, J., Benyamin, B., McEvoy, B. P., Gordon, S.,
Henders, A. K., Nyholt, D. R., … Visscher, P. M.
(2010). Common SNPs explain a large proportion of the
heritability for human height. Nature Genetics, 42(7),
565–569. https://doi.org/10.1038/ng.608
Zhang, J.-P., Robinson, D., Yu, J., Gallego, J.,
Fleischhacker, W. W., Kahn, R. S., … Lencz, T. (2019).
Schizophrenia Polygenic Risk Score as a Predictor of
Antipsychotic Efficacy in First-Episode Psychosis.
American Journal of Psychiatry, 176(1), 21–28.
https://doi.org/10.1176/appi.ajp.2018.17121363
Zhang, X., Veturi, Y., Verma, S., Bone, W., Verma, A.,
Lucas, A., … Ritchie, M. D. (2019). Detecting potential
pleiotropy across cardiovascular and neurological
diseases using univariate, bivariate, and multivariate
methods on 43,870 individuals from the eMERGE
network. Pacific Symposium on Biocomputing. Pacific
Symposium on Biocomputing, 24, 272–283. Retrieved
from http://www.ncbi.nlm.nih.gov/pubmed/30864329
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
130
