
The Sensitivity Evaluation of mt-DNA Genes; NADH Dehydrogenase
Sub Unit 5 (ND5), D-Loop, and Cytochrome-b (Cty-b) to Detect Pork
(Sus scrofa) DNA Isolate and DNA Fragment in Meatball
using PCR Technique
Joni Kusnadi
1,2
and Noval Audi Ashari
1
1
Department of Agricultural Product Technology, Faculty of Agricultural Technology, University of Brawijaya,
Jl. Veteran, Malang, Indonesia
2
Central Laboratory of Life Science, University of Brawijaya Malang, Jl. Veteran, Malang, Indonesia
Keywords: Meatball, mt-DNA, Sensitivity, Primer, Pork.
Abstract: The utilization of mt-DNA primers on previous study specific to detect DNA pork fragments. This study aims
were to evaluate the sensitivity of mt-DNA primers (ND5, D-Loop, and Cyt-b) in pork DNA isolates and its
meatballs product. The sensitivity analysis was conducted in pork DNA isolates with concentrations (10, 1,
10
-1
, 10
-2
, 10
-3
, 10
-4
, 10
-5
, and 10
-6
ng/µl) and its meatballs product with variation of pork content (0%, 0.01%,
0.05%, 0.1%, 0.5%, 1%, and 5%). Furthermore, the DNA fragment amplification process was carried out
using PCR technique. The amplification results showed that ND5 and Cyt-b primers were more sensitive
because they were able to amplify DNA with concentrations up to 10
-3
ng/µl compared to D-Loop which was
only able to amplify with concentrations up to 10
-2
ng/µl. The sensitivity results using meatballs showed that
ND5 was the most sensitive primer in detecting meatballs with concentration of pork up to 0.01%. It can be
concluded that ND5, Cyt-b, and D-Loop primers are able to detect pork DNA fragment with high sensitivity.
The ND5 primer gave the most sensitive amplification results because it was able to detect pork DNA
fragments with the lowest concentration and meatball with the lowest pork content.
1 INTRODUCTION
There were 1,300 cases of food adulteration between
1980 till 2010 (Moore et al., 2012). Some food
falsification case in various regions in Indonesia are
using pork in resembles beef and applying pork in the
process making of beef meatballs (Bempah, 2017;
Harianmerapi, 2018; Nuryanti, 2020). Meatballs are
food product that often being falsification in the
purpose of economic gain in the anticipation of high
price of beef. Food falsification is defined as an
attempt to replace, imitate, increase, change, or
misrepresent a food product, food packaging, and
fraudulent label information for economic gain
purposes (Hariyadi, 2015).
Identification and prevention of falsification food
is important to protect consumers and prevent
unhealthy competition for food producers especially
processed meat products. Therefore, we need a
detection method that simple and fast for everyday
application (Kesmen et al., 2010). In the analysis of
meatball samples, the meat has undergone high
temperature processing and treatment. Denaturation
of meat protein during heating treatment or some
specific technology applications in the processing
stage may decrease the success of the analytical
method. Therefore, the method is no longer able to
distinguish the species that related closely and
unsuitable for use as an everyday analysis, such as
species-specific detection becomes difficult and long
time (Hofman, 1987; Jemmi and Schlosser, 1992;
Koh et al., 1998; Kesmen et al., 2010).
DNA hybridization and PCR methods have been
widely used for the identification of meat processed
in meat products (Fei et al., 1996; Matsunaga et al.,
1999). Polymerase Chain Reaction (PCR) technique
is widely applied for the analysis of processed meat-
based food products because it is fast, simple,
specific, and sensitive (Matsunaga et al., 1999;
Kesmen et al., 2007; Haunshi et al., 2008; Yahya et
al., 2017). PCR technique is often used as a
technique for fast detection of pork content using
12
Kusnadi, J. and Ashari, N.
The Sensitivity Evaluation of mt-DNA Genes; NADH Dehydrogenase Sub Unit 5 (ND5), D-Loop, and Cytochrome-b (Cty-b) to Detect Pork (Sus scrofa) DNA Isolate and DNA Fragment in
Meatball using PCR Technique.
DOI: 10.5220/0010507000003108
In Proceedings of the 6th Food Ingredient Asia Conference (6th FiAC 2020) - Food Science, Nutrition and Health, pages 12-19
ISBN: 978-989-758-540-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

mt-DNA primers in the amplification process.
Mitochondrial DNA (mt-DNA) is DNA derived from
mitochondrial organelles with nucleotide that
similar to its parent and lots present in cells (Felk et
al., 2017). Species-specific primers are designed
based on the mt-DNA sequences of various animal
species used in species identification.
The study related to the primer species mt-DNA,
namely NADH dehydrogenase subunit 5 (ND5) with
a target sequence of 227 bp has been used in skeletal
muscle tissue samples from pigs and applied to
detect pork content in typical turkey meat products,
namely Sucuk (Kesmen et al., 2010). Research on
the use of mt-DNA Cyt-b species primers used the
multiplex PCR method to detect the content of pork
which pork had undergone heating treatment at
temperatures of 100°C and 120°C (Matsunaga et al.,
1999). Previous research, mt-DNA primers (ND5, D-
Loop, and Cyt-b) using conventional PCR
techniques applied to beef, goat, pork, lamb, and
chicken have been successfully applied to detect
specific pork content. In this study, mt-DNA primers
(ND5, D-Loop, and Cyt-b) were used to evaluate the
primary sensitivity to various variations of pork
DNA concentrations and the content of pork DNA
fragments in meatballs. This research was conducted
to support the mt-DNA primers specificity data
(ND5, D-Loop, and Cyt-b) in detecting the content
of pig DNA fragments using conventional PCR
techniques and for further research in detecting
cases of counterfeiting food processed products.
2 RESEARCH METHODS
The sensitivity primer test was carried out based on
variations in the DNA concentration of pork and pork
meatballs. The sample of pork DNA isolates had been
obtained and analyzed quantitatively using Nanodrop
(spectrophotometer) produced a DNA concentration
of 132.21 ng/µl and DNA purity of 1.98 (Kusnadi et
al., 2019). Pork DNA isolation samples were diluted
into several concentrations, namely 10 ng/µl, 1 ng/µl,
10
-1
ng/µl, 10
-2
ng/µl, 10
-3
ng/µl, 10
-4
ng/µl, 10
-5
ng/µl,
and 10
-6
ng/µl. In addition, pork meatballs are made
with a mixture consisting of spices (garlic, onion),
tapioca flour, monosodium glutamate, ice cubes, and
STTP (Sodium tripolipospat). The proportion of each
dough ingredient is shown on Table 1. The producing
of meatballs were mix all ingredients based on
predetermined proportions. The meatball dough was
boiled in water with a temperature of 80°C for 15
minutes until the meatball expands and is cooked. The
meatballs were drained and boiled again at 100°C for
5 minutes, removed, and drained. Furthermore, pork
meatballs with various pork content of 0% (negative
control), 0.01%, 0.05%, 0.1%, 0.5%, 1%, and 5%
(w/w) were DNA isolated using the alkaline-lysis
method with modified procedures.
PCR reactions consist of Go Taq Green Master
Mix (Promega), BSA (Bovine Serum Albumine),
Forward and reverse mt-DNA Primers (ND5, D-
Loop, and Cyt-b), pig DNA with various
concentrations and DNA pork meatballs. The PCR
program consisted of hot start at 95°C for 5 minutes,
denaturation at 95°C for 1 minute, annealing at 54°C
for 1 minute, extension at 72°C for 1 minute, and
post extension at 72°C for 7 minutes. There were 3
mt-DNA primers used, namely ND5, D-Loop, and
Cyt-b with target DNA sequence lengths, namely
227 bp, 835 bp, and 398 bp (Table 2). The
electrophoresis process was used 1.5% agarose gel,
loading dye, 1x TBE, EtBr (Ethidium Bromide), and
1 Kb of DNA ladder. Electrophoresis results were
visualized using gel doc imaging.
Table 1: Meatball ingredient formulation with pork substitution in beef.
Raw Material
Concentration of Beef Substitution with Pork
0% 0.01% 0.05% 0.1% 0.5% 1% 5%
Beef (gr) 40 39.99 39.98 39.96 39.8 39.6 38
Pork (gr) - 0.004 0.02 0.04 0.2 0.4 2
Instant flour for meatballs (gr) 16 16 16 16 16 16 16
Ice Cube (gr) 3.2 3.2 3.2 3.2 3.2 3.2 3.2
STTP (gr) 0.8 0.8 0.8 0.8 0.8 0.8 0.8
Total (gr) 60
The Sensitivity Evaluation of mt-DNA Genes; NADH Dehydrogenase Sub Unit 5 (ND5), D-Loop, and Cytochrome-b (Cty-b) to Detect Pork
(Sus scrofa) DNA Isolate and DNA Fragment in Meatball using PCR Technique
13
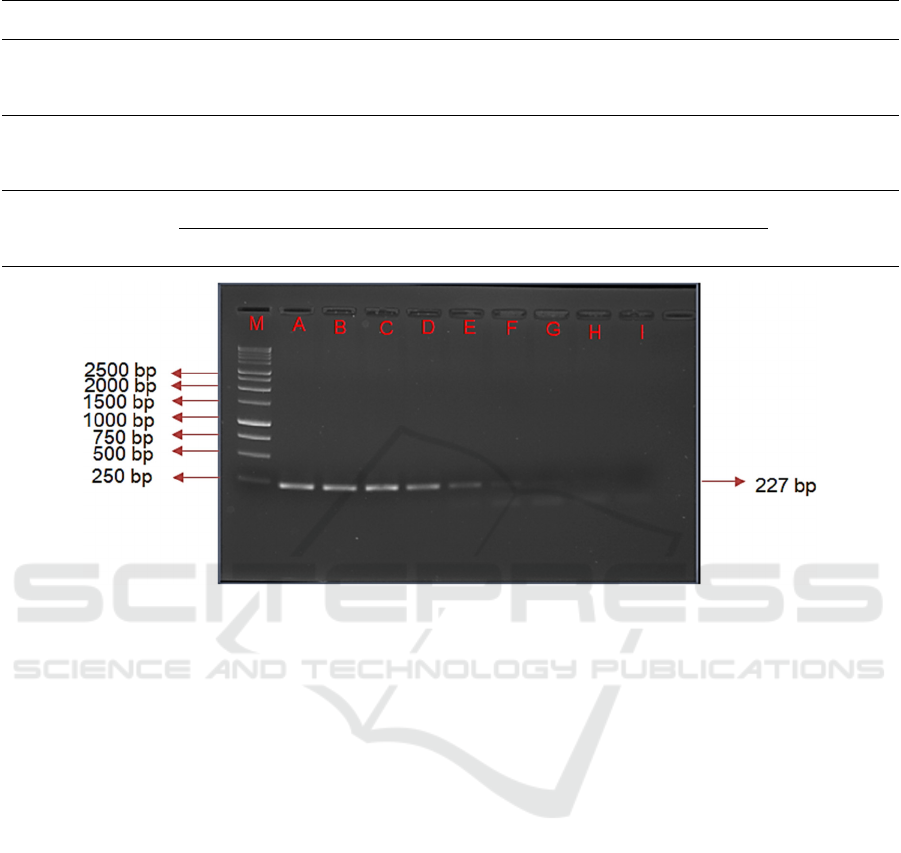
Table 2: Primers used in PCR stages.
Primer Sequence Position (5’- 3’) DNA Length (bp)
ND5 (Kesmen et al.,
2007)
F: 5’-CAT TCG CCT CAC TCA CAT TAA CC-3’
227
R: 5’-AAG AGA GAG TTC TAC GGT CTG TAG-3’
D-loop (Haunshi et al.,
2008)
F:5’-TAC TTC AGG ACC ATC TCA CC-3’
835
R:5’-TAT TCA GAT TGT GGG CGT AT-3’
Cyt-b (Matsunaga et
al., 1999)
F:5’-GAC CTC CCA GCT CCA TCA AAC ATC TCA TCT TGA TGA AA-3’
398
R: 5’-GCT GAT AGT AGA TTT GTG ATG ACC GTA-3’
Figure 1: Sensitivity Test using ND5 Primer in Various Pork DNA. Note: M: DNA ladder 1000 bp, A: initial concentration
(132,05 ng/μl), B: 10 ng/μl, C: 1 ng/μl, D: 10
-1
ng/μl, E: 10
-2
ng/μl, F: 10
-3
ng/μl, G: 10
-4
ng/μl, H: 10
-5
ng/μl, and I: 10
-6
ng/μl.
3 RESULTS
3.1 Sensitivity Test of Primer with
Various Pork DNA Concentrations
The sensitivity analysis aims to determine the primer
ability to detect fragment DNA with the smallest
concentration. In this study, the three species-specific
mt-DNA primers (ND5, D-Loop, and Cyt-b) were
tested at various concentrations of isolated pork
DNA. Previous research results showed that pork
DNA isolates were 132.05 ng/µl with a purity of 1.98
(Kusnadi et al., 2020). The pork DNA isolates has
been used with concentration of 132.05 ng/µl and
purity of 1.98 produced in previous research (Kusnadi
et al., 2020). The amplification results of the three mt-
DNA primers on pork DNA isolates showed that the
lower the DNA concentration used in the
amplification, the thinner the DNA bands produced
(Figures 1, 2, and 3).
Visualization of amplification results using
primers ND5, D-loop, and Cyt-b at various
concentrations of pork were showed that ND5 primers
were able to amplify DNA at DNA concentrations of
10 ng/μl, 1 ng/μl, 10
-1
ng/μl, 10
-2
ng/μl, and 10
-3
ng/μl.
However, the amplification results using the ND5
primer appeared to be thinner than the DNA band
amplified using the Cyt-b primer (Figure 1). Cyt-b
primer was able to amplify DNA with concentration
of 10 ng/μl, 1 ng/μl, 10
-1
ng/μl, 10
-2
ng/μl, and 10
-3
ng/μl constructs with clearer results than D-Loop and
ND5 primers (Figure 3). D-Loop primer was lowest
sensitivity to amplify of DNA samples with
concentrations of 10 ng/μl, 1 ng/μl, 10
-1
ng/μl, 10
-2
ng/μl (Figure 2).
The results of the primer sensitivity analysis have
been carried out before, ND5 primer sensitivity was
able to detect DNA concentrations of 10
-2
ng/µl
(Kesmen et al., 2007), while Cyt-b primer was able to
detect DNA concentrations of 0.25 ng/µl (Matsunaga
et al., 1999), and D-Loop primer has not been studies.
Primers are one of the components that determine the
success of the PCR technique. Primer specificity and
sensitivity analysis are very necessary to support
primary applications to detect contamination in food
processed products.
6th FiAC 2020 - The Food Ingredient Asia Conference (FiAC)
14

Figure 2: Sensitivity Test using D-Loop Primer in Various Pork DNA. Note: M: DNA ladder, A: initial concentration (132,05
ng/μl), B: 10 ng/μl, C: 1 ng/μl, D: 10
-1
ng/μl, E: 10
-2
ng/μl, F: 10
-3
ng/μl, G: 10
-4
ng/μl, H: 10
-5
ng/μl, dan I: 10
-6
ng/μl.
Figure 3: Sensitivity Test using Cyt-b Primer in Various Pork DNA. Note: M: DNA ladder, A: initial concentration (132,05
ng/μl), B: 10 ng/μl, C: 1 ng/μl, D: 10
-1
ng/μl, E: 10
-2
ng/μl, F: 10
-3
ng/μl, G: 10
-4
ng/μl, H: 10
-5
ng/μl, dan I: 10
-6
ng/μl.
Tabel 3: Concentration and Purity of Pork Meatball DNA Isolate.
Sample
Purity (λ 260/280)
Average
Concentration (ng/μl)
Average
Replicate Replicate
1 2 3 1 2 3
0% 2.12 1.65 1.89 1.89 55.26 142.95 133.05 110.42
0.01% 1.68 1.64 2.01 1.78 26.06 275.82 141.07 147.65
0.05% 2.30 1.92 1.90 2.04 46.75 151.33 90.21 96.0967
0.1% 1.87 1.99 1.93 1.93 63.02 154.03 85.68 100.91
0.5% 2.27 1.87 2.02 2.05 72.90 96.79 128.68 99.4567
1% 2.19 1.86 1.93 1.99 61.85 106.83 83.28 83.9867
5% 2.39 1.87 2.06 2.10 64.60 133.34 144.37 114.103
3.2 Sensitivity Test of Primer in Pork
Meatball Samples
The results of DNA isolation of pork meatballs with
concentrations of pork content of 1%, 0.01%, 0.05%,
0.1%, 0.5%, 1%, and 5% with 3 replications each
resulted in an average concentration of 83.99 to
147.65 ng/µl and the mean purity of 1.78 to 2.10
(Table 3). Based on the DNA concentration that has
been obtained, it shows that the isolation of pork
The Sensitivity Evaluation of mt-DNA Genes; NADH Dehydrogenase Sub Unit 5 (ND5), D-Loop, and Cytochrome-b (Cty-b) to Detect Pork
(Sus scrofa) DNA Isolate and DNA Fragment in Meatball using PCR Technique
15

meatballs using the alkaline-lysis method produces
high DNA concentrations, however, some samples
still contain RNA and protein contaminants.
Meatball is a processed meat product that has
undergone high temperature treatment and the
addition of various other ingredients. Other
ingredients added to the meatball making, namely
flour, STTP, and salt are added to represent meatball
products on the market. The potential for meatballs to
experience more contamination will be higher and
will result in lowering the concentration of DNA
produced. Based on the results of DNA isolation, it
shows that the alkaline-lysis method is effective for
use as a method for isolating pork meatball DNA, a
slight modification with the addition of Pro-K and
RNAse is needed to reduce protein and RNA
contamination.
The results of DNA isolation of pork meatballs
with pork content of 5%, 1%, 0.5%, and 0.1% were
amplified using three mt-DNA primers ND5, D-Loop,
and Cyt-b primers which were able to produce DNA
bands. D-Loop primer is able to amplify pork
meatballs with pork content of 5% to 0.1% with
different appearance levels, pork meatballs with a
concentration of 5% and 1% can appear for 2
repetitions, while concentrations of 0.05% and 0.01%
appears 1 time. The results of amplification of pork
meatballs with a concentration of 5%, 1%, 0.5%, and
0.1% using Cyt-b primer produced DNA bands with
2 appearances. The amplification results using D-
Loop primer was able to amplify pork meatball
samples at concentrations of 5%, 1%, 0.5%, and
0.1%, however, the resulting DNA bands were not as
good as the amplification results using Cyt-b primer.
The results of DNA isolation of pork meatballs using
ND5 primers were able to amplify DNA up to a
concentration of 5%, 1%, 0.5%, 0.1%, and 0.01%
with an appearance rate of 2 to 3 times. Based on the
results of the primary sensitivity test on meatball
samples, the ND5 primer was the most sensitive
compared to Cyt-b and D-Loop, it was shown that the
primer was able to amplify meatball samples with the
lowest pork concentration of 0.01%. Pork samples
have undergone a processing process (milling and
heating) and it is possible to experience degradation
which can reduce the quality of the sample DNA. The
results showed that the meatball sample with the
lowest concentration of pork DNA content of 0.01%
could be amplified using ND5 primer. The
amplification results using ND5 primer produced 227
bp of target DNA strands. Compared to D-Loop and
Cyt-b primers, ND5 primer produced the shortest
length of the target DNA sequence. The results
showed that primers with a short target DNA length
such as ND5 with 227 bp length to detect specific
species in preheated foods gave the most sensitive
results. The results of amplification of pork meatballs
with D-Loop primer were able to detect the content of
pork meatballs with a concentration of 5% to 0.1% in
meatballs. The D-Loop primer has a target DNA
sequence length of 835 bp (Figure 5). Amplification
using D-Loop primers results in lower sensitivity
compared to ND5 primers.
The amplification results using Cyt-b primer
showed that the primer was able to detect the content
of pork with concentration of 5% to 0.1% in
meatballs. The Cyt-b primer has a target DNA
sequence with length of 398 bp. Based on the results
of the sensitivity analysis, it shows that D-Loop and
Cyt-b primers are sensitive in detecting pork content
Figure 4: Sensitivity Test Using ND5 Primer in Pork Meatballs. Note: A. Pork Meat Concentration 5%, B. Pork Meat
Concentration 1%, C. Pork Concentration 0.5%, D. Pork Concentration 0.1%, E. Pork Meat Concentration 0.05%, F. Pork
Concentration 0.01%, G. Pork Concentration 0%, (-) Negative Control, (+) Positive Control, M. DNA Ladder.
6th FiAC 2020 - The Food Ingredient Asia Conference (FiAC)
16
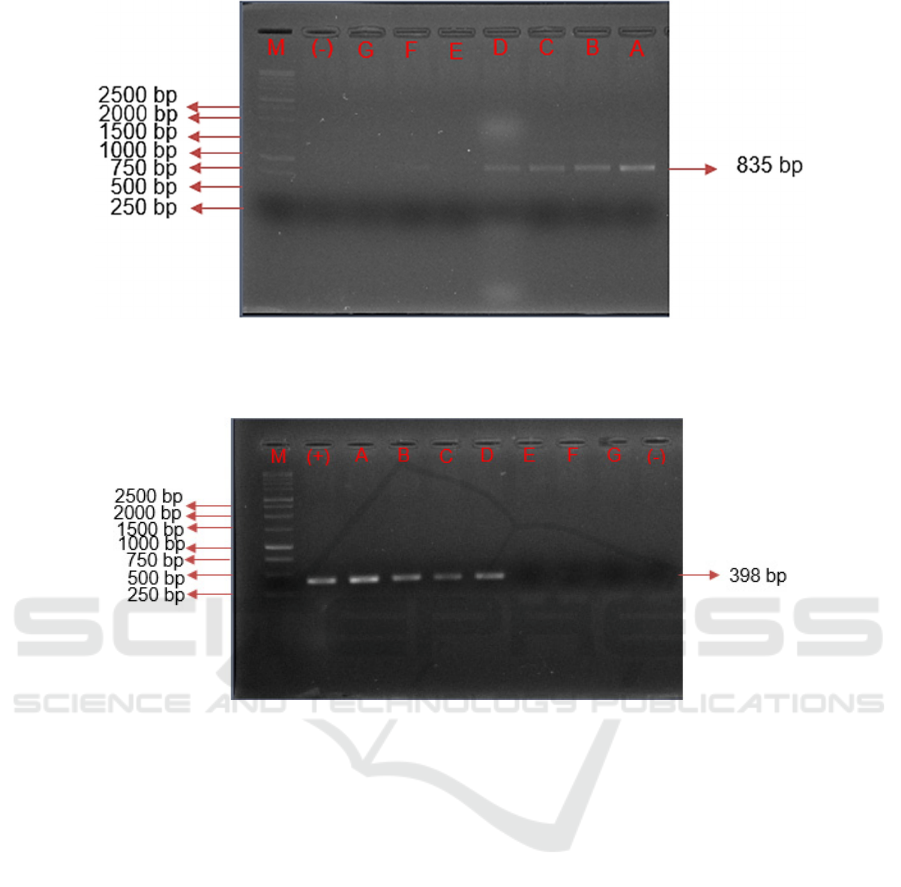
Figure 5: Sensitivity Test using D-Loop Primer in Pork Meatballs. Note: A. Pork Concentration 5%, B. Pork Meat
Concentration 1%, C. Pork Concentration 0.5%, D. Pork Concentration 0.1%, E. Pork Meat Concentration 0.05%, F. Pork
Concentration 0.1%, G. Pork Concentration 0%, (-) Negative Control, (+) Positive Control, M. DNA Ladder.
Figure 6: Sensitivity Test Using Cyt-b Primer in Pork Meatballs. Note: A. Pork Concentration 5%, B. Pork Meat
Concentration 1%, C. Pork Meat Concentration 0.5%, D. Pork Meat Concentration 0.1%, E. Pork Meat Concentration 0.05%,
F. Pork Concentration 0.1%, G. Pork Concentration 0%, (-) Negative Control, (+) Positive Control, M. DNA Ladder.
with concentration of rok content 5% to 0.1% in
meatballs. The overall results of primer sensitivity
analysis showed that the ND5 primer was the most
sensitive in detecting pork content in meatballs
compared to D-Loop and Cyt-b primers. ND5 primers
had the shortest target gene with sequence length of
227 bp compared to both D-Loop and Cyt-b primers,
this was thought to cause the primer to have the
highest sensitivity.
In this study, the results of the ND5 primer
sensitivity test showed the most sensitive primer was
able to detect up to 0.01% pork concentration in
meatballs. Previous research results of ND5 primer
had low sensitivity in detecting pork content, namely
a concentration of 0.1% in sausage samples (Kesmen,
2007). Meanwhile, in another study, the ND5 primer
had low sensitivity and was able to detect the content
of pork with a concentration of 0.1% in a sample of a
mixture of chicken meat and lard (Felk et al., 2017).
In this study, Cyt-b primer had lower sensitivity
compared to ND5 primer. Another study, Cyt-b
primer had low sensitivity in detecting pork with a
concentration of 1% in a mixture of cooked pork and
beef.
The high sensitivity of the ND5 primer can be
caused by the reason that the orimer has a short DNA
sequence target so that it is sensitive in detecting
meatball samples. PCR testing uses a primer with a
short amplicon target which aims to detect specific
species in foods given heat treatment, is better and
more stable than using primers with a long amplicon
target (Ali et al., 2016). This is supported by Rahman
et al (2014), that primers with short amplicon targets
can be amplified with samples processed by high
pressure autoclave. Yoshida et al. (2009) who tested
primers with a target of 126 bp and 83 bp with their
samples given heating and pressure treatments, also
showed that primers with shorter amplicon targets
were more sensitive for detecting DNA fragments.
The Sensitivity Evaluation of mt-DNA Genes; NADH Dehydrogenase Sub Unit 5 (ND5), D-Loop, and Cytochrome-b (Cty-b) to Detect Pork
(Sus scrofa) DNA Isolate and DNA Fragment in Meatball using PCR Technique
17

The presence of a mixture of flour, spices, salt,
and STTP can potentially be a contaminant in DNA
isolates. The presence of these contaminants can
interfere with the success of the PCR process and are
not repeatable. The results showed that the
occurrence rate of DNA bands from 3 repetitions
varied. The high content of polysaccharides in
isolates originating from flour, contamination of
organic compounds such as polysaccharides may
interfere with the enzymatic process of DNA
polymerase by mimicking the structure of nucleic
acids (Schrader et al., 2012). According to Bergallo
et al (2006), there are several ways to remove these
contaminants, for example, such as giving Tween 20
to remove polysaccharides.
4 CONCLUSION
The mt-DNA ND5 and Cyt-b primers were more
sensitive because they were able to amplify DNA up
to concentration of 10
-3
ng/µl compared to D-Loop
primer which were only able to amplify at
concentrations up to 10
-2
ng/µl. In addition, ND5
primer was the most sensitive to detect meatballs with
pork content up to 0.01%. Thus, the three mt-DNA
ND5, Cyt-b, and D-Loop primers were able to detect
specific and sensitive pork DNA fragments. The ND5
primer gave the most sensitive amplification results
because it was able to detect pig DNA fragments with
the lowest concentration of 10
-3
ng/µl and meatballs
with the lowest pork content of 0.01%.
ACKNOWLEDGEMENTS
Thank you very much to Brawijaya University for
supporting research funding through the Doctoral
Grant Program in 2019 and 2020. Thanks also to the
Central Laboratory of Life Sciences (LSIH) which
has facilitated the implementation of research.
REFERENCES
Ali, M. E., Al Amin, M., Razzak, M. A., Abd Hamid, S. B.,
Rahman, M. M., & Rashid, N. A. (2016). Short
amplicon-length PCR assay targeting mitochondrial
cytochrome b gene for the detection of feline meats in
burger formulation. Food analytical methods, 9(3),
571-581.
Bempah, R. T. 2017. Polres Bogor Amankan 7 Orang
Terkait Bakso Oplosan Daging Babi. https://regional.
kompas.com/read/2017/05/30/14192801/polres.bogor.
amankan.7.orang.terkait.bakso.oplosan.daging.babi.
Accessed January 2020.
Bergallo, M., Costa, C., Gribaudo, G., Tarallo, S., Baro, S.,
Ponzi, A. N., & Cavallo, R. (2006). Evaluation of six
methods for extraction and purification of viral DNA
from urine and serum samples. New Microbiologica,
29(2), 111-119.
Fei, S., Okayama, T., Yamanoue, M., Nishikawa, I.,
Mannen, H., & Tsuji, S. (1996). Species identification
of meats and meat products by PCR. Animal Science
and Technology (Japan).
Felk, G. S., Marinho, R. S., Montanhini, M. T. M.,
Rodrigues, S. A., & Bittencourt, J. V. (2017). Detection
limit of polymerase chain reaction technique for species
authentication in meat products. International Food
Research Journal, 24(3).
Hariyadi, P. 2015. Ancaman Serius Pemalsuan Pangan.
https://aipi.or.id/frontend/opinion/read/556a35524d67
3d3d. Accessed January 2020.
Haunshi, S., Pattanayak, A., Bandyopadhaya, S., Saxena, S.
C., & Bujarbaruah, K. M. (2008). A simple and quick
DNA extraction procedure for rapid diagnosis of sex of
chicken and chicken embryos. The Journal of Poultry
Science, 45(1), 75-81.
Harianmerapi. 2018. Campur Daging Sapi dan Babi-
Pedagang Nakal Kena Tipiring. https://www.
harianmerapi.com/ news/2018/05/24/17510/campur-
daging-sapi-dan-babi-pedagang-nakal-kena-tipiring.
Accessed January 2020.
Hofmann, K. (1986). Fundamental problems in identifying
the animal species of muscle meat using electrophoretic
methods. Fleischwirtschaft, 66(1), 91-98.
Kesmen, Z., Yetim, H., & Sahin, F. (2010). Identification
of different meat species used in sucuk production by
PCR assay. Gida, 35(2), 81-87.
Kesmen, Z. Ü. L. A. L., Sahin, F., & Yetim, H. (2007). PCR
assay for the identification of animal species in cooked
sausages. Meat science, 77(4), 649-653.
Koh, M. C., Lim, C. H., Chua, S. B., Chew, S. T., & Phang,
S. T. W. (1998). Random amplified polymorphic DNA
(RAPD) fingerprints for identification of red meat
animal species. Meat Science, 48(3-4), 275-285.
Kusnadi, J., Ashari, N. A., & Arumingtyas, E. L. (2020).
Specificity of Various Mitochondrial DNA (mtDNA),
ND5, D-Loop, and Cty-b DNA Primers in Detecting
Pig (Sus scrofa) DNA Fragments. American Journal of
Molecular Biology, 10(3), 141-147.
Kusnadi, J., Azhari, V., Martati, E. & Saparianti, E. (2019).
Comparison of the 3 DNA Isolation Methods (Alkaline,
Conventional and Commercial KIT) and Specificity
Analysis of mt-ND5 Gene for Detecting Pork (Sus
scrofa) Contamination in Sausages. International Halal
Festival Universitas Brawijaya (Tidak duiterbitkan).
Kusnadi. J. (2019). Analisa Cemaran Fragmen Babi pada
Produk Olahan Dgaing (Sosis) di Kota Malang
menggunaka Metode Alkali, Konvensional. Dan KIT
Komersial. Laporan Penelitian PNBP Fakultas (Tidak
Diterbitkan). Fakultas Teknologi Hasil Pertanian,
Universitas Brawijaya.
Kusnadi, J. (2019). Spesifitas dan Sensitivitas berbagai
6th FiAC 2020 - The Food Ingredient Asia Conference (FiAC)
18

Macam Primer untuk Deteksi Fragmen DNA Babi pada
Berbagai Produk Oahan Makanan. Laporan Penelitian
Program Hibah Doktor Lektor/ Asisten Ahli (Tidak
Diterbitkan). Fakultas Teknologi Hasil Pertanian,
Universitas Brawijaya.
Matsunaga, T., Chikuni, K., Tanabe, R., Muroya, S.,
Shibata, K., Yamada, J., & Shinmura, Y. (1999). A
quick and simple method for the identification of meat
species and meat products by PCR assay. Meat science,
51(2), 143-148.
Moore, J. C., Spink, J., & Lipp, M. (2012). Development
and application of a database of food ingredient fraud
and economically motivated adulteration from 1980 to
2010. Journal of Food Science, 77(4), R118-R126.
Nuryanti. 2020. UPDATE Kasus Pemalsuan Daging Sapi
Ternyata Babi: Bandung Jadi Target Penjualan, 25
Pasar Diperiksa. https://www.tribunnews.com/
regional/2020/05/12/update-kasus-pemalsuan-daging-
sapi-ternyata-babi-bandung-jadi-target-penjualan-25-
pasar-diperiksa?page=2. Accessed January 2020.
Rahman, M. M., Ali, M. E., Abd Hamid, S. B., Mustafa, S.,
Hashim, U., & Hanapi, U. K. (2014). Polymerase chain
reaction assay targeting cytochrome b gene for the
detection of dog meat adulteration in meatball
formulation. Meat Science, 97(4), 404-409.
Sambrook, J., & Russell, D. W. (2006). The condensed
protocols from molecular cloning: a laboratory manual
(No. Sirsi) i9780879697723).
Sambrook, J. E. F., Fritsch, dan T. Miniatis. (1989).
Molecular Clooning: A Laboratory Manual. New
York: Cold Spring Harbor Laboratory Press
Schrader, C., Schielke, A., Ellerbroek, L., & Johne, R.
(2012). PCR inhibitors–occurrence, properties and
removal. Journal of applied microbiology, 113(5),
1014-1026.
Yahya, A., Firmansyah, M., Arlisyah, A., & Risandiansyah,
R. (2017). Comparison of DNA Extraction Methods
Between Conventional, Kit, Alkali and Buffer-Only for
PCR Amplification on Raw and Boiled Bovine and
Porcine Meat. The Journal of Experimental Life
Science, 7(2), 110-114.
Yoshida, T., Nomura, T., Shinoda, N., Kusama, T.,
Kadowaki, K., and Sugiura, K. (2009). Development of
PCR Primers for the Detection of Porcine DNA in Feed
Using mtATP6 as the Target Sequence. Journal Food
Hygene. 50 (2): 89-92.
The Sensitivity Evaluation of mt-DNA Genes; NADH Dehydrogenase Sub Unit 5 (ND5), D-Loop, and Cytochrome-b (Cty-b) to Detect Pork
(Sus scrofa) DNA Isolate and DNA Fragment in Meatball using PCR Technique
19
