
A Recurrent Neural Network and Differential Equation based
Spatiotemporal Infectious Disease Model with Application to COVID-19
Zhijian Li
1
, Yunling Zheng
1
Jack Xin
1
and Guofa Zhou
2
1
Department of Mathematics, University of California, Irvine, U.S.A.
2
College of Health Science, University of California, Irvine, U.S.A.
Keywords:
COVID-19, Recurrent Neural Network, Discrete Epidemic Model, Spatiotemporal Deep Learning.
Abstract:
The outbreaks of Coronavirus Disease 2019 (COVID-19) have impacted the world significantly. Modeling the
trend of infection and real-time forecasting of cases can help decision making and control of the disease spread.
However, data-driven methods such as recurrent neural networks (RNN) can perform poorly due to limited
daily samples in time. In this work, we develop an integrated spatiotemporal model based on the epidemic
differential equations (SIR) and RNN. The former after simplification and discretization is a compact model
of temporal infection trend of a region while the latter models the effect of nearest neighboring regions. The
latter captures latent spatial information. We trained and tested our model on COVID-19 data in Italy, and
show that it out-performs existing temporal models (fully connected NN, SIR, ARIMA) in 1-day, 3-day, and
1-week ahead forecasting especially in the regime of limited training data.
1 INTRODUCTION
Susceptible-Infected-Removed (SIR) is a classical
differential equation model of infectious diseases
(Anderson and May, 1992). It divides the total popu-
lation into three compartments and models their evo-
lution by the system of equations
dS
dt
= −β I S
dI
dt
= β I S − γI
dR
dt
= γ I
where β and γ are two positive parameters. SIR is
a simple and efficient model of temporal data for a
given region, see also (Hethcote, 2000) for related
compartment models with social structures.
Yet the infectious disease data are often spatio-
temporal as in the case of COVID-19, see (Italian re-
gion, 2020). A natural question is how to extend SIR
to a space time model of suitable complexity so that it
can be quickly trained from the available public data
sets and applied in real-time forecasts. See (Roosa
et al., 2020) for temporal model real-time forecasts
on cumulative cases of China in Feb 2020.
In this paper, we explore spatial infectious dis-
ease information to model the latent effect due to
the in-flow of the infected people from the geograph-
ical neighbors. The in-flow data is not observed.
To this end, machine learning tools such as regres-
sion and neural network models are more convenient.
Auto-regressive model (AR) and its variants are lin-
ear statistical models to forecast time-series data. The
Long Short Term Memory (LSTM) neural networks,
originally designed for natural language processing
(Hochreiter and Schmidhuber, 1997), have more rep-
resentation power and can be applied to disease time-
series data as well. With spatial structures added, the
graph-structured LSTM models can achieve state-of-
the-art performance on spatiotemporal influenza data
(Li et al., 2019), crime and traffic data (Wang et al.,
2019; Wang et al., 2018). However, they require a
large enough supply of training data. For COVID-19,
we only have limited daily data since the outbreaks
began in early 2020. Applying space-time LSTM
models (Li et al., 2019; Wang et al., 2018) directly
to COVID-19 turns out to produce poor results.
In view of the limited COVID-19 data, we shall
propose a hybrid SIR-LSTM model where a discrete
time equation for the I-component of SIR model will
be derived and coupled to time dependent features of
neighboring regions through LSTMs.
Li, Z., Zheng, Y., Xin, J. and Zhou, G.
A Recurrent Neural Network and Differential Equation based Spatiotemporal Infectious Disease Model with Application to COVID-19.
DOI: 10.5220/0010130000930103
In Proceedings of the 12th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2020) - Volume 1: KDIR, pages 93-103
ISBN: 978-989-758-474-9
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
93

2 RELATED WORK
In (Yang et al., 2015), the authors designed a vari-
ant of AR, the AutoRegression with Google search
data (ARGO), that utilizes external feature of google
search data to forecast influenza data from Centers for
Disease Control and Prevention (CDC). Google Cor-
relate is google search trend data that represents the
popularity of an item and its correlated items searched
in Google engine. It is a weekly data that normalized
to range [0, 100]. Based on Google search trend of
influenza and its correlated items, ARGO is a linear
model that processes historical observations and ex-
ternal features. The prediction of influenza activity
level at time t, defined as ˆy
t
, is given by:
ˆy
t
= u
t
+
52
∑
j=1
α
j
y
t− j
+
100
∑
i=1
β
i
X
i,t
.
ARGO is optimized as:
min
µ
y
,
~
α,
~
β
y
t
− u
t
−
52
∑
j=1
α
j
y
t− j
−
100
∑
i=1
β
i
X
i,t
2
+λ
a
||
~
α||
1
+ η
a
||
~
β||
1
+ λ
b
||
~
α||
2
2
+ η
b
||
~
β||
2
2
where
~
α = (α
1
,··· , α
52
) and
~
β = (β
1
,··· , β
100
). The
y
t− j
’s, 1 ≤ j ≤ 52, are historical observations of pre-
vious 52 weeks and X
i,t
are the google search trend
measures of top 100 terms that are most correlated
to influenza at time t. Essentially, ARGO is a lin-
ear regression with regularization terms. In (Yang
et al., 2015), ARGO is shown to outperform stan-
dard machine learning models such as LSTM, AR,
and ARIMA.
In (Li et al., 2019), graph structured recurrent
neural network (GSRNN) further improved ARGO
in the forecasting accuracy of CDC influenza activ-
ity level. The CDC partitions the US into 10 Health
and Human Services (HHS) regions for reporting.
GSRNN treats the 10 regions as a graph with nodes
v
1
,··· , v
10
, and E be the collection of edges (i.e E =
{(v
i
,v
j
)|v
i
,v
j
are adjacent}). Based on the average
history of activity levels, the 10 HHS regions are di-
vided into two groups, relatively active group, H , and
relatively inactive group, L . There are 3 types of
edges, L − L , H − L , and H − H , and each edge
type has a corresponding RNN to train the edge fea-
tures. There are also two node RNNs for each group
to output the final prediction. Given a node (region) v,
suppose v ∈ H . GSRNN generates the edge features
of v at time t, e
t
v,H
and e
t
v,L
, by averaging the history
of neighbors of v in the corresponding groups. Next,
the edge features are fed into the corresponding edge
RNNs:
f
t
v
= edgeRNN
H −L
(e
t
v,L
), h
t
v
= edgeRNN
H −H
(e
t
v,L
)
Then, the outputs of edgeRNNs are fed into the
nodeRNN of group H together with the node feature
of v at time t, denoted as v
t
, to output the prediction
of the activity level of node v at time t + 1, or y
t+1
v
:
y
t+1
v
= nodeRNN
H
(v
t
, f
t
v
,h
t
v
).
3 OUR CONTRIBUTION: IeRNN
MODEL
We propose a novel spatiotemporal model integrating
LSTM (Hochreiter and Schmidhuber, 1997) with a
discrete time I-equation derived from SIR differen-
tial equations. The LSTM is utilized to model la-
tent spatial information. The I-equation models the
observed temporal information. Our model, named
IeRNN, differs from (Li et al., 2019; Wang et al.,
2019; Wang et al., 2018) in that a difference equa-
tion with 3 parameters (the I-equation) fits the limited
temporal data. The I-equation is far more compact
than LSTM.
3.1 Derivation of Discrete-time
I-Equation
Based on SIR model, we add an additional feature I
e
that represents the external infection influence from
the neighbors of a region. Then the SIR nonlinear
system with I
e
as external forcing becomes
dS
dt
= −β
1
S I − β
2
S I
e
(1)
dI
dt
= β
1
S I + β
2
S I
e
− γ I (2)
dR
dt
= γI (3)
which conserves the total mass (normalized to 1): S +
I + R = 1. It follows from (3) that
R(t) = R(t
0
) +
Z
t
t
0
γI dτ
Hence,
S(t) = 1 − I(t) − R(t
0
) − γ
Z
t
t
0
I dτ
Substituting S(t) into (2) we have:
dI
dt
= (β
1
I +β
2
I
e
)
1 − I(t) − R(t
0
) − γ
Z
t
t
0
I(τ)dτ
−γI
Combining forward Euler method and Riemann sum
approximation of the integral, we have a discrete ap-
proximation:
I(t + 1) = (1 − γ)I(t) +
β
1
I(t) + β
2
I
e
(t)
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
94
·
1 − I(t) − R(t
0
) − γ
t − t
0
p + 1
p
∑
j=0
I(t − j)
As we model I(t) from the beginning of the infection,
we have t
0
= 0 and R(t
0
) = 0. We arrive at the follow-
ing discrete time I-equation:
I(t + 1) = (1 − γ)I(t) +
β
1
I(t) + β
2
I
e
(t)
·
1 − I(t) − γ
t
p + 1
p
∑
j=0
I(t − j)
(4)
Note that if we let I
e
(t) = 0, then we have an approx-
imation of I(t) for the original SIR model, which is a
solely temporal model (named I-model):
I(t + 1) = (1 − γ − β)I(t) − βI
2
(t)
− β γ
t
p + 1
I(t)
p
∑
j=0
I(t − j) (5)
In reality, it is hard to know how a population of a
region interacts with populations of neighboring re-
gions. As a result, I
e
(t) is a latent information that is
difficult to model by a mathematical formula or equa-
tion. In order to retrieve latent spatial information,
we employ recurrent neural networks made of LSTM
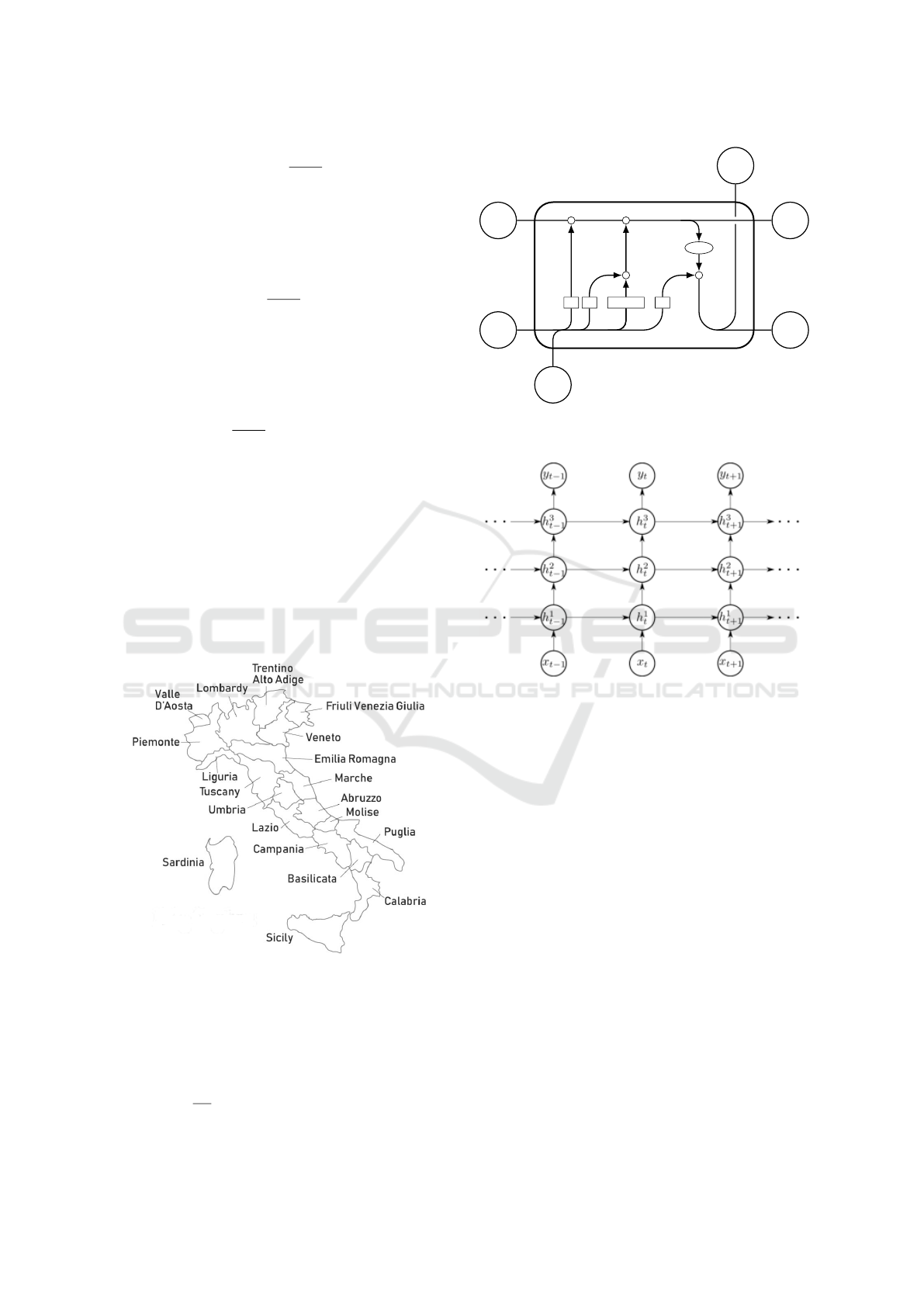
cells (Hochreiter and Schmidhuber, 1997), see Fig. 2.
3.2 Generating Edge Feature and
Computing Latent I
e
Figure 1: Italian Region Map.
We utilize the spatial information based on the Italy
region map, Fig. 1. In order to learn the latent in-
formation I
e
of a region v, we first generate the edge
feature of v. Let C be the collection of neighbors of v.
Then, the edge feature of v at time t is formulated as:
f
t
e
=
1
|C|
∑
i:v
i
∈C
I
i
(t − 1),· ·· , I
i
(t − p)
σ σ
Tanh
σ
×
+
× ×
Tanh
c
ht−1i
h
ht−1i
x
hti
Input
c
hti
h
hti
h
hti
Output
Figure 2: LSTM cell: input x
t
, output: h
t
; σ is a sigmoid
function.
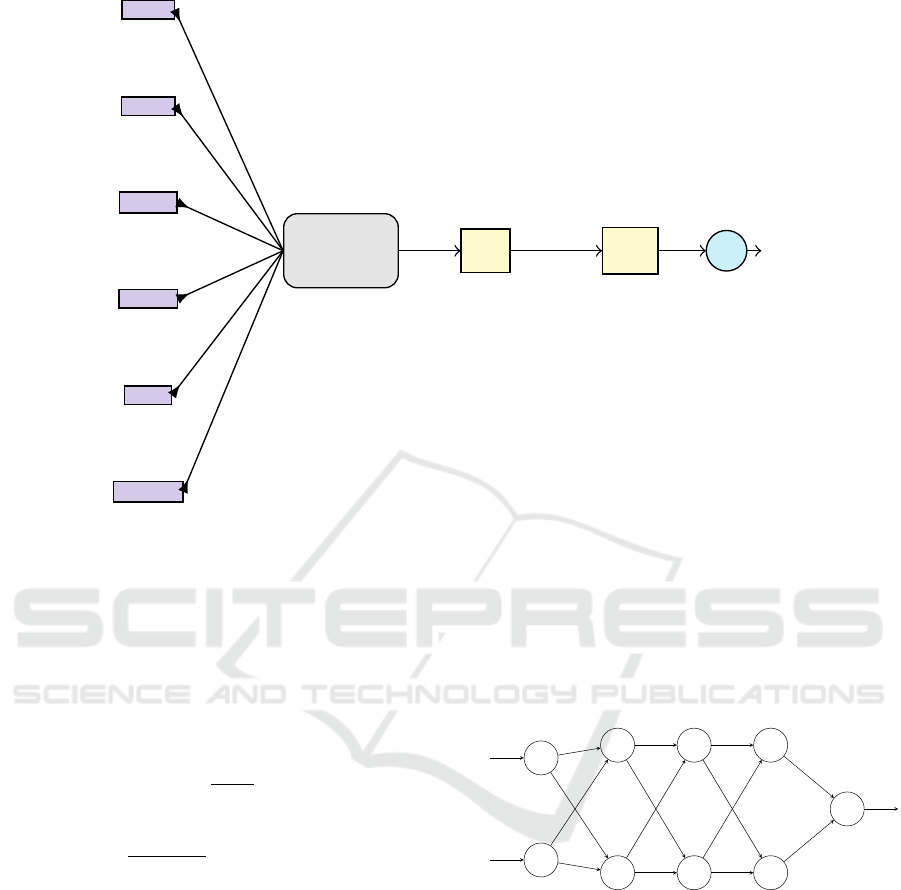
Figure 3: Edge RNN consisting of 3 stacked LSTM cells.
where I
i
(t) is the infection population percentage of
region v
i
at time t. Then, we feed f
t
e
into an Edge
RNN, an RNN with 3 stacked LSTM cells (see Fig.
3), followed by a dense layer for computing I
e
. The
activation function of the dense layer is hyperbolic
tangent function. Figure 4 illustrates the procedure
of computing I
e
for Lazio as an illustration. We hence
call our model IeRNN due to its integrated design of
I-equation and edge RNN.
4 EXPERIMENT
To calibrate our model IeRNN, we use the Italy
COVID-19 data (Italian region, 2020) for training and
testing. Although the US has the most infected cases,
the recovered cases are largely missing. On the other
hand, the Italian COVID-19 data is more accurately
reported and better maintained, reflecting a nearly
complete duration of the rise and fall of infection.
We collect the data of daily new (current) cases from
2020-02-24 to 2020-06-18 of 20 Italian regions. We
A Recurrent Neural Network and Differential Equation based Spatiotemporal Infectious Disease Model with Application to COVID-19
95
Tuscany
Umbria
Marche
Abruzzo
Molise
Campania
Edge
Feature
Edge
RNN
Dense
Layer
tanh
I
e
Figure 4: Computing I
e
of Lazio region. Edge RNN is as shown in Fig. 3. Dense layer is fully connected (see Fig. 5).
set p = 3 in (4) based on experimental performance.
As a result, we have the current data for 113 days,
with 81 days to train our model and 32 days to test our
model (or 70%/30% training/testing data split). Our
training loss function is the mean squared error of the
model output and training data:
ˆy(t) = (1 − γ)y(t − 1) +
β
1
y(t − 1) + β
2
I
e
(t − 1)
·
1 − y(t − 1) − γ
t
p + 1
p+1
∑
j=1
y(t − j)
Loss =
1
T − p − 1
T
∑
t=p+1
y(t) − ˆy(t)
2
Since the training is minimization of the above
loss fucntion over parameters in both I-equation and
RNN, the two components of IeRNN are coupled while
learning from data. We use Adam gradient descent to
learn the weights of LSTM and the dense layer, as
well as I-equation parameters β
1
, β
2
, and γ.
To evaluate the performance of our model, we
compare IeRNN, I-model (5), a fully-connected neu-
ral network (fcNN, Fig. 5) with hyperbolic tan-
gent activation function, and auto-regression model
(ARIMA). As the standard setting of ARIMA is 1-
day ahead prediction, we shall only compare with it in
such a very short-term case. Since infectious disease
evolution is intrinsically nonlinear, we shall compare
nonlinear models for 3-day and 1-week ahead fore-
casting. Based on experimental performance, we set
the number of hidden units to be 100, 150, and 100
for the three layers of fcNN respectively.
.
.
.
.
.
.
.
.
.
.
.
.
y
t−1
y
t−1
y
t−1
y
t−p
y
t−p
y
t−p
w
(1)
1
w
(1)
1
w
(1)
1
w
(1)
n
w
(1)
n
w
(1)
n
w
(2)
1
w
(2)
1
w
(2)
1
w
(2)
n
w
(2)
n
w
(2)
n
w
(3)
1
w
(3)
1
w
(3)
1
w
(3)
n
w
(3)
n
w
(3)
n
tanh
tanh
tanh
y
t
y
t
y
t
Input
layer
Hidden
layer
Output
layer
Figure 5: Schematic of fcNN for modeling time series.
4.1 One-day Ahead Forecast
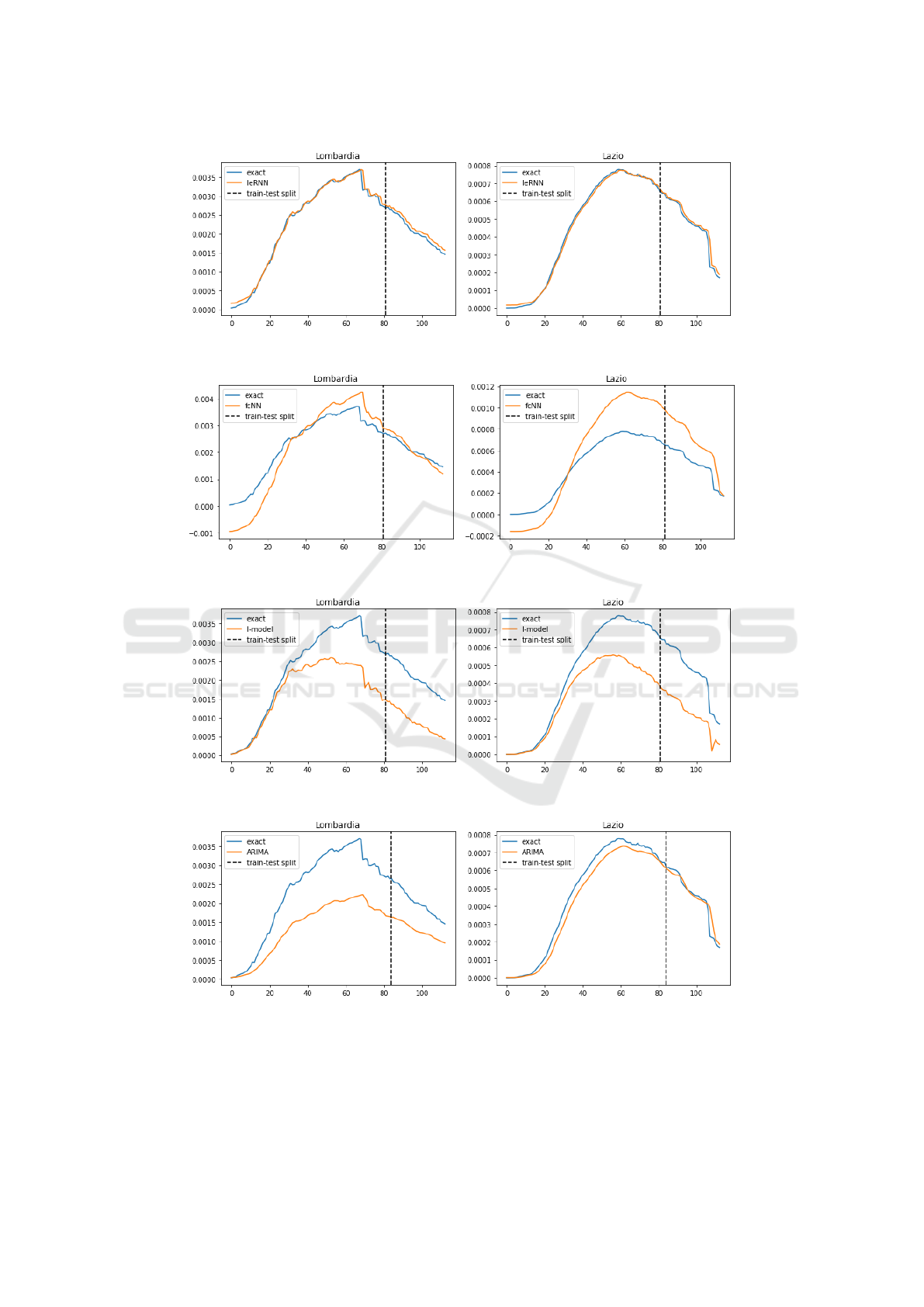
As we see in Fig. 6, fcNN can perform poorly. This
is not a surprise, as both (Li et al., 2019) and (Yang
et al., 2015) relied on hundreds of historical observa-
tions to train their models. The I-model based on only
sequential data in time of one region merely follows
the trend of the true data but cannot provide accurate
predictions. Our IeRNN model, with the help of ad-
ditional spatial information, is able to make accurate
predictions and outperform other models. We also
test the IeRNN with training data reduced to 40% (46
days). IeRNN is still able to track the general trend of
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
96
(a) Lombardy (b) Lazio
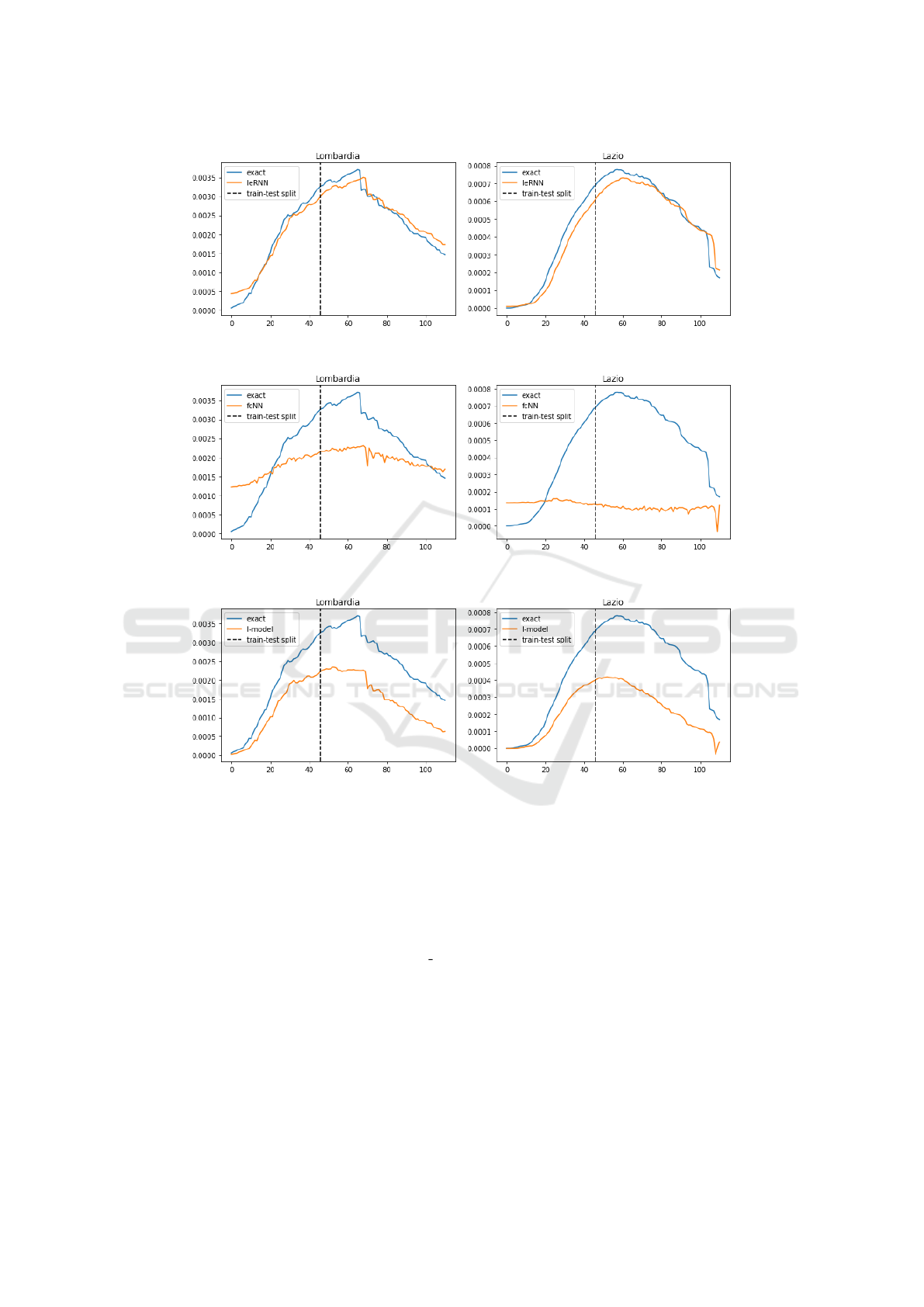
(c) Lombardy (d) Lazio
(e) Lombardy (f) Lazio
(g) Lombardy (h) Lazio
Figure 6: Training and 1-day ahead forecast of 4 models (IeRNN, fcNN, I-model, and ARIMA) in 4 rows respectively. The
vertical axis is fraction of newly infected people in the population. The horizontal axis is time in unit of days.
A Recurrent Neural Network and Differential Equation based Spatiotemporal Infectious Disease Model with Application to COVID-19
97
(a) Lombardy (b) Lazio
(c) Lombardy (d) Lazio
(e) Lombardy (f) Lazio
(g) Lombardy (h) Lazio
Figure 7: Training and 1-day ahead forecast of 4 models with reduced (40%) training data. The 4 rows are IeRNN, fcNN,
I-model, and ARIMA respectively. The vertical axis is fraction of newly infected people in the population. The horizontal
axis is time in unit of days.
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
98
the infected population percentage.
We measure the test accuracy with the Root Mean
Square Error (RMSE) averaged over a few trials in
training. In Tables 1 and 2 on 1-day ahead forecast,
IeRNN achieves the smallest RMSE errors, and I-
model has the largest errors. The compact I-model
with 2 parameters cannot do 1-day ahead prediction
as accurately. ARIMA outperforms I-model and does
better on Emilia-Romagna and Lazio regions than
fcNN. ARIMA, a linear model, has simpler structure
than fcNN whose nonlinearity does not play out in
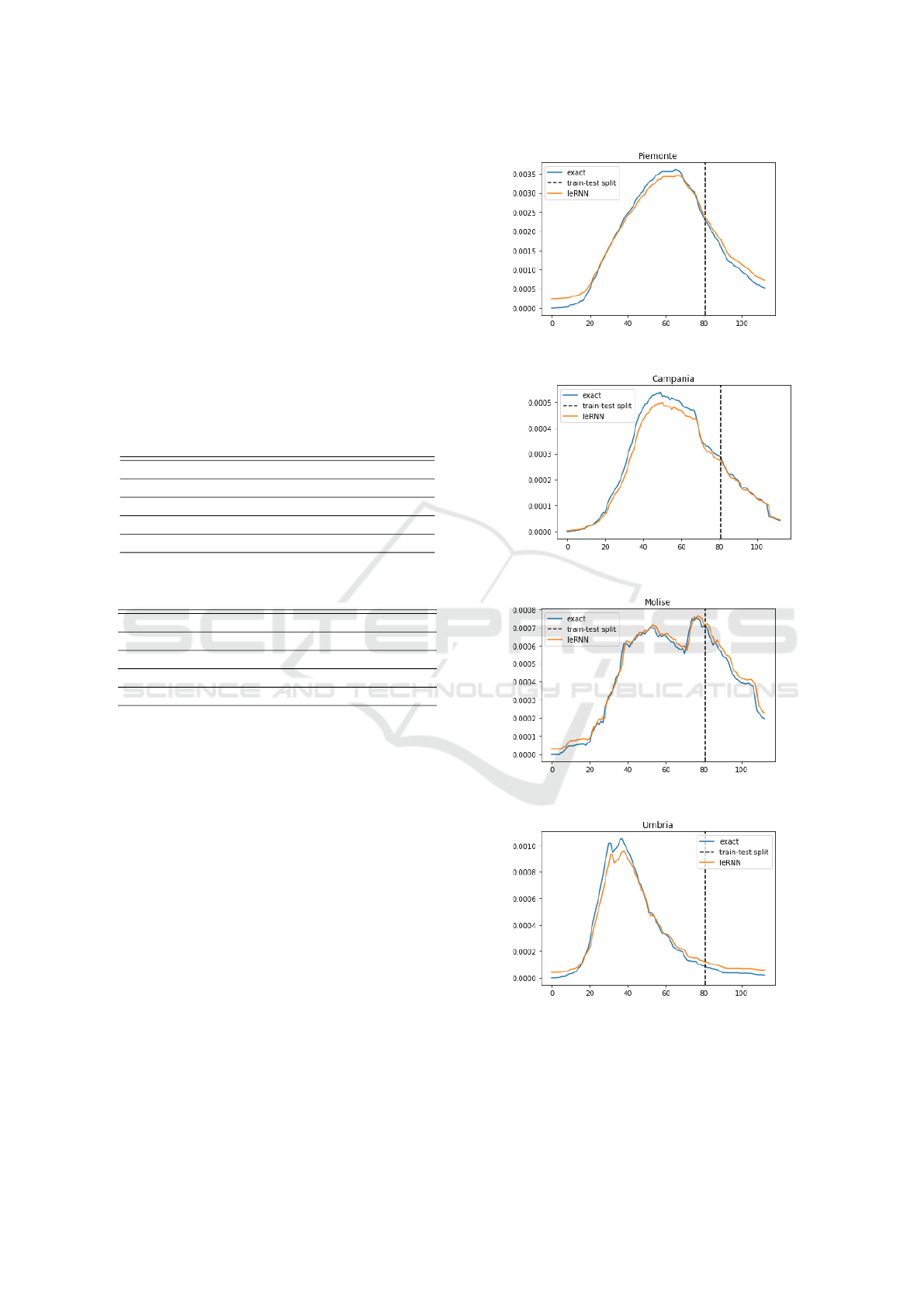
such a short time task. Fig. 8 shows 1-day ahead
forecast of IeRNN model on other regions with the
learned latent external forcing I
e
in Fig. 9.
Table 1: RMSE test errors in 1-day ahead forecast trained
with 70 % of data. E-R= Emilia-Romagna.
Model Lombardy E-R Lazio
IeRNN 1.027e-04 6.333e-05 3.251e-05
I-model 1.175e-03 3.284e-04 2.439e-04
fcNN 1.580e-04 4.614e-04 2.294e-04
ARIMA 9.789e-04 3.627e-04 4.365e-05
Table 2: RMSE test errors in 1-day ahead forecast trained
with reduced (40 % of) data. E-R= Emilia-Romagna.
Model Lombardy E-R Lazio
IeRNN 9.850e-05 1.778e-04 3.617e-05
I-model 1.871e-03 1.252 e-03 5.443e-04
fcNN 3.364e-04 6.204e-04 8.030e-04
ARIMA 1.277e-03 1.082e-03 4.018e-05
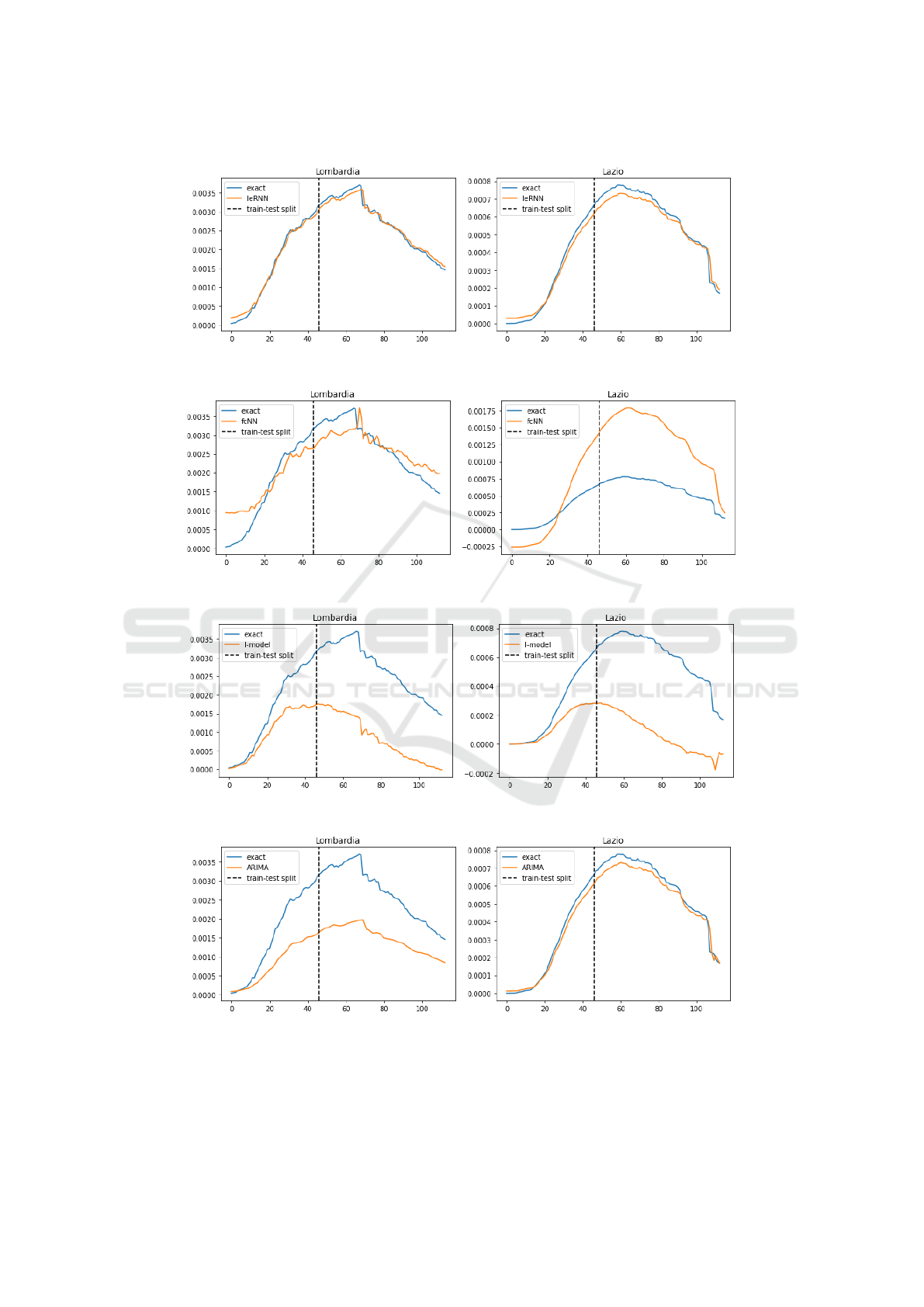
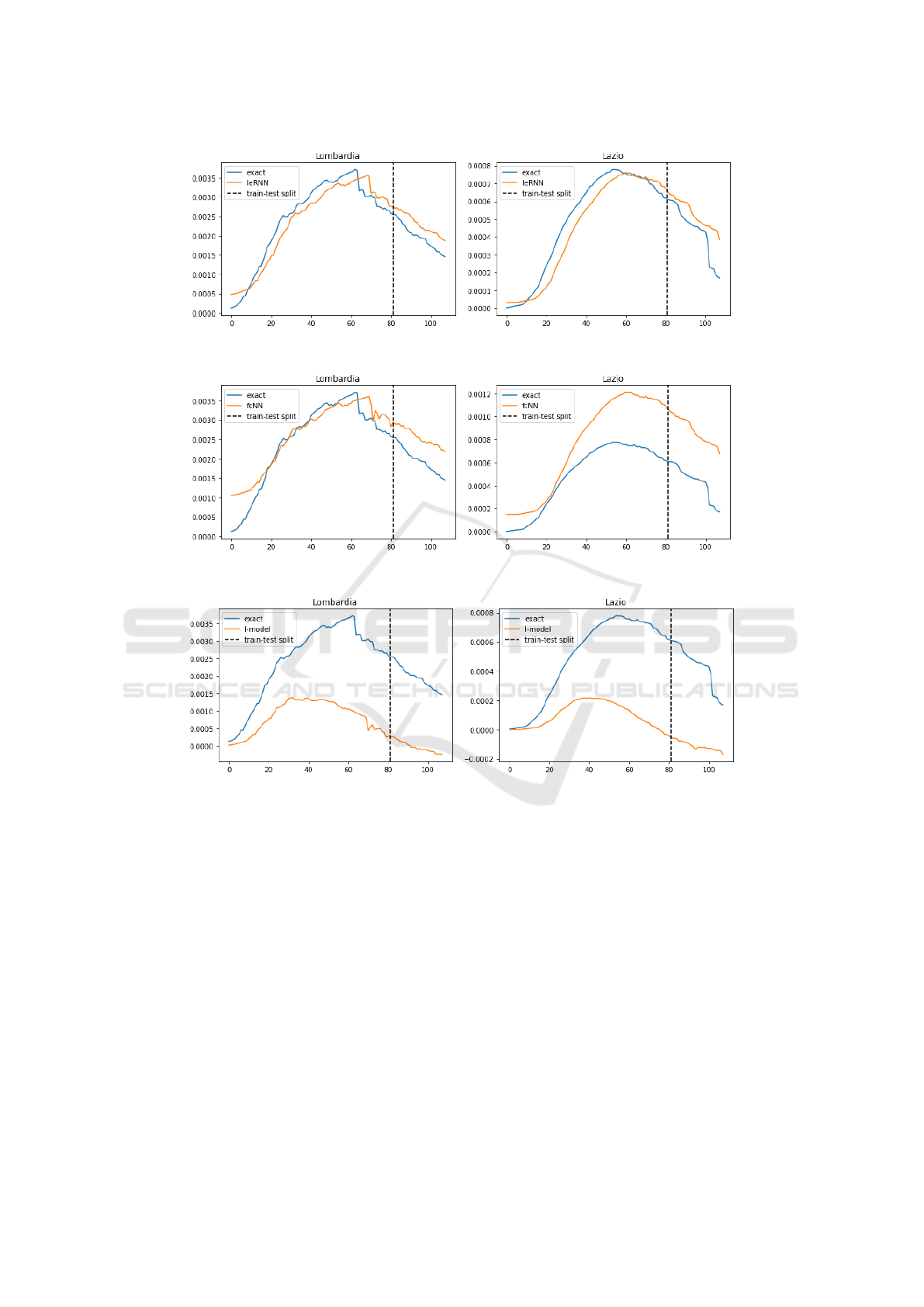
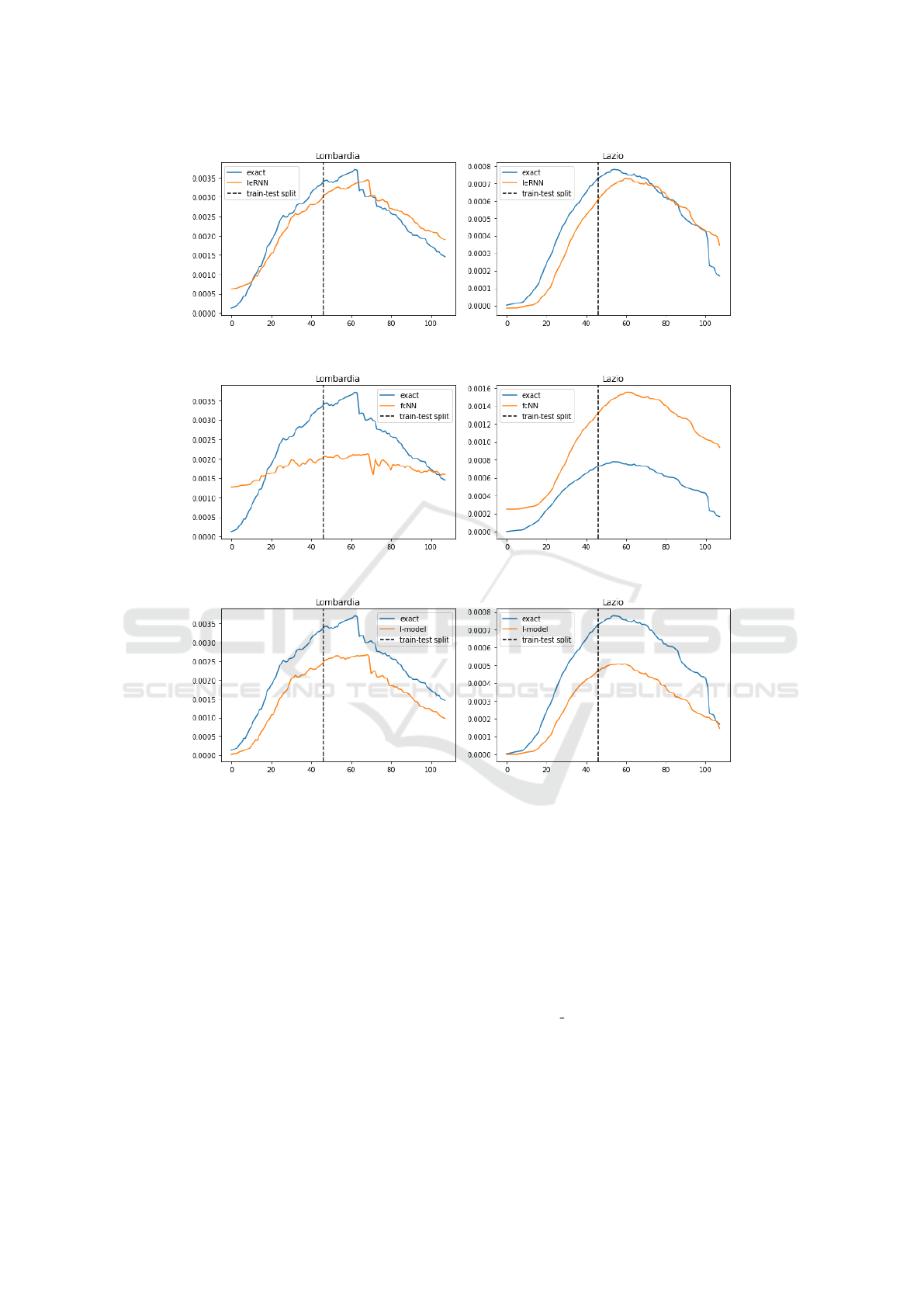
4.2 Multi-day Ahead Forecast
In model training for multi-day ahead forecast, the
training loss function is modified so that the model
input comes from multiple days in the past. In 7-day
ahead forecast, IeRNN leads the other two nonlinear
models especially in the 40% training data case, by
as much as a factor of 7 in Lombardy. In the 3-day
ahead forecast, IeRNN leads fcNN by a factor of 4
in the 40% training data case, as much as a factor of
10 in Lazio. Figs. 10-12 show model comparison in
training and forecast phases for Lombardy and Lazio.
4.3 Model Size and Computing Time
IeRNN (fcNN) has about 16400 (1800) parameters.
The optimized (β
1
,β
2
,γ) = (0.685, 0.158, 0.044) in
Lombardy, similarly in other regions. Table 7 lists
average model training and inference times.
(a) Piemonte
(b) Campania
(c) Molise
(d) Umbria
Figure 8: IeRNN training and 1-day ahead forecast on four
additional regions.
A Recurrent Neural Network and Differential Equation based Spatiotemporal Infectious Disease Model with Application to COVID-19
99
(a) Piemonte
(b) Campania
(c) Molise
(d) Umbria
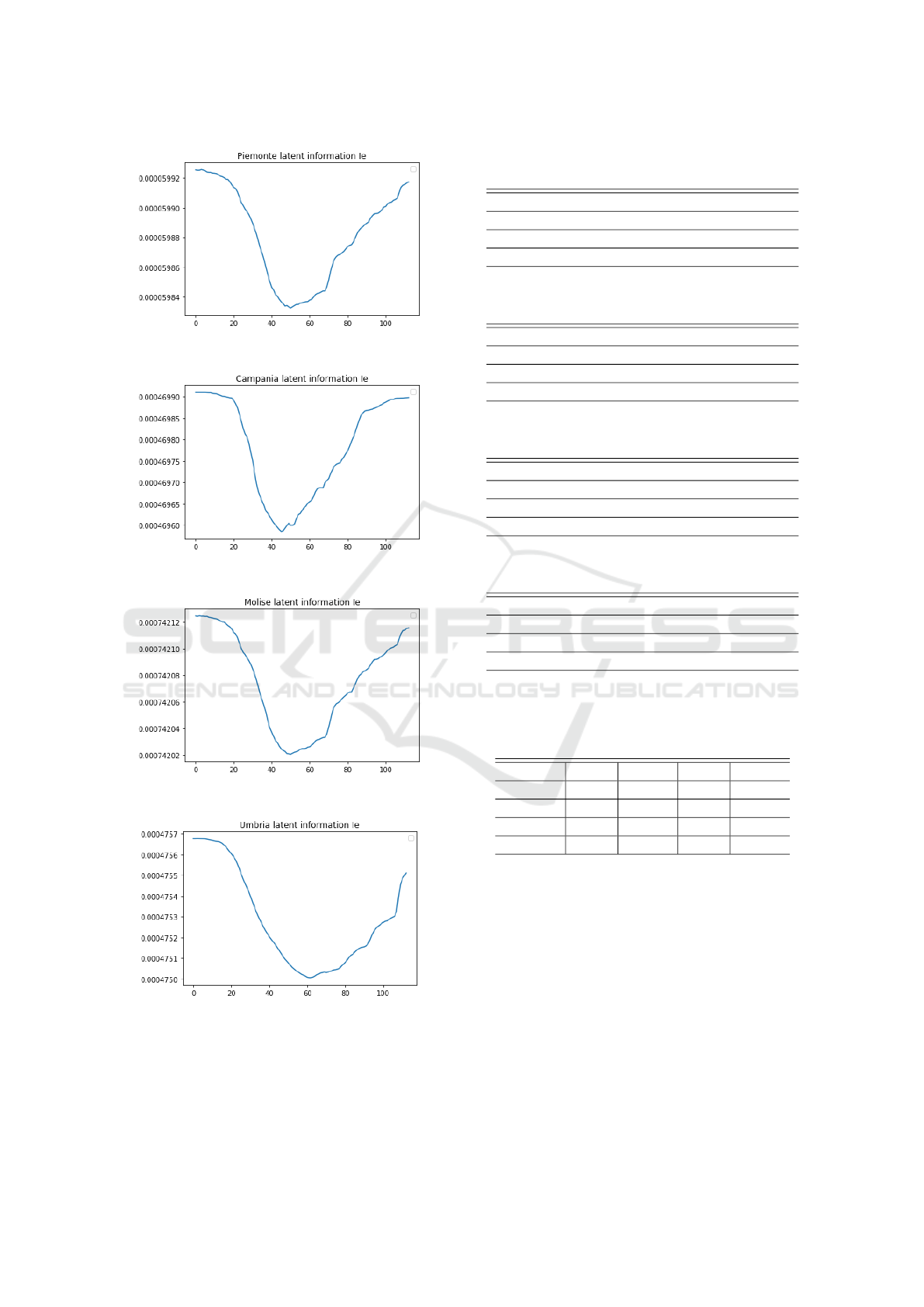
Figure 9: Visualization of the latent information I
e
in Fig. 8
learned by IeRNN.
Table 3: RMSE test errors in 7-day ahead forecast trained
with 70 % of data. E-R= Emilia-Romagna.
Model Lombardy E-R Lazio
IeRNN 3.513e-04 4.423e-04 1.161e-04
I-model 2.004e-03 6.627e-04 5.586e-04
fcNN 6.608e-04 4.804e-04 4.508e-04
Table 4: RMSE test errors in 7-day ahead forecast trained
with reduced (40 % of) data. E-R= Emilia-Romagna.
Model Lombardy E-R Lazio
IeRNN 3.061e-04 4.324e-04 7.754e-05
I-model 2.196e-03 1.167e-03 6.011e-04
fcNN 2.224e-03 6.889e-04 1.851e-04
Table 5: RMSE test errors in 3-day ahead forecast trained
with 70 % of data. E-R= Emilia-Romagna.
Model Lombardy E-R Lazio
IeRNN 2.479e-04 3.668e-04 5.979e-05
I-model 5.609e-04 1.724e-04 1.383e-04
fcNN 8.165e-04 6.757e-04 1.689e-04
Table 6: RMSE test errors in 3-day ahead forecast trained
with reduced (40 % of) data. E-R= Emilia-Romagna.
Model Lombardy E-R Lazio
IeRNN 1.987e-04 3.256e-04 5.297e-05
I-model 1.114e-03 7.337e-04 3.507e-04
fcNN 8.611e-04 1.374e-03 5.290e-04
Table 7: Average model training (tr) and inference (inf)
times in seconds on Macbook Pro with Intel i5 CPU. The
first two columns are for 70 % training (tr70) data and the
last two columns are for 40 % training (tr40) data.
Model tr70 inf70 tr40 inf40
IeRNN 0.58s 0.018s 0.51s 0.02s
I-model 0.14s 0.004s 0.11s 0.004s
fcNN 0.09s 0.003s 0.09s 0.003s
ARIMA 0.23s 0.014s 0.19s 0.015s
5 CONCLUSIONS AND FUTURE
WORK
We developed a novel spatiotemporal infectious dis-
ease model consisting of a discrete epidemic equation
for the region of interest and RNNs for interactions
with nearest geographic regions. Our model can be
trained under 1 second. Its inference takes a fraction
of a second, suitable for real-time forecasting and as-
sisting governments in decision making. Our model
out-performs temporal models in one-day and multi-
day ahead forecasts in limited training data regime.
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
100
(a) Lombardy (b) Lazio
(c) Lombardy (d) Lazio
(e) Lombardy (f) Lazio
Figure 10: Training and 7-day ahead forecast of 3 models (IeRNN, fcNN, and I-model) in 3 rows respectively. The vertical
axis is fraction of newly infected people in the population. The horizontal axis is time in unit of days.
In future work, we shall study COVID-19 data in
other regions of the world and more sophisticated dif-
ferential equation models such as SEIR
(Anderson and May, 1992; Hethcote, 2000) in our
framework here. It is interesting to derive similar dis-
crete time I-equations. We shall also include a con-
trol mechanism in our model to evaluate various lock-
down and social distancing measures on the evolu-
tion of infections. This will provide a useful tool for
policymakers to assess the impact of their proposed
measures. The first step in this direction is to replace
the interaction parameter β
1
by a parameterized func-
tion of time (Morris et al., 2020) to model government
policies.
Another future direction is to study: 1) traffic data
for understanding interactions and transmissions be-
yond nearest geographic neighbors; 2) social mech-
anism (Albi et al., 2020) to distinguish interactions
among and within different age groups.
ACKNOWLEDGEMENTS
The work was partially supported by NSF grants IIS-
1632935, and DMS-1924548. The authors thank
Prof. Frederic Wan for helpful communications on
disease modeling, and the anonymous reviewers of
KDIR 2020 for their constructive comments.
A Recurrent Neural Network and Differential Equation based Spatiotemporal Infectious Disease Model with Application to COVID-19
101
(a) Lombardy (b) Lazio
(c) Lombardy (d) Lazio
(e) Lombardy (f) Lazio
Figure 11: Training and 7-day ahead forecast of 3 models (IeRNN, fcNN, and I-model) with reduced (40%) training data in 3
rows respectively. The vertical axis is fraction of newly infected people in the population. The horizontal axis is time in unit
of days.
REFERENCES
Albi, G., Pareschi, L., and Zanella, M. (2020). Con-
trol with uncertain data of socially structured com-
partmental epidemic models. ArXiv Preprint ArXiv,
2004.13067v1:1–26.
Anderson, R. and May, R. (1992). Infectious Diseases of
Humans: Dynamics and Control. Oxford University
Press, Oxford.
Hethcote, H. (2000). The mathematics of infectious dis-
eases. SIAM Review, 42:599 – 653.
Hochreiter, S. and Schmidhuber, J. (1997). Long short-term
memory. Neural computation, 9(8):1735–1780.
Italian region (accessed March-July, 2020). Italian COVID-
19 Data. https://github.com/Akaza994/COVID-19-
Data/blob/master/covid-19/italy.csv.
Li, Z., Luo, X., Wang, B., Bertozzi, A., and Xin, J. (2019).
A study on graph-structured recurrent neural networks
and sparsification with application to epidemic fore-
casting. In World Congress on Global Optimization,
pages 730–739, https://doi.org/10.1007/978–3–030–
21803–4 80. Springer.
Morris, D., Rossine, F., Plotkin, J., and Levin, S. (2020).
Optimal, near-optimal, and robust epidemic control.
ArXiv Preprint ArXiv, 2004.02209v2:1–28.
Roosa, K., Lee, Y., Luo, R., Kirpich, A., Rothenberg, R.,
Hyman, J., Yan, P., and Chowell, G. (2020). Real-
time forecasts of the COVID-19 epidemic in China
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
102
(a) Lombardy (b) Lazio
(c) Lombardy (d) Lazio
(e) Lombardy (f) Lazio
Figure 12: Training and 3-day ahead forecast of 3 models (IeRNN, fcNN, and I-model) with reduced (40 %) training data in
3 rows respectively. The vertical axis is fraction of newly infected people in the population. The horizontal axis is time in unit
of days.
from February 5th to February 24th, 2020. Infectious
Disease Modelling, 5:256 – 263.
Wang, B., Luo, X., Zhang, F., Yuan, B., Bertozzi, A., and
Brantingham, J. (2018). Graph-based deep model-
ing and real time forecasting of sparse spatio-temporal
data. MiLeTS ’18, London, UK, DOI: 10.475/123 4;
arXiv preprint arXiv:1804.00684.
Wang, B., Yin, P., Bertozzi, A., Brantingham, J., Osher, S.,
and Xin, J. (2019). Deep learning for real-time crime
forecasting and its ternarization. Chinese Annals of
Mathematics, Series B, 40(6):949–966.
Yang, S., Santillana, M., and Kou, S. C. (2015). Accu-
rate estimation of influenza epidemics using Google
search data via ARGO. Proceedings of the National
Academy of Sciences, 112(47):14473–14478.
A Recurrent Neural Network and Differential Equation based Spatiotemporal Infectious Disease Model with Application to COVID-19
103
