
Complexity vs. Performance in Granular Embedding Spaces for Graph
Classification
Luca Baldini
a
, Alessio Martino
b
and Antonello Rizzi
c
Department of Information Engineering, Electronics and Telecommunications, University of Rome “La Sapienza”,
via Eudossiana 18, 00184 Rome, Italy
Keywords:
Structural Pattern Recognition, Supervised Learning, Embedding Spaces, Granular Computing, Graph Edit
Distances.
Abstract:
The most distinctive trait in structural pattern recognition in graph domain is the ability to deal with the
organization and relations between the constituent entities of the pattern. Even if this can be convenient and/or
necessary in many contexts, most of the state-of the art classification techniques can not be deployed directly
in the graph domain without first embedding graph patterns towards a metric space. Granular Computing is a
powerful information processing paradigm that can be employed in order to drive the synthesis of automatic
embedding spaces from structured domains. In this paper we investigate several classification techniques
starting from Granular Computing-based embedding procedures and provide a thorough overview in terms of
model complexity, embedding space complexity and performances on several open-access datasets for graph
classification. We witness that certain classification techniques perform poorly both from the point of view
of complexity and learning performances as the case of non-linear SVM, suggesting that high dimensionality
of the synthesized embedding space can negatively affect the effectiveness of these approaches. On the other
hand, linear support vector machines, neuro-fuzzy networks and nearest neighbour classifiers have comparable
performances in terms of accuracy, with second being the most competitive in terms of structural complexity
and the latter being the most competitive in terms of embedding space dimensionality.
1 INTRODUCTION
The possibility of solving pattern recognition prob-
lems in the graph domain challenged computer sci-
entists and machine learning engineers alike for more
than two decades. That is because graphs are able
to encode both topological information (namely, re-
lationship between entities) and semantic information
(whether nodes and/or edges are equipped with suit-
able attributes). In turn, this high level of abstrac-
tion and customization made graphs suitable mathe-
matical objects for modelling several real-world sys-
tems in fields such as biology, social networks anal-
ysis, computer vision and image analysis. The draw-
back when dealing with graph-based pattern recogni-
tion relies on the computational complexity required
in order to measure the (dis)similarity between two
graphs, which exponentially grows with respect to
their size (Bunke, 2003). This inevitably results in
a
https://orcid.org/0000-0003-4391-2598
b
https://orcid.org/0000-0003-1730-5436
c
https://orcid.org/0000-0001-8244-0015
an heavy computational burden when it comes to per-
form pattern recognition in the graph domain, espe-
cially when also node and/or edge attributes have to
be taken into account.
One of the mainstream approaches when dealing
with graph-based pattern recognition relies on embed-
ding spaces: in short, the pattern recognition problem
is cast from the structured input domain towards the
Euclidean space in which classification is performed.
Nonetheless, building the embedding space is a deli-
cate issue that must fill the informative and semantic
gap between the two domains (Baldini et al., 2019a).
This paper follows two previous works (Baldini
et al., 2019a; Baldini et al., 2019b) on the definition
of embedding spaces for graph classification thanks
to the Granular Computing paradigm (Bargiela and
Pedrycz, 2006; Bargiela and Pedrycz, 2003; Pedrycz,
2001). According to the latter, structured domains
can be analyzed by means of suitable information
granules, namely small data aggregates that endow
similar functional and/or structural characteristics.
Specifically, we investigate several classification sys-
338
Baldini, L., Martino, A. and Rizzi, A.
Complexity vs. Performance in Granular Embedding Spaces for Graph Classification.
DOI: 10.5220/0010109503380349
In Proceedings of the 12th International Joint Conference on Computational Intelligence (IJCCI 2020), pages 338-349
ISBN: 978-989-758-475-6
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

tems working on a properly-synthesized embedding
space. In fact, in our previous works (Baldini et al.,
2019a; Baldini et al., 2019b) we used a plain nearest
neighbours decision rule for the sake of simplicity.
Notwithstanding that, a nearest neighbours decision
rule suffers from several drawbacks, including high
computational burden, sensitivity to the classes distri-
bution and the number of classes. Hence, in this work,
we consider five different classifiers operating in the
embedding space: two support vector machines vari-
ants (with linear and radial basis kernels), two neuro-
fuzzy approaches and K-nearest neighbours. We pro-
vide a general three-fold overview in terms of perfor-
mances, embedding space complexity and structural
complexity of the adopted classification system.
The remainder of this paper is structured as fol-
lows: in Section 2 we provide an overview of possible
strategies for solving pattern recognition problems in
structured domains, with a major emphasis on Granu-
lar Computing-based systems; in Section 3 we intro-
duce GRALG, the Granular Computing-based graph
classification system at the basis of this work; in Sec-
tion 4 we introduce the datasets used for analysis,
along with the proper computational results; finally,
Section 5 concludes the paper.
2 CURRENT APPROACHES FOR
PATTERN RECOGNITION ON
THE GRAPH DOMAIN
In the literature, several mainstream strategies can
be found in order to solve pattern recognition prob-
lems in structured domains. According to the macro-
taxonomy presented in (Martino et al., 2018a), five
main strategies can be found. A first strategy consists
in engineering numerical features to be drawn from
the structured data at hand, to be concatenated in a
vector form. Examples of feature engineering tech-
niques involve entropy measures (Han et al., 2011;
Ye et al., 2014; Bai et al., 2012), centrality measures
(Mizui et al., 2017; Martino et al., 2018b; Leone Scia-
bolazza and Riccetti, 2020; Martino et al., 2020a),
heat trace (Xiao and Hancock, 2005; Xiao et al.,
2009) and modularity (Li, 2013). Whilst this ap-
proach is straightforward and allows to move the pat-
tern recognition problem towards the Euclidean space
in which any pattern recognition algorithm can be
used without alterations, designing the mapping func-
tion (i.e., enumerating the set of numerical features to
be extracted) requires a deep knowledge of both the
problem and the data at hand: indeed, the input spaces
being equal, specific subsets of features allow to solve
different problems.
A dual strategy consists in defining ad hoc dissim-
ilarity measures tailored to the input space under anal-
ysis. In this manner, the pattern recognition problem
can directly be solved in the input space without any
explicit cast towards the Euclidean space. On the plus
side, the input space might not be metric altogether,
yet this is not a prerogative; on the negative side, this
limits the range of pattern recognition algorithms that
can be employed. Indeed, the possibility of the input
space being non-metric reflects the non-metric pecu-
liarity of the dissimilarity measure itself: as such, the
pattern recognition algorithm must not rely on any
algebraic structures (e.g., mean, median, inner prod-
uct) and leverage pairwise dissimilarities only. Ex-
amples of custom dissimilarity measures include the
so-called edit distances, defined both on graph (Bal-
dini et al., 2019a; Baldini et al., 2019b) and sequence
(Levenshtein, 1966; Cinti et al., 2020) domains.
A further family is composed by embedding tech-
niques, which can either be explicit or implicit.
Amongst the implicit embedding techniques, ker-
nel methods emerge (Schölkopf and Smola, 2002;
Shawe-Taylor and Cristianini, 2004): kernel methods
exploit the so-called kernel trick (i.e., the inner prod-
uct in a reproducing kernel Hilbert space) in order
to measure similarity between patterns. In the litera-
ture, have been proposed several graph kernels (Kon-
dor and Lafferty, 2002; Borgwardt and Kriegel, 2005;
Shervashidze et al., 2009; Shervashidze and Borg-
wardt, 2009; Vishwanathan et al., 2010; Shervashidze
et al., 2011; Livi and Rizzi, 2013; Neumann et al.,
2016; Ghosh et al., 2018; Bacciu et al., 2018) that,
for example, consider substructures such as (random)
walks, trees, paths, cycles in order to measure simi-
larity or exploit propagation/diffusion schemes. Con-
versely, as explicit embedding techniques are con-
cerned, dissimilarity spaces are one of the main ap-
proaches (P˛ekalska and Duin, 2005). A dissimilar-
ity space can be built by first defining an ad-hoc dis-
similarity measure and then evaluating the pairwise
dissimilarity matrix which can be considered as em-
bedded in a Euclidean space: in other words, each
original (structured) pattern is represented by a real-
valued vector containing the distances with respect to
all other patterns (P˛ekalska and Duin, 2005) or with
respect to a properly selected subset of prototypes
(P˛ekalska et al., 2006). Alongside embedding meth-
ods, deep learning approaches are emerging as effec-
tive frameworks for different tasks like graph classifi-
cation, link prediction and node classification (Zhang
et al., 2019; Wu et al., 2020). In this context, the neu-
ral architectures typically implement a convolution
layer resembling the well-know convolutional neural
Complexity vs. Performance in Granular Embedding Spaces for Graph Classification
339

networks, with the scope to learn nodes representa-
tion taking into account neighbours features. Two
distinct approaches are typically employed: spectral-
based methods perform convolution operation in the
spectral domain by means of Graph Fourier Trans-
form (Kipf and Welling, 2016; Bianchi et al., 2019),
whilst spatial-based approaches used to aggregates
nodes attributes by directly exchanging information
amongst neighbours (Niepert et al., 2016; Hamilton
et al., 2017).
As anticipated in Section 1, Granular Comput-
ing is a powerful information processing paradigm
for building explicit embedding spaces and has been
successfully applied for synthesizing effective and in-
terpretable advanced pattern recognition systems for
structured data (see e.g. (Rizzi et al., 2012; Baldini
et al., 2019a; Baldini et al., 2019b; Martino et al.,
2019a; Martino et al., 2020b; Bianchi et al., 2014a;
Bianchi et al., 2014b; Bianchi et al., 2016) and ref-
erences therein). In short, Granular Computing is of-
ten described as a human-centered information pro-
cessing paradigm based on formal mathematical en-
tities known as information granules (Bargiela and
Pedrycz, 2006). The human-centered computational
concept in soft computing and computational intel-
ligence was initially developed by Lotfi A. Zadeh
through fuzzy sets (Zadeh, 1979) that exploits human-
inspired approaches to deal with uncertainties and
complexities in data. The process of ‘granulation’,
intended as the extraction of meaningful aggregated
data, mimics the human mechanism needed to or-
ganize complex data from the surrounding environ-
ment in order to support decision making activities
and to describe the world around (Pedrycz, 2016). For
this reason, Granular Computing can be described as
a framework for analyzing data in complex systems
aiming to provide human interpretable results (Mar-
tino et al., 2020b). The importance of information
granules resides in the ability to underline properties
and relationships between data aggregates. Specifi-
cally, their synthesis can be achieved by following the
indistinguishability rule, according to which elements
that show enough similarity, proximity or functional-
ity shall be grouped together (Zadeh, 1997; Pedrycz
and Homenda, 2013). With this approach, each gran-
ule is able to show homogeneous semantic informa-
tion from the problem at hand. Furthermore, data
at hand can be represented using different levels of
‘granularity’ and thus different peculiarities of the
considered system can emerge. Depending on this
resolution, an unknown computational process to be
modelled may exhibit different properties and differ-
ent atomic units that show different representations
of the system as a whole. Clearly, an efficient and
automatic procedure to select the most suitable level
of abstraction according to both the problem at hand
and the data description is of utmost importance. Due
to the tight link between ‘information granules’ and
‘groups of similar data’, the most straightforward ap-
proach in order to synthesize a possibly meaning-
ful set of information granules can be found in data
clustering (Frattale Mascioli et al., 2000; Pedrycz,
2005; Ding et al., 2015). Since the clustering proce-
dure is usually employed to discover groups of sim-
ilar data aggregates, it operates in the input (struc-
tured) domain: to this end, not only the (dis)similarity
measure, but also the cluster representative shall be
tailored accordingly. In order to represent clusters
in structured domains, the medoid (also called Min-
SOD) is usually employed (Martino et al., 2019b),
due to the fact that its evaluation relies only on pair-
wise dissimilarities between patterns belonging to the
cluster itself, without any algebraic structures that can
not be defined in non-geometric spaces. The clus-
ter representatives from the resulting partition can
be considered as symbols belonging to an alphabet
A = {s
1
, ..., s
n
}: these symbols are the pivotal gran-
ules on the top of which the embedding space can
be built thanks to the symbolic histograms paradigm.
According to the latter, each pattern can be described
as an n-length integer-valued vector which counts in
position i the number of occurrences of the i
th
symbol
drawn from the alphabet within the patter itself. The
embedding space can finally be equipped with alge-
braic structures such as the Euclidean distance or the
dot product and standard classification systems can be
used without alterations.
3 GRALG
The classification system considered in this work
is called GRALG (GRanular computing Approach
for Labeled Graphs) originally proposed in (Bianchi
et al., 2014a). GRALG aims to be a general purpose
classifier for labelled graphs embracing the Granular
Computing paradigm. The main core of the classi-
fier is a graph embedding procedure via symbolic his-
tograms, the geometric representation of graphs built
upon the symbols in an alphabet A which, in turn,
are synthesized in an unsupervised way through an
ensemble of clustering algorithms. These two stages
rely on a suitable dissimilarity measure, which al-
lows the pattern comparison in the graph domain. In
this section, we first introduce the dissimilarity mea-
sure involved in the atomic operations between graphs
(Section 3.1) and then we provide a brief description
of the building blocks that make up the stages needed
NCTA 2020 - 12th International Conference on Neural Computation Theory and Applications
340

to perform the explicit graph mapping towards a suit-
able embedding space (Sections 3.2–3.5), along the
way in which they cooperate for training (Section 3.6)
and testing (Section 3.7) the overall system.
3.1 Graph Edit Distance
As anticipated in Section 2, the Graph Edit Distance
(GED) is a common approach to evaluate the dissim-
ilarity between graphs. GEDs define the distance be-
tween two graph G
1
and G
2
as the minimum cost
path needed to turn G
1
into G
2
, by applying a se-
ries of atomic operations defined on both vertices and
edges: substitution, insertion and deletion. Besides
the costs, the edit operations may also be associated
to some weights in order to reflect the importance
of the aforementioned operations. Generally speak-
ing, the dissimilarity measure d : G ×G → R between
two graphs can be cast as the following minimization
problem:
d(G
1
, G
2
) = min
(ε
1
,...,ε
k
)∈O(G
1
,G
2
)
∑
k
i=1
c(ε
i
) (1)
where c (ε
i
) is the cost associated to the generic
1
edit
operation ε
i
and O(G
1
, G
2
) is the set of prospective
operations that gives an isomorphism between the two
graphs. The main drawback when using a GED is the
unfeasible computational complexity needed to com-
pute an exact solution of Eq. (1) and, for this reason,
many heuristics were proposed in order to solve the
graph matching problem in a sub-optimal manner, yet
with reasonable computational burden. Given these
aspects, in GRALG we employ a greedy heuristic
called node Best Match First (nBMF) (Bianchi et al.,
2016) in order to measure the dissimilarity between
graphs. Formally, let G
1
= (V
1
, E
1
, L
v
, L
e
) and G
2
=
(V
2
, E
2
, L
v
, L
e
) be two graphs labelled on both edges
and vertices. Furthermore, let d
π
v
v
: L
v
× L
v
→ R and
d
π
e
e
: L
e
× L
e
→ R be the dissimilarities defined on
nodes’ and edges’ attributes (L
v
and L
e
, respectively).
For the sake of generalization, d
π
v
v
and d
π
e
e
might be
parametric with respect to π
v
and π
e
, namely sets of
real-valued parameters required to evaluate d
π
v
v
and
d
π
e
e
, respectively. In nBMF, the substitution cost on
nodes and edges is evaluated with the corresponding
dissimilarity function (d
π
v
v
and d
π
e
e
). Accordingly, it
is possible to define the overall substitution costs on
nodes and edges (c
sub
nodes
and c
sub
edge
) by summing up the
dissimilarity values of all the nodes or edges substi-
tuted during the procedure. Conversely, insertion and
deletion costs c
ins
node
, c
ins
edge
, c
del
node
, c
del
edge
depend on the
difference between the two graphs in terms of nodes
1
Regardless on whether it is a substitution, deletion or
insertion on nodes or edges.
and edges set cardinality. The interested reader can
found detailed pseudo-codes describing the nBMF in
(Baldini et al., 2019a). Additionally, each operation
is weighted by w
sub
node
, w
sub
edge
, w
ins
node
, w
ins
edge
, w
del
node
, w
del
edge
individually bounded in [0, 1]. At the end of the pro-
cedure, the dissimilarities between nodes and edges,
say d
V
(V
1
, V
2
) and d
E
(E
1
, E
2
), are evaluated as:
d
V
(V
1
, V
2
) = w
sub
node
c
sub
node
+ w
ins
node
c
ins
node
+ w
del
node
c
del
node
d
E
(E
1
, E
2
) = w
sub
edge
c
sub
edge
+ w
ins
edge
c
ins
edge
+ w
del
edge
c
del
edge
(2)
Each dissimilarity is normalized by taking into ac-
count the different orders between the two graphs:
d
0
V
(V
1
, V
2
) =
d
V
(V
1
, V
2
)
max{o
1
, o
2
}
d
0
E
(E
1
, E
2
) =
d
E
(E
1
, E
2
)
1
2
(min{o
1
, o
2
} · (min{o
1
, o
2
} − 1))
(3)
with o
1
= |V
1
| and o
2
= |V
2
|. Finally, the dissimilar-
ity between the two graphs reads as:
d(G
1
, G
2
) =
1
2
d
0
V
(V
1
, V
2
) + d
0
E
(E
1
, E
2
)
(4)
3.2 Extractor
The subgraph extraction block aims at breaking down
a graph into its constituent parts following a stochas-
tic subsampling approach operating in a class-aware
fashion. These improvements have been thoroughly
described and tested in (Baldini et al., 2019a) and
(Baldini et al., 2019b), to which the interested reader
is referred to for further details, in order to overcome
the main limitation of GRALG: indeed, in its origi-
nal implementation (Bianchi et al., 2014a), GRALG
used to exhaustively extract all subgraphs up to a
given user-defined order o from any input graph, re-
sulting in a non-negligible running time and memory
footprint. In (Baldini et al., 2019a), we show how a
stochastic subsampling can lead to comparable per-
formances with respect to the original (exhaustive)
implementation with remarkably lower running times
and memory usage. Indeed, this procedure builds a
subgraph set with a fixed cardinality W by extract-
ing subgraphs uniformly at random (replacements are
allowed) from a set of graphs using either one of
two well-known traversal strategies, namely Breadth
First Search and Depth First Search. Furthermore, in
(Baldini et al., 2019b), we show how the stochastic
subsampling granulation procedure can be performed
in a class-aware fashion: this allows the exploita-
tion of the ground-truth class labels in the training
data, resulting in a more effective alphabet synthe-
sis. The latter, being competitive both in terms of per-
Complexity vs. Performance in Granular Embedding Spaces for Graph Classification
341

formances and computational complexity, is the strat-
egy we used in this work. Let S be a set of graphs,
each of which is associated to its ground-truth class
label L with L ∈ {1, . . . , N} and N being the number
of classes. The class-aware stochastic extraction ran-
domly draws a graph G := {V , E} from S and then
randomly draws a vertex v ∈ V . Then, starting from
the node v, a suitable traversal strategy explores G
collecting a subgraph g = {V
g
, E
g
} with V
g
⊂ V ver-
tices and E
g
⊂ E edges. The subgraph g is inserted
in a subgraph set S
L
g
according to the graph’s label L.
In this way, it is also possible to the define the num-
ber of subgraphs W to be sampled from the starting
set by fixing how many times the procedure must oc-
cur, since each extraction outputs a single graph. The
maximum order o of the extracted subgraph, namely
the maximum number of vertices for all subgraphs g,
is still a user-defined parameter. Conversely to the
original work (Bianchi et al., 2014a), this procedure
allows to arbitrary choose the cardinality W of the
subgraphs set
S
N
L=1
S
L
g
and consequently relieves the
memory issues and improves the wall-clock runtime
of the algorithm.
3.3 Granulator
The Granulator defines a procedure able to synthe-
size information granules by building the alphabet
A = {s
1
, . . . , s
n
}, namely a set that collects the rele-
vant substructures s
i
starting from a given subgraphs
set. The core method in charge of returning suit-
able information granules is the Basic Sequential Al-
gorithmic Scheme (BSAS) clustering algorithm (Bal-
dini et al., 2019a; Baldini et al., 2019b) that works
directly in the graph domain thanks to the dissimilar-
ity measure described in Section 3.1. BSAS needs
two parameters: the dissimilarity threshold θ below
which a pattern is included into the closest cluster and
the maximum number of allowed clusters Q, in or-
der to reasonably bound the number of resulting clus-
ters which can grow indefinitely (especially for small
values of θ). According to the Granular Computing
paradigm, the granulator shall extract symbols by ex-
ploring different levels of abstraction for the problem
at hand: indeed, the granulation performs a clustering
ensemble procedure by generating different partitions
of the input data using different θ values. Then, for
every cluster C in the resulting partitions, a cluster
quality index F(C) is defined as:
F(C) = η · Φ(C) + (1 − η) · Θ(C) (5)
where the two terms Φ(C) and Θ(C) are respectively
the compactness and the cardinality of cluster C, de-
fined in turn as:
Φ(C) =
1
|C| − 1
∑
i
d(g
∗
, g
i
) (6)
Θ(C) = 1 −
|C|
|S
g
|
(7)
where g
∗
is the representative of cluster C and g
i
the
i
th
pattern in the cluster. In this way, we evaluate each
cluster by considering its compactness Φ and cardi-
nality Θ, whose importance is weighted by a trade-off
parameter η ∈ [0, 1]. Since the BSAS operates in the
graph domain, as discussed in Section 2, the repre-
sentative g
∗
is defined as the medoid of the cluster,
namely the element that minimizes the pairwise sum
of distances among all patterns in the cluster (Martino
et al., 2019b). Finally, by thresholding F(C) with
a value τ
F
, only well-formed clusters (i.e., compact
and populated) are promoted to be symbols s belong-
ing to the alphabet A. Since the symbol synthesis oc-
curs on a labelled training set and since the Extrac-
tor operates in a class-aware fashion (Section 3.2),
the clustering ensemble is performed individually on
the S
L
g
sets, hence collecting class-related symbols in
class-aware alphabets A
L
, which are finally merged
together in A =
S
N
L=1
A
L
.
3.4 Embedder
The Embedder block plays the key role necessary to
map patterns from the graph domain towards a ge-
ometric (Euclidean) space. Formally, let G be the
graph domain, the Embedder is in charge of build-
ing the function φ : G → D that maps the graph do-
main into an n-dimensional space D ⊆ R
n
. The em-
bedding function relies on the symbolic histograms
paradigm, exploiting the relevant substructures syn-
thesized by the Granulator (i.e., the alphabet). In or-
der to describe this process, let G
exp
be the set made
up of the subgraphs in G ∈ G, i.e. the expanded rep-
resentation of G in subgraphs. Given the alphabet set
A = {s
1
, . . . , s
n
}, the vectorial representation h (i.e.,
the symbolic histogram) of G is a vector whose com-
ponents represents the number of occurrences of each
symbol s ∈ A in G
exp
:
h = φ
A
(G
exp
) = [occ(s
1
, G
exp
), . . . , occ(s
n
, G
exp
)]
(8)
The function occ : A → N compares a symbol s
j
with
j = 1 . . . n and all the subgraphs g ∈ G
exp
. If the
resulting dissimilarity measure is below a symbol-
dependent threshold value τ
j
= Φ(C
j
) · ε, this counts
as a match and the counter is increased by 1. The ε
value is a user-defined tolerance parameter and C
j
is
the cluster whose MinSOD is s
j
. It is worth remark-
ing here that the computational complexity needed to
NCTA 2020 - 12th International Conference on Neural Computation Theory and Applications
342

expand exhaustively G in the set of its subgraph G
exp
may lead to memory issues, since the number of sub-
graphs grows exponentially with respect to the order
of graph. Furthermore, the number of matches needed
to build the embedding space can be very large since
all symbols have to be compared with the subgraphs
in G
exp
. This could lead to unfeasible running times
even for medium size datasets. For this reason, the set
G
exp
is evaluated with a deterministic algorithm that
tries to partially expand G = {V , E}: starting from
a seed node v ∈ V , the same traverse strategy em-
ployed in the Extractor drives the exploration and the
extraction of subgraphs from G, starting from order 1
up until a user-defined value o and collects these sub-
structures in G
exp
. Since the goal of the procedure is
to limit the cardinality of G
exp
, if a node v ∈ V al-
ready appears in one of the previously-extracted sub-
graphs, it will not be eligible to be a seed node for
further traversals. The procedure goes on as long as a
vertex v ∈ V can be legally used as root node for the
traversal strategy.
3.5 Classifier
The three blocks in Sections 3.2–3.4 allow us to build
an embedding space D ⊆ R
n
, where each graph is
mapped with an appropriate vector h ∈ D, i.e. its
symbolic histogram. Further, such approach allows
us to use any classifier designed in R
n
without limita-
tions. In this work, we considered different classifica-
tion systems:
Support Vector Machines (SVMs) aim at estimat-
ing the decision boundary between two classes by
a maximal-margin hyperplane. We employed the
well-known ν-SVM (Schölkopf et al., 2000) using
two different kernels: the linear kernel endowing
the dot product between patterns K (x, x
0
) = hx, x
0
i
and a non-linear radial basis function (RBF) ker-
nel, i.e. K (x, x
0
) = exp{−γkx − x
0
k
2
}
Neuro-Fuzzy Min-Max classifier relying on the
construction of hyperboxes, which serve as
atomic geometric structures needed to define
the decision boundary in the training phase.
Specifically, we tested two algorithms for training
the model (i.e., building hyperboxes): Adaptive
Resolution Classifier (ARC) and Pruning Adap-
tive Resolution Classifier (PARC) (Rizzi et al.,
2002).
K-Nearest Neighbours (K-NN) in which, when an
unseen pattern is considered for classification, its
K nearest patterns are selected and the decision is
taken according to the most voted class amongst
the K patterns (Cover and Hart, 1967).
3.6 Training Phase
After individual and detailed descriptions of the build-
ing blocks that made up the GRALG classification
system, here we describe how those blocks cooper-
ate together in order to synthesize the final model. To
this end, let S be a dataset of labelled graphs on nodes
and/or edges and let S
tr
, S
vs
and S
ts
be the training,
validation and test set drawn from S:
Extractor: takes as input S
tr
and the parameter W
and, according to the description given in Section
3.2, extracts N different class-specific subgraphs
sets S
L
g,tr
Granulator: each class-aware subgraphs set S
L
g,tr
is
granulated and consequently A
L
class-specific al-
phabets are constructed
Embedder: the overall alphabet A =
S
N
L=1
A
L
, with
n = |A| symbols, enters the embedding block.
First, all graphs in S
tr
and S
vs
are expanded as de-
scribed in Section 3.4. Then, for each G
tr
i
∈ S
tr
and G
vs
i
∈ S
vs
, we can construct the associated
vector representation h
tr
i
∈ D and h
vs
i
∈ D such
that D ⊆ R
n
Classifier: given the embedded version of training
and validation set (h
tr
and h
vs
), the quality of
the mapping function is evaluated by considering
a given classifier (amongst the ones presented in
Section 3.5) which is trained on h
tr
and later eval-
uated on h
vs
.
The procedures involved to complete a graph classi-
fication are very sensitive to specific parameters that
can strongly affect the overall performances. In or-
der to face this issue, two different optimizations take
place separately.
3.6.1 Alphabet Optimization Phase
A first stage of optimization aims to synthesize an op-
timal set of symbols A
∗
and properly tune the hyper-
parameters of the chosen classifier, by deploying an
evolutive strategy, i.e. a genetic algorithm. In order to
purse this goal, the following parameters (and corre-
sponding procedures) are considered:
GED: the dissimilarity measure in the graph domain
requires a fine tuning of the 6 weights for the
edit operations: W = {w
sub
node
, w
sub
edge
, w
ins
node
, w
ins
edge
,
w
del
node
, w
del
edge
}. Depending on the dataset, both
vertices and edges can require a parametric dis-
similarity measure d
π
v
v
and d
π
e
e
with parameters
Π = {π
v
, π
e
} whose values are optimized as well.
Granulator: in order to synthesize an optimal alpha-
bet, we optimize Q (maximum number of allowed
Complexity vs. Performance in Granular Embedding Spaces for Graph Classification
343

clusters for BSAS), τ
F
(MinSOD quality thresh-
old for being promoted to a symbol) and η (trade-
off parameter that weights compactness and car-
dinality in Eq. (5)).
Classifier: the set of hyperparameters C that depends
on the considered classifier:
• ν-SVM: only the regularization term ν ∈ (0, 1].
• RBF ν-SVM: along with ν ∈ (0, 1], we also
tune the kernel shape γ ∈ (0, 100].
• ARC/PARC: we optimize the λ ∈ (0, 1) pa-
rameter used as trade-off between the error on
training set and the network complexity. Fur-
thermore, we optimize the type of membership
function (to be chosen between Trapezoidal and
Simpson’s) and the decay parameter µ ∈ (0, 1)
associated with it.
• K-NN: the number K ∈ [1, 2
p
|S
tr
|] of nearest
patterns involved in the voting.
To summarize, the genetic code reads as:
[Q τ
F
η W Π C] (9)
whereas the fitness function J
al ph
reads as the classi-
fier accuracy ω ∈ [0, 1] on the validation set:
J
al ph
= ω(S
vs
) =
∑
|S
vs
|
i=1
δ(y
i
, ˆy
i
)
|S
vs
|
(10)
with
δ(y
i
, ˆy
i
) =
(
1 if y
i
= ˆy
i
0 otherwise
(11)
and where, in turn, ˆy
i
and y
i
are (respectively) the pre-
dicted label and the ground-truth label for i-th pattern
in S
vs
. Standard genetic operators (mutation, selec-
tion, crossover and elitism) take care of moving from
one generation to the next. The best individual is re-
tained at the end of the evolution, specifically the por-
tions of the genetic code W
?
and Π
?
, along with the
alphabet A
?
synthesized using its genetic code.
3.6.2 Feature Selection Phase
The cardinality of A
?
and consequently the embed-
ding space D ⊆ R
|A
?
|
may be too large, impacting
the classification performance and the interpretabil-
ity of the final model. To this end, we implement
a feature selection phase still based on genetic opti-
mazation. Formally, let m ∈ {0, 1}
|A
?
|
be a projection
mask, then:
1. we perform the component-wise product between
the mask and the embedded vectors of the training
set:
h
tr
= m h
tr
(12)
by further neglecting components corresponding
to 0’s in h
tr
. Hence, it is possible to figure h
tr
as lying in a (possibly) reduced embedding space
D ⊆ R
m
with m ≤ n
2. according to the mask m employed in step 1, the
validation set h
vs
∈ D is reduced in the same way,
leading to h
vs
∈ D
3. the classification system is trained on h
tr
using the
hyperparameters set C . The accuracy on the (re-
duced) validation set h
vs
is finally computed.
Following this scheme, a genetic algorithm drives the
optimization of the mask m and, at the same time, the
classifier hyperparameters C by maximizing a fitness
function J
f s
that reads as the linear convex combina-
tion between the classifier accuracy on the validation
set and the cost of the mask weighted by a trade-off
value α ∈ [0, 1]:
J
f s
= α ·ω (S
vs
) + (1 − α) · µ (13)
where the cost of the mask µ ∈ [0, 1] is defined as:
µ = 1 −
|{i : m
i
= 1}|
|m|
(14)
When the evolution is completed, the best individual
is retained, as it encodes the optimal projection mask
m
?
, able to return the reduced alphabet A
?
, and the
optimal set of parameters C
?
used to train the classi-
fier in the embedding space D.
3.7 Synthesized Classification Model
After the two optimizations stages are over, the final
classification performances can be evaluated on the
test set. First, the test set S
ts
is embedded in the vec-
tor space D. To this aim, the embedder block specif-
ically equipped with parameters W
?
and Π
?
for the
GED dissimilarity measure, outputs the vector set h
ts
by taking advantage of the alphabet A
?
. The classifier
returned by the optimization phase (i.e., trained on the
projected vectors h
tr
with the hyperparameters C
∗
) is
tested on the embedded test set h
ts
∈ D, returning the
overall accuracy of the GRALG system.
4 EXPERIMENTS
Five different datasets from the IAM repository
(Riesen and Bunke, 2008) have been considered for
testing:
Letter: a triad of datasets where each graph repre-
sents a letter drawing with different level of distor-
tions: low (L), medium (M) and high (H). Match-
NCTA 2020 - 12th International Conference on Neural Computation Theory and Applications
344
AIDS GREC Letter-L Letter-M Letter-H
40
50
60
70
80
90
100
110
Accuracy [%]
(a) ARC
AIDS GREC Letter-L Letter-M Letter-H
40
50
60
70
80
90
100
110
Accuracy [%]
(b) PARC
AIDS GREC Letter-L Letter-M Letter-H
40
50
60
70
80
90
100
110
Accuracy [%]
(c) ν-SVM
AIDS GREC Letter-L Letter-M Letter-H
40
50
60
70
80
90
100
110
Accuracy [%]
(d) RBF ν-SVM
AIDS GREC Letter-L Letter-M Letter-H
40
50
60
70
80
90
100
110
Accuracy [%]
(e) K-NN
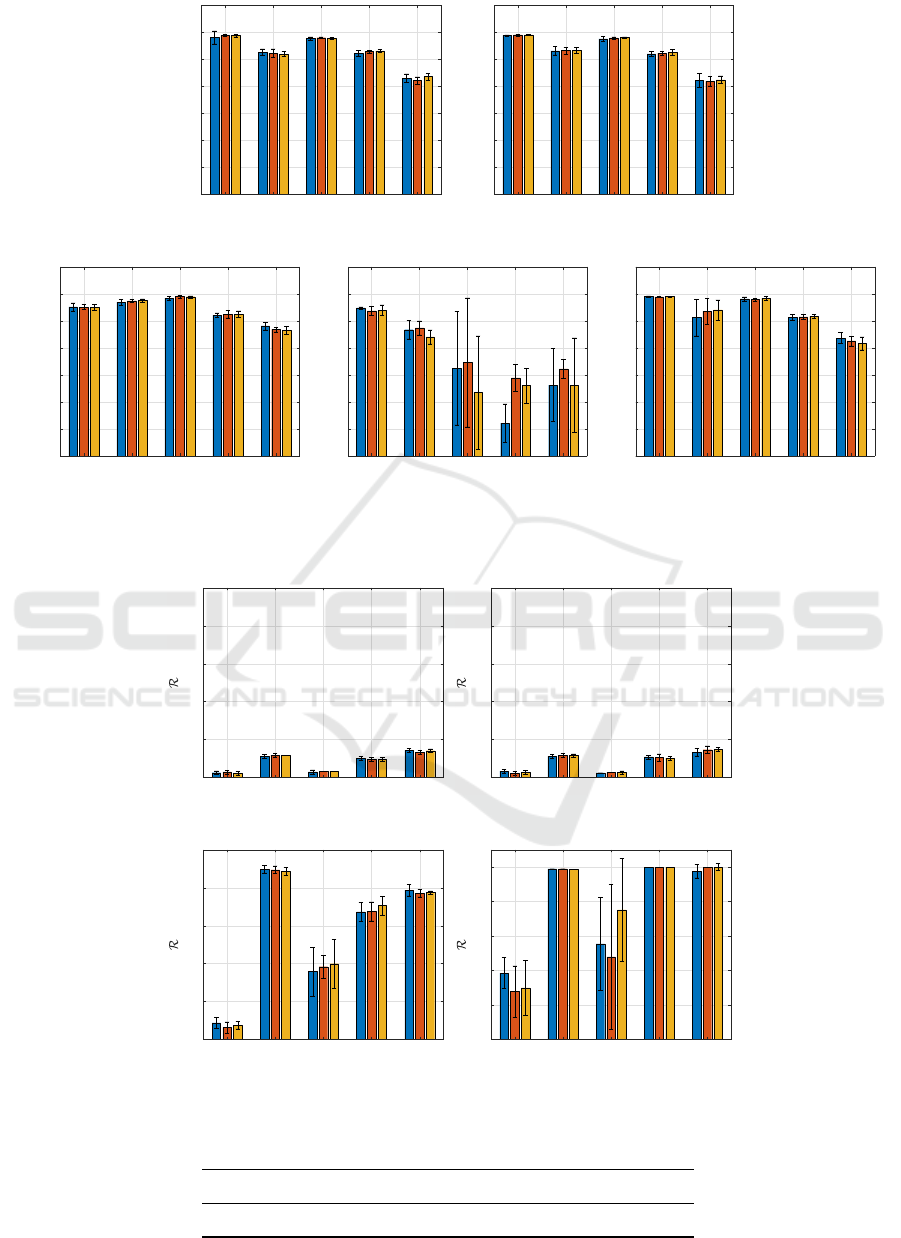
Figure 1: Accuracy comparison for the 5 classifiers. Blue, red and yellow bars correspond to subsampling rates W =
10%, 30%, 50%, respectively. Whiskers indicate the standard deviation.
AIDS GREC Letter-L Letter-M Letter-H
0
0.2
0.4
0.6
0.8
1
(a) ARC
AIDS GREC Letter-L Letter-M Letter-H
0
0.2
0.4
0.6
0.8
1
(b) PARC
AIDS GREC Letter-L Letter-M Letter-H
0
0.2
0.4
0.6
0.8
1
(c) ν-SVM
AIDS GREC Letter-L Letter-M Letter-H
0
0.2
0.4
0.6
0.8
1
(d) RBF ν-SVM
Figure 2: Complexity ratio for SVM and Min-Max classifiers. For bar legend, see caption of Fig. 1.
Table 1: Number of subgraphs extracted from S
tr
(o = 5) by the exhaustive procedure.
Letter-L Letter-M Letter-H GREC AIDS
8193 8582 21165 27119 35208
Complexity vs. Performance in Granular Embedding Spaces for Graph Classification
345
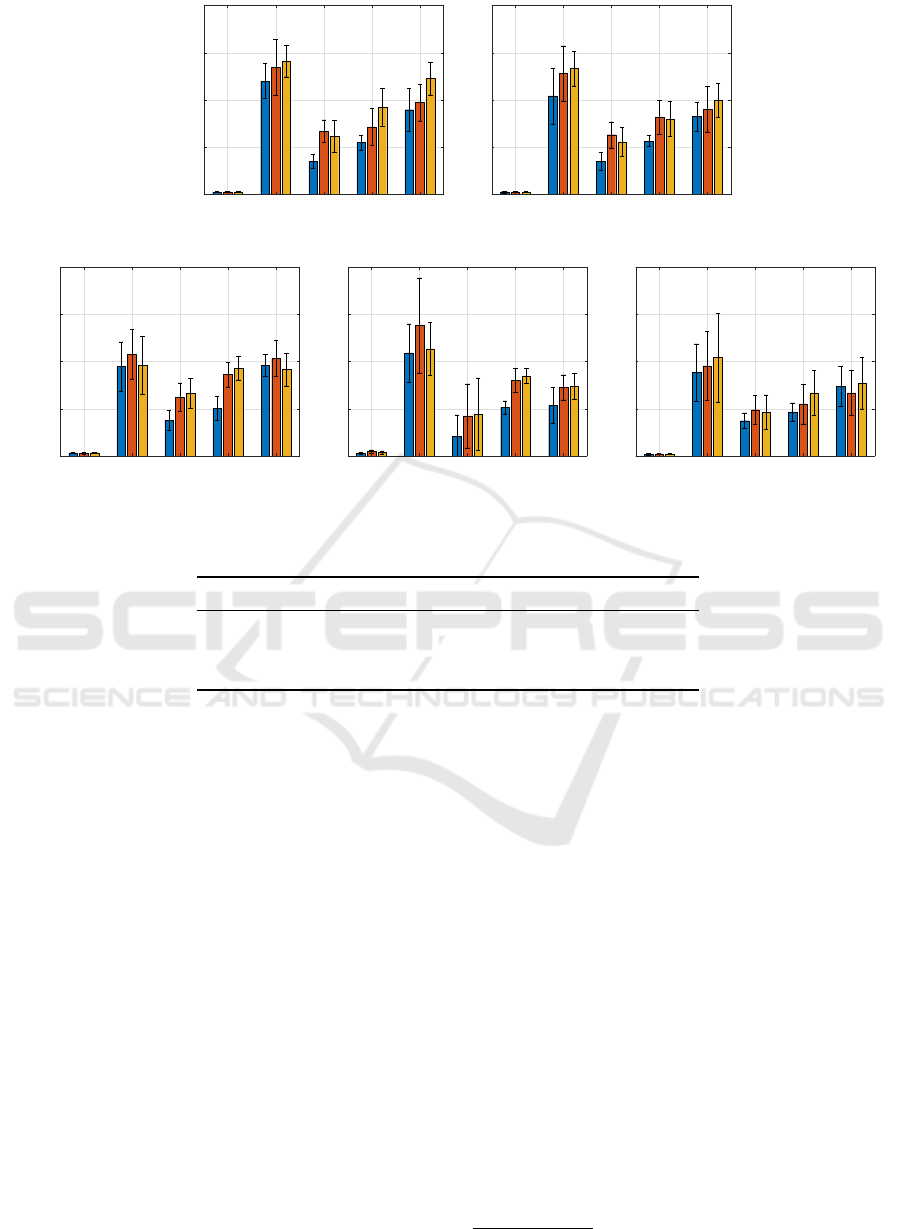
AIDS GREC Letter-L Letter-M Letter-H
0
200
400
600
800
Number of Selected Features
(a) ARC
AIDS GREC Letter-L Letter-M Letter-H
0
200
400
600
800
Number of Selected Features
(b) PARC
AIDS GREC Letter-L Letter-M Letter-H
0
200
400
600
800
Number of Selected Features
(c) ν-SVM
AIDS GREC Letter-L Letter-M Letter-H
0
200
400
600
800
Number of Selected Features
(d) RBF ν-SVM
AIDS GREC Letter-L Letter-M Letter-H
0
200
400
600
800
Number of Selected Features
(e) K-NN
Figure 3: Selected number of symbols comparison for the 5 classifiers. For bar legend, see caption of Fig. 1.
Table 2: Average K value for K-NN classifier, with standard deviation.
Sample % AIDS GREC Letter1 Letter2 Letter3
10 5 ± 4 3 ± 2 4 ± 4 1 ± 0 3 ± 3
30 8 ± 5 2 ± 1 5 ± 4 1 ± 0 4 ± 2
50 7 ± 4 2 ± 1 4 ± 4 1 ± 0 5 ± 5
ing between vertices is evaluated with a plain Eu-
clidean distance, whereas edge matching is a sim-
ple delta distance, being unlabelled.
GREC: patterns in this dataset are graph representa-
tion of symbols taken from architectural and elec-
tronic drawings. Since node labels are composed
by different data structures, a custom dissimilar-
ity measure is involved in the vertex matching, as
well as the edge dissimilarity function. In both
cases, dissimilarity measures are parametric with
respect to five real-valued parameters bounded in
[0, 1] whose values are stored in the set Π (cf. Eq.
(9)).
AIDS: graph instances in this dataset are molecular
compounds where atoms are represented as nodes
and covalent bonds as edges. Nodes dissimilarity
is a custom (non parametric) dissimilarity mea-
sure.
Further properties of these datasets and formal defini-
tion of nodes and vertices dissimilarities can be found
in (Baldini et al., 2019b).
The software is developed in C++ using the
SPARE library (Livi et al., 2014). Additional depen-
dancies include LibSVM (Chang and Lin, 2011) for
ν-SVMs and the Boost Graph Library
2
for handling
graph data structures. In all tests, we adopt the Class-
Aware granulator (Baldini et al., 2019b) along with
the stochastic sampling method (Baldini et al., 2019a)
described in Section 3.2, where a Breadth First Search
strategy has been employed for graph traversing. In
this way, the cardinality of the set
S
N
L=1
S
L
g,tr
needed to
the granulator has been fixed to a given number W as
percentage of the number of subraphs returned by the
exhaustive procedure (Bianchi et al., 2014a), reported
in Table 1. In order to take into account the intrinsic
randomness in the model synthesis procedure, we run
GRALG 10 times (both training and testing phases),
showing the results on the test set in terms of average
and standard deviation. For each of the five datasets,
training, validation and test splits are kept unchanged
with respect to the ones available in the IAM reposi-
tory.
The system parameters have been chosen as fol-
lows:
• W = 10%, 30%, 50% of the maximum number of
2
http://www.boost.org/
NCTA 2020 - 12th International Conference on Neural Computation Theory and Applications
346

subgraphs that can be drawn from the training set
(Table 1)
• Q ∈ [1, 500]
• o = 5 maximum subgraphs vertices
• 20 individuals per population of both genetic al-
gorithms
• 20 generations for the first genetic algorithm (al-
phabet optimization)
• 50 generations for the second genetic algorithm
(feature selection)
• α = 0.99 in the fitness function for the second ge-
netic algorithm (cf. Eq. (13))
• ε = 1.1 as tolerance value for the symbolic his-
tograms evaluation.
As the model complexity is concerned, we define R
as the ratio between complexity of the synthesized
model and its maximal attainable complexity:
• for SVMs, R reads as the ratio between the num-
ber of support vectors and |S
tr
| (Martino et al.,
2020a);
• for ARC/PARC, one takes the ratio between the
number of hyperboxes and |S
tr
| (Rizzi et al.,
2002);
• K-NN notably has the maximum complexity,
which equals |S
tr
|, since all pairwise distances
with respect to the training data must be evalu-
ated for classification. Notwithstanding that, for
the sake of completeness, we report the resulting
number of neighbours in Table 2.
In Fig. 1, we show the accuracy (both in terms
of average and standard deviation) obtained by the
five classifiers on the embedded version of the test
set S
ts
, alongside the cardinality of the optimized al-
phabet A
∗
in Fig. 3. The latter simply reads as the
number of selected features after the second genetic
optimization and it can be considered as a measure
of model interpretability (the lower, the better). The
good performances obtained by ν-SVM are striking:
this classifier outperforms the competitors especially
for GREC and Letter-H. On the other hand, its kernel-
ized counterpart shows generally poor performances
with respect to the rest of competitors. Notably, the
accuracy achieved on the three Letter datasets are
far from being comparable with the other methods.
This could be explained by considering that even if
a feature selection mechanism is employed, the re-
sulting embedding space is generally large, making
unnecessary or, as in this case, disadvantageous the
projection mapping due to kernel functions (Martino
et al., 2019a; Martino et al., 2020b). By observing
the number of selected features, the K-NN classifier
emerges as the one that generally shows the smallest
number of symbols in the final alphabet, especially
for Letter datasets, while keeping comparable perfor-
mances in terms of accuracy with the linear ν-SVM.
When we compare the classifiers in terms of struc-
tural complexity via the R score, as can be seen in
Fig. 2, Min-Max networks clearly show remarkable
behaviour with respect to SVMs. Indeed, even if ν-
SVMs have (slightly) better performances in terms
of accuracy, Min-Max networks strikingly outperform
them in terms of structural complexity. Further, it is
worth noting that RBF ν-SVMs in three cases (GREC,
Letter-M and Letter-H) tend to elect all patterns as
support vectors: a clear sign that they strive in dis-
criminating patterns in a Hilbert space.
5 CONCLUSIONS
In this paper, we proposed a comparison between dif-
ferent supervised learning algorithms for classifying
graphs in geometric spaces thanks to a graph embed-
ding procedure. Specifically, starting from GRALG,
whose embedding strategy relies on symbolic his-
tograms, we considered a ν-SVM equipped with lin-
ear and Gaussian kernels, a Min-Max neuro-fuzzy
network with two different training methods (namely,
ARC and PARC) and a simple K-NN decision rule.
The classifiers are evaluated by taking into account
both the accuracy on the test set and the R score for
the structural complexity. If on one hand these two
indices can summarize their classification and gen-
eralization abilities, they do not consider the com-
plexity (dimensionality) of the underlying embedding
space. Consequently, the number of selected features
are considered as a futher performance measure of the
overall classification system. At least for the con-
sidered datasets, our tests returned linear ν-SVM as
generally the most performing method and its kernel-
ized counterpart as the least performing one: this sug-
gests that non-linear kernels that implicitly map pat-
terns into higher dimensions may not work properly
with graph embedding strategies based on symbolic
histograms since the corresponding embedding vec-
tors are likely to reside in an already high dimensional
space. Notwithstanding their good performance, if
compared with ARC and PARC classifiers, SVMs
tend to have higher structural complexity. Under the
model interpretability viewpoint, K-NN seems to be
the most promising classifier.
Complexity vs. Performance in Granular Embedding Spaces for Graph Classification
347

REFERENCES
Bacciu, D., Micheli, A., and Sperduti, A. (2018). Gen-
erative kernels for tree-structured data. IEEE trans-
actions on neural networks and learning systems,
29(10):4932–4946.
Bai, L., Hancock, E. R., Han, L., and Ren, P. (2012). Graph
clustering using graph entropy complexity traces. In
Proceedings of the 21st International Conference on
Pattern Recognition (ICPR2012), pages 2881–2884.
Baldini, L., Martino, A., and Rizzi, A. (2019a). Stochastic
information granules extraction for graph embedding
and classification. In Proceedings of the 11th Inter-
national Joint Conference on Computational Intelli-
gence - Volume 1: NCTA, (IJCCI 2019), pages 391–
402. INSTICC, SciTePress.
Baldini, L., Martino, A., and Rizzi, A. (2019b). Towards
a class-aware information granulation for graph em-
bedding and classification. In Computational Intel-
ligence: 11th International Joint Conference, IJCCI
2019 Vienna, Austria, September 17-19, 2019 Revised
Selected Papers. To appear in.
Bargiela, A. and Pedrycz, W. (2003). Granular comput-
ing: an introduction. Kluwer Academic Publishers,
Boston.
Bargiela, A. and Pedrycz, W. (2006). The roots of granular
computing. In 2006 IEEE International Conference
on Granular Computing, pages 806–809.
Bianchi, F. M., Grattarola, D., Alippi, C., and Livi, L.
(2019). Graph neural networks with convolutional
arma filters. arXiv preprint arXiv:1901.01343.
Bianchi, F. M., Livi, L., Rizzi, A., and Sadeghian, A.
(2014a). A granular computing approach to the de-
sign of optimized graph classification systems. Soft
Computing, 18(2):393–412.
Bianchi, F. M., Scardapane, S., Livi, L., Uncini, A., and
Rizzi, A. (2014b). An interpretable graph-based im-
age classifier. In 2014 International Joint Conference
on Neural Networks (IJCNN), pages 2339–2346.
Bianchi, F. M., Scardapane, S., Rizzi, A., Uncini, A.,
and Sadeghian, A. (2016). Granular computing tech-
niques for classification and semantic characterization
of structured data. Cognitive Computation, 8(3):442–
461.
Borgwardt, K. M. and Kriegel, H. P. (2005). Shortest-path
kernels on graphs. In Fifth IEEE International Con-
ference on Data Mining (ICDM’05), pages 8 pp.–.
Bunke, H. (2003). Graph-based tools for data mining and
machine learning. In Perner, P. and Rosenfeld, A., ed-
itors, Machine Learning and Data Mining in Pattern
Recognition, pages 7–19, Berlin, Heidelberg. Springer
Berlin Heidelberg.
Chang, C.-C. and Lin, C.-J. (2011). Libsvm: A library for
support vector machines. ACM transactions on intel-
ligent systems and technology (TIST), 2(3):27.
Cinti, A., Bianchi, F. M., Martino, A., and Rizzi, A. (2020).
A novel algorithm for online inexact string matching
and its fpga implementation. Cognitive Computation,
12(2):369–387.
Cover, T. and Hart, P. (1967). Nearest neighbor pattern clas-
sification. IEEE transactions on information theory,
13(1):21–27.
Ding, S., Du, M., and Zhu, H. (2015). Survey on granularity
clustering. Cognitive neurodynamics, 9(6):561–572.
Frattale Mascioli, F. M., Rizzi, A., Panella, M., and Mar-
tinelli, G. (2000). Scale-based approach to hierarchi-
cal fuzzy clustering. Signal Processing, 80(6):1001 –
1016.
Ghosh, S., Das, N., Gonçalves, T., Quaresma, P., and
Kundu, M. (2018). The journey of graph kernels
through two decades. Computer Science Review,
27:88 – 111.
Hamilton, W., Ying, Z., and Leskovec, J. (2017). In-
ductive representation learning on large graphs. In
Advances in neural information processing systems,
pages 1024–1034.
Han, L., Hancock, E. R., and Wilson, R. C. (2011). Charac-
terizing graphs using approximate von neumann en-
tropy. In Vitrià, J., Sanches, J. M., and Hernández,
M., editors, Pattern Recognition and Image Analysis,
pages 484–491, Berlin, Heidelberg. Springer Berlin
Heidelberg.
Kipf, T. N. and Welling, M. (2016). Semi-supervised clas-
sification with graph convolutional networks. arXiv
preprint arXiv:1609.02907.
Kondor, R. I. and Lafferty, J. (2002). Diffusion kernels on
graphs and other discrete structures. In Proceedings of
the 19th international conference on machine learn-
ing, volume 2002, pages 315–322.
Leone Sciabolazza, V. and Riccetti, L. (2020). Diffusion de-
lay centrality: Decelerating diffusion processes across
networks. Available at SSRN 3653030.
Levenshtein, V. I. (1966). Binary codes capable of correct-
ing deletions, insertions, and reversals. Soviet physics
doklady, 10(8):707–710.
Li, W. (2013). Modularity embedding. In Lee, M., Hirose,
A., Hou, Z.-G., and Kil, R. M., editors, Neural Infor-
mation Processing, pages 92–99, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Livi, L., Del Vescovo, G., Rizzi, A., and Frattale Mascioli,
F. M. (2014). Building pattern recognition applica-
tions with the SPARE library. CoRR, abs/1410.5263.
Livi, L. and Rizzi, A. (2013). Graph ambiguity. Fuzzy Sets
and Systems, 221:24–47.
Martino, A., De Santis, E., Giuliani, A., and Rizzi, A.
(2020a). Modelling and recognition of protein contact
networks by multiple kernel learning and dissimilarity
representations. Entropy, 22(7).
Martino, A., Giuliani, A., and Rizzi, A. (2018a). Gran-
ular computing techniques for bioinformatics pat-
tern recognition problems in non-metric spaces. In
Pedrycz, W. and Chen, S.-M., editors, Computational
Intelligence for Pattern Recognition, pages 53–81.
Springer International Publishing, Cham.
Martino, A., Giuliani, A., and Rizzi, A. (2019a). (hy-
per)graph embedding and classification via simplicial
complexes. Algorithms, 12(11).
Martino, A., Giuliani, A., Todde, V., Bizzarri, M., and
Rizzi, A. (2020b). Metabolic networks classification
NCTA 2020 - 12th International Conference on Neural Computation Theory and Applications
348

and knowledge discovery by information granulation.
Computational Biology and Chemistry, 84:107187.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M. (2018b).
Supervised approaches for protein function prediction
by topological data analysis. In 2018 International
Joint Conference on Neural Networks (IJCNN), pages
1–8.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M.
(2019b). Efficient approaches for solving the large-
scale k-medoids problem: Towards structured data.
In Sabourin, C., Merelo, J. J., Madani, K., and War-
wick, K., editors, Computational Intelligence: 9th In-
ternational Joint Conference, IJCCI 2017 Funchal-
Madeira, Portugal, November 1-3, 2017 Revised Se-
lected Papers, pages 199–219. Springer International
Publishing, Cham.
Mizui, Y., Kojima, T., Miyagi, S., and Sakai, O. (2017).
Graphical classification in multi-centrality-index di-
agrams for complex chemical networks. Symmetry,
9(12).
Neumann, M., Garnett, R., Bauckhage, C., and Kersting,
K. (2016). Propagation kernels: efficient graph ker-
nels from propagated information. Machine Learning,
102(2):209–245.
Niepert, M., Ahmed, M., and Kutzkov, K. (2016). Learning
convolutional neural networks for graphs. In Interna-
tional conference on machine learning, pages 2014–
2023.
Pedrycz, W. (2001). Granular computing: an introduction.
In Proceedings Joint 9th IFSA World Congress and
20th NAFIPS International Conference, volume 3,
pages 1349–1354. IEEE.
Pedrycz, W. (2005). Knowledge-based clustering: from
data to information granules. John Wiley & Sons.
Pedrycz, W. (2016). Granular computing: analysis and de-
sign of intelligent systems. CRC press.
Pedrycz, W. and Homenda, W. (2013). Building the
fundamentals of granular computing: A principle
of justifiable granularity. Applied Soft Computing,
13(10):4209 – 4218.
P˛ekalska, E. and Duin, R. P. (2005). The dissimilarity rep-
resentation for pattern recognition: foundations and
applications. World Scientific.
P˛ekalska, E., Duin, R. P., and Paclík, P. (2006). Prototype
selection for dissimilarity-based classifiers. Pattern
Recognition, 39(2):189–208.
Riesen, K. and Bunke, H. (2008). Iam graph database
repository for graph based pattern recognition and ma-
chine learning. In Joint IAPR International Work-
shops on Statistical Techniques in Pattern Recognition
(SPR) and Structural and Syntactic Pattern Recogni-
tion (SSPR), pages 287–297. Springer.
Rizzi, A., Del Vescovo, G., Livi, L., and Frattale Mascioli,
F. M. (2012). A new granular computing approach
for sequences representation and classification. In The
2012 International Joint Conference on Neural Net-
works (IJCNN), pages 1–8.
Rizzi, A., Panella, M., and Frattale Mascioli, F. M. (2002).
Adaptive resolution min-max classifiers. IEEE Trans-
actions on Neural Networks, 13(2):402–414.
Schölkopf, B. and Smola, A. J. (2002). Learning with ker-
nels: support vector machines, regularization, opti-
mization, and beyond. MIT press.
Schölkopf, B., Smola, A. J., Williamson, R. C., and Bartlett,
P. L. (2000). New support vector algorithms. Neural
computation, 12(5):1207–1245.
Shawe-Taylor, J. and Cristianini, N. (2004). Kernel methods
for pattern analysis. Cambridge University Press.
Shervashidze, N. and Borgwardt, K. M. (2009). Fast subtree
kernels on graphs. In Advances in neural information
processing systems, pages 1660–1668.
Shervashidze, N., Schweitzer, P., Van Leeuwen, E. J.,
Mehlhorn, K., and Borgwardt, K. M. (2011).
Weisfeiler-lehman graph kernels. Journal of Machine
Learning Research, 12(9).
Shervashidze, N., Vishwanathan, S. V. N., Petri, T.,
Mehlhorn, K., and Borgwardt, K. M. (2009). Effi-
cient graphlet kernels for large graph comparison. In
van Dyk, D. and Welling, M., editors, Proceedings of
the Twelfth International Conference on Artificial In-
telligence and Statistics, volume 5 of Proceedings of
Machine Learning Research, pages 488–495. PMLR.
Vishwanathan, S. V. N., Schraudolph, N. N., Kondor, R.,
and Borgwardt, K. M. (2010). Graph kernels. Journal
of Machine Learning Research, 11(Apr):1201–1242.
Wu, Z., Pan, S., Chen, F., Long, G., Zhang, C., and Philip,
S. Y. (2020). A comprehensive survey on graph neural
networks. IEEE Transactions on Neural Networks and
Learning Systems.
Xiao, B. and Hancock, E. R. (2005). Graph clustering using
heat content invariants. In Marques, J. S., Pérez de la
Blanca, N., and Pina, P., editors, Pattern Recognition
and Image Analysis, pages 123–130, Berlin, Heidel-
berg. Springer Berlin Heidelberg.
Xiao, B., Hancock, E. R., and Wilson, R. C. (2009). Graph
characteristics from the heat kernel trace. Pattern
Recognition, 42(11):2589–2606.
Ye, C., Wilson, R. C., and Hancock, E. R. (2014). Graph
characterization from entropy component analysis.
In 2014 22nd International Conference on Pattern
Recognition, pages 3845–3850.
Zadeh, L. A. (1979). Fuzzy sets and information granu-
larity. Advances in fuzzy set theory and applications,
11:3–18.
Zadeh, L. A. (1997). Toward a theory of fuzzy information
granulation and its centrality in human reasoning and
fuzzy logic. Fuzzy sets and systems, 90(2):111–127.
Zhang, S., Tong, H., Xu, J., and Maciejewski, R. (2019).
Graph convolutional networks: a comprehensive re-
view. Computational Social Networks, 6(1):11.
Complexity vs. Performance in Granular Embedding Spaces for Graph Classification
349
