
Fast EvaGreen Real-time Duplex PCR for the Individual Detection
of Staphylococcus aureus and Bacillus cereus using a Uniform
Amplification Strategy
Nur Thaqifah Salihah
1
, Mohammad Mosharraf Hossain
2
and Minhaz Uddin Ahmed
1
1
Biosensors and Nanobiotechnology Laboratory, Integrated Science Building, Faculty of Science,
Universiti Brunei Darussalam, Jalan Tungku Link, Gadong BE 1410, Brunei
2
Institute of Forestry and Environmental Sciences, University of Chittagong, Chittagong 4331, Bangladesh
Keywords: Bacillus cereus, DNA, EvaGreen®, Foodborne Pathogens, Multiplex, Real-time PCR, Staphylococcus
aureus.
Abstract: Reliable and sensitive detection of Bacillus cereus (B. cereus) and Staphylococcus aureus (S. aureus) is
needed to limit the outbreak of food poisoning thereof. This paper reports the development of two individual
duplex real-time PCR assays with subsequent melting curve analyses based on EvaGreen
®
dye for dual gene
detections of two bacteria under uniform amplification condition. The duplex assays targeted thermostable
nuclease gene (nuc) and heat-shock protein gene (htrA) of S. aureus and non-haemolytic enterotoxin gene
(nhe) and cereolysin A gene (cerA) for B. cereus detection. The assays successfully detected both the species
with high specificity and sensitivity in genomic DNA samples and in simulated real milk samples. The
selectivity was also confirmed against a wide range of background microflora. Sensitivity of 500 cell/mL and
25 cell/mL of milk was obtained respectively for S. aureus and B. cereus. The proposed methodology allowed
for fast, inexpensive, selective and sensitive multi-targets detections of both bacteria in a single amplification
run on multiple genes to detect S. aureus and B. cereus in milk product by using dsDNA binding EvaGreen
®
dye.
1 INTRODUCTION
S. aureus and B. cereus – prevalent foodborne
pathogens – cause overlapping symptoms of
diarrhoea, vomiting, and abdominal pains (Bennett
and Monday, 2003; Rajkowski and Bennett, 2003)
and are potent to become epidemic. Culture-based
techniques, while inexpensive in confirming the
presence of these pathogens, are labourious and time
consuming – taking up to 5 or 8 days respectively for
S. aureus and B. cereus (Bennett and Lancette, 2012;
Tallent et al., 2012). It may also help pathogens to
reenter the environment because of false negative
results of viable but non-culturable pathogens that
retained their virulence (Gunasekera et al., 2002).
In recent years, alternate bacterial pathogens
detection techniques are developed to overcome the
issues presented by conventional culture-based
method such electrochemical, biosensor and real-time
PCR based detections (Ahmed et al., 2013; Safavieh
et al., 2012; 2013; 2014a; 2014b; Salihah et al., 2018;
2019; Tlili et al., 2013; Tolba et al., 2012). Real-time
PCR-based detection of infectious agents, especially
bacterial enteric pathogens, is becoming popular over
conventional culture-based and even gel-based PCR
techniques due to higher sensitivity, shorter
turnaround time, and enhanced environmental and
analyst safety. Faster detection is crucial to identify
the source to limit the spread of pathogens while
delays in detection may lead to their outbreaks. The
higher sensitivity of PCR-based detection reduces the
need for pre-enrichment or enrichment of bacteria and
limits the possibilities of re-introduction of the
bacterial pathogens into the environment (Martínez-
Blanch et al., 2009; Salihah et al., 2018; 2019).
However, PCR-based detection based on a single
marker gene has limited scope in identifying bacterial
pathogens because of the varying occurrence of genes
in different strains of targeted bacterial species
(Stenfors Aresen et al., 2008; Guinebretiére et al.,
2010) as well as due to the variations or deletions of
a marker gene at primer-binding sites
(Klaassen et al.,
2003), which may produce false-negatives results.
Salihah, N., Hossain, M. and Ahmed, M.
Fast EvaGreen Real-time Duplex PCR for the Individual Detection of Staphylococcus aureus and Bacillus cereus using a Uniform Amplification Strategy.
DOI: 10.5220/0009985100002964
In Proceedings of the 16th ASEAN Food Conference (16th AFC 2019) - Outlook and Opportunities of Food Technology and Culinary for Tourism Industry, pages 197-204
ISBN: 978-989-758-467-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
197

Detecting more than a single gene marker can address
this. However, detection of multiple gene markers
individually requires more PCR consumables,
samples and time. On the other hand, multiplex
reactions allow the amplification of two or more
genes in a single tube with less amount of
consumables and samples. As with any real-time PCR
assays, multiplex real-time PCR reactions utilize
either sequence specific probe-based or the non-
specific dsDNA binding dyes fluorescence detection
chemistries
(Klaassen et al., 2003). While probe-
based chemistry is more specific and allow for
quantitative multiplex analysis, it is also more
expensive and difficult to design
(Salihah et al.,
2016). In contrary, the non-specific dsDNA (double-
stranded DNA) binding dyes offer a cheaper
alternative for multiplex reactions. However, due to
their unspecific affinity to any dsDNA, a post-
amplification melting curve analysis is required to
differentiate different targets’ amplicons which
makes multiplexing possible (Postollec et al., 2011;
Salihah et al., 2016).
This report describes the development of two
multiplex assays targeting two gene markers each for
S. aureus and B. cereus by using the EvaGreen
®
dye
chemistry. EvaGreen
®
dsDNA binding dye was
selected because it produces higher melting curve
resolution and unlike SYBR Green I, it does not bind
preferentially to GC-rich amplicons which can
adversely affect the multiplexing detection (Hu et al.,
2014; Giglio et al., 2003; Eischeid et al., 2011).
However, as a dsDNA binding dye, EvaGreen
®
fluoresce in the presence of any dsDNA, giving off
the same fluorescent signal. Therefore, all the
amplicons of multi-targets real-time PCR
amplification are differentiated by mean of the
amplicons’ unique T
m
. Several studies have proven
that multiplexed real-time PCR with dsDNA binding
dyes are possible by differentiating the amplicons of
the different targets by their T
m
values (Hu et al.,
2014; Safdar et al., 2015) which can be obtained
immediately and automatically after amplification
with zero additional handling with the current real-
time PCR instruments
(Salihah et al., 2016). Since
the post-PCR melting curve analysis further increases
the detection time, the protocol in this study used a
uniform set of conditions to amplify both S. aureus
and B. cereus’ multi-genes multiplex reactions in a
single run to reduce the detection time on both
bacteria. The use of rapid cycle amplification
protocol further reduced the detection time. Hence,
this study successfully developed a fast, sensitive, and
specific real-time multiplex PCR method to amplify
two gene targets for specific detection of S. aureus
and B. cereus with EvaGreen
®
dye chemistry under a
single amplification condition without pre-
enrichment step.
2 MATERIALS AND METHODS
2.1 Genomic DNA of Bacterial Strains
This study used Genomic DNA purchased from
American Type Culture Collection (ATCC,
Manassas, USA) listed in Table 1, both as reference
strains and cross-reactivity analysis. The
concentration and purity of the genomic DNA was
measured by NanoPhotometer
TM
P-Class (Implen,
Munchen, Germany) spectrophotometer by reading
off the absorbance at 260 nm and the absorbance
A
260
/A
280
ratio, respectively. The genomic DNAs
were then diluted with 1 × TE buffer to appropriate
concentrations before use.
2.2 Bacterial Cultivations and Cell
Counting
The S. aureus ATCC 25923 and B. cereus ATCC
14579 live bacterial strains were obtained from
Microbiologics Inc (Minnesota, USA). They were
cultured in LB broth, Miller (Fisher Scientific,
Pittsburgh, USA) at 30
o
C for 48 hours. The total cell
count of the culture was determined with a Neubauer
haemocytometer (Hausser Scientific, Horsham,
USA) before inoculating food products with them and
their subsequent extractions. The culture broth was
concentrated by centrifuging followed by removal of
the supernatant broth and addition of 10 mL of 1 ×
PBS. 1 mL of the cultured broth was heat treated at
100
o
C for 10 minutes for safe handling and counting.
The heat-treated culture was then serially diluted with
1 × PBS buffer and counted with haemocytometer for
at least three times. The non-treated cultured broth
was then diluted with 1 × PBS buffer to appropriate
concentration before used to inoculate real-food
sample.
2.3 Oligonucleotides Design and
Selections
The oligonucleotides designed and selected for this
study are listed on Table 2. The two primer pairs were
selected to target nhe and cerA of B. cereus, and nuc
and htrA genes for S. aureus. To ensure that the
duplex assays for S .aureus and B. cereus are specific,
in-silico
analysis with Primer-Blast (National Centre
16th AFC 2019 - ASEAN Food Conference
198
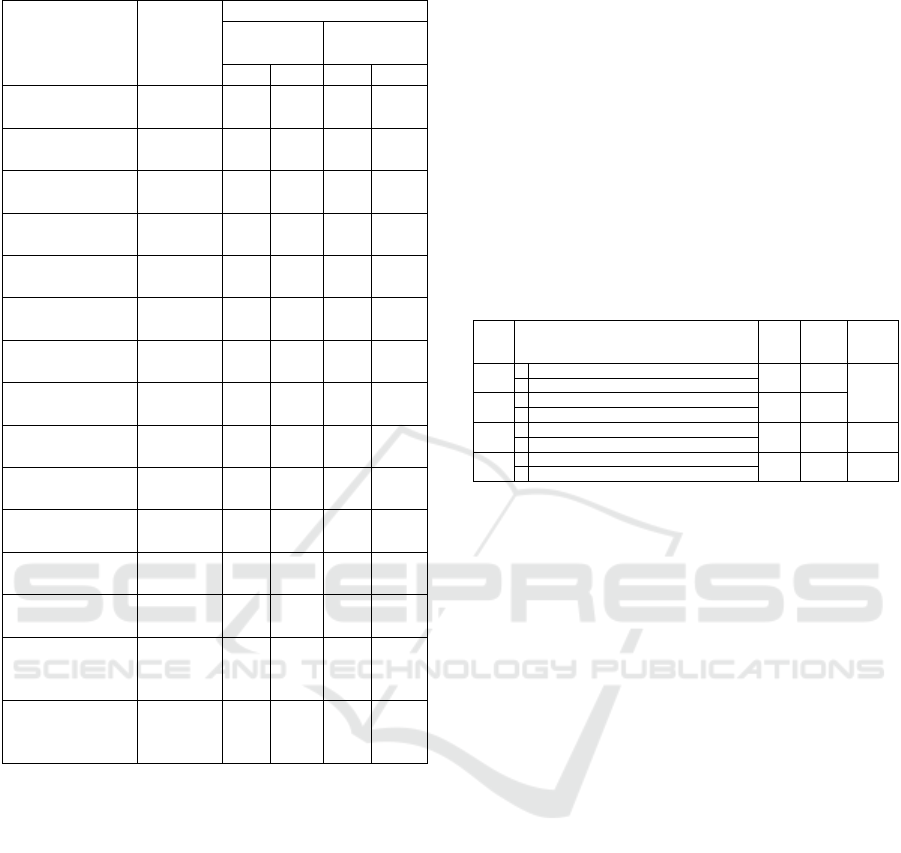
Table 1: Bacterial strains used in this study.
Bacteria Strain
no.
Cross-reactivit
y
anal
y
sis
S. aureus
duplex
B. cereus
duplex
nuc htrA nhe cerA
Staphylococcus
aureus
ATCC
25923
+ + - -
Bacillus cereus ATCC
14579
- - + +
Legionella
pneumophila
ATCC
33152
- - - -
Bacillus
s
ubtilis
ATCC
23857
- - - -
Salmonella
enterica
ATCC
13311
- - - -
Escherichia
coli
ATCC
25922
- - - -
Clostridium
perfringens
ATCC
35401
- - - -
Shigella
f
lexneri
ATCC
13124
- - - -
Campylobacter
j
e
j
uni
ATCC
33292
- - - -
Yersinia
enterocolitoca
ATCC
27739
- - - -
Aeromonas
hydrophila
ATCC
7966
- - - -
Plesiomonas
s
hi
g
elloides
ATCC
51903
- - - -
Streptococcus
pyogens
ATCC
19615
- - - -
Cronobacter
sakazakii
ATCC
BAA-
894
- - - -
Mycobacterium
avium
ATCC
BAA-
968
- - - -
for Biotechnology Information, http://www.ncbi.nlm.
nih.gov/) and OligoAnalyzer Tool (IDT) was carried
out.
The suitability of the assays was first analyzed by
singleplex real-time PCR. Briefly, the assays were
reacted in a 25 µL PCR mixture that contained
Ultrapure MilliQ water, 1 of Buffer II, 250 nM of
both the forward and reverse primers, 1.5 mM MgCl
2
,
0.2 mM of dNTP mix (Invitrogen
™
Lifetechnologies,
Van Allen Way, U.S.A.), 0.1 ROX reference dye
(Invitrogen
™
Life technologies), 1 EvaGreen
®
dye,
0.625U of AmpliTaq DNA polymerase (Applied
Biosystem
™
Life technologies, Van Allen Way,
U.S.A.) and 3 µL of DNA template and were run in
duplicates. The amplifications were carried out on the
7500 Fast real-time PCR system (Applied
Biosystem
™
Lifetechnologies, Van Allen Way,
U.S.A.). Fast cycle amplifications were conducted
with the singleplex analysis with the initial
denaturation at 95
o
C for 20 seconds, and 40 cycles of
denaturation at 95
o
C for 3 seconds followed by
Annealing/extension for 30 seconds at 60
o
C.
A step-hold melting curve analysis was also
performed after amplifications by heating the PCR
mixture at 95
o
C for 15 seconds, and then lowering to
60
o
C for 1 minute. The temperature was then
increased to 95
o
C for 30 seconds and the
fluorescence signal was monitored at this
temperature. The PCR mixtures were then cooled to
60
o
C for 15 seconds.
Table 2: List of primer pairs and probes designed and
selected.
Primer
name
Sequence
(5’-3’)
Product
size
(bp
)
Product
GC (%)
Reference
BCcera F TGGAACTGGAAAGGTACG 200 42.5 This
study
R GTAACACGTTGTGCATC
C
BCnhe F GCATCCAAGAGAGTATGG 186 32.2
R GTTCAGCTTGAATTTCC
C
SAnuc F AATATGGACGTGGCTTAGCG 196 35.7 Salihah et
al., 2019
R TGACCTGAATCAGCGTTGT
C
SAhtra F CGTAAGCGTCGTGAATTCTTC
C
208 30 This
study
R CTTCAGCTTTATTCTCATTAACATCACG
2.4 Development of Duplex Real-time
PCR Assays
The duplex real time PCR reactions were
subsequently reacted in 25 µL of PCR master mix
prepared with Ultrapure MilliQ water containing 1
of Buffer II, 100 nM of each primer pairs for B. cereus
duplex reaction whereas 80 nM and 100 nM for nuc
and htrA primer pairs respectively for S. aureus
duplex reaction, 4 mM MgCl
2
, 0.4 mM of dNTP mix
(Invitrogen
™
Lifetechnologies, Van Allen Way,
U.S.A.), 0.1 ROX reference dye (Invitrogen
™
Life
technologies), 1 EvaGreen
®
dye, 1.25 U of
AmpliTaq DNA polymerase (Applied Biosystem
™
Life technologies, Van Allen Way, U.S.A.) with 6 µL
or 8 µL of DNA template for S. aureus and B. cereus
respectively. The duplex real-time PCR
amplifications were performed on the same 7500 Fast
real-time PCR system (Applied Biosystem
™
Lifetechnologies, Van Allen Way, U.S.A.) in fast
cycle amplification. Step-hold melting curve analysis
was performed after the amplification, as previously
described. All real-time reactions were performed in
either duplicates or triplicates.
2.5 Qualitative Detection in Milk
A 10-fold serial dilution of B. cereus ATCC 14579
and S. aureus ATCC 25923 cultures with 1 PBS
Fast EvaGreen Real-time Duplex PCR for the Individual Detection of Staphylococcus aureus and Bacillus cereus using a Uniform
Amplification Strategy
199
buffer yielded 1 to 1 × 10
3
cells/μL. For B. cereus and
S. aureus detection in real samples, 200 μL milk
samples were artificially contaminated with 1 μL
serial dilutions of B. cereus ATCC 14579 and S.
aureus ATCC 25923 cultures DNA was extracted
from the milk matrix DNeasy Blood and Tissue kit
(Qiagen GmbH, Hilden, Germany) previously
described in Salihah et al. (2019). Genomic DNA
from food matrix was extracted by a combination of
boiling method and DNeasy Blood and Tissue kit
(Qiagen GmbH, Hilden, Germany). The protocol was
modified as follows: 200 μL of the sample was
centrifuged for 30 minutes at 14,000 rpm. The pellet
was washed twice with 500 μL of 1 × TE buffer (pH
8.0) before re-suspending in 200 µL of 1 × TE buffer.
It was then incubated at 99
o
C for 15 minutes before
lysis with 200 µL of AL buffer (containing guanidium
chloride, supplied by the kit) and 25 µL Qiagen
Proteinase K at 70
o
C for 30 minutes. After heating,
200 μL of 99.8 % ethanol (Sigma-Aldrich, Singapore)
was added to the sample and vortexed thoroughly.
The mixture was then pipetted into the DNeasy Mini
spin column (supplied by the kit) with 2 mL
collection tube attached. The column was then
centrifuged at 8,000 rpm for 1 minute, collection tube
and flow-through were then discarded and replaced
with clean new collection tubes (supplied by the kit).
Then the column and centrifuge at 8,000 rpm for 1
minute after addition of 500 μL of AW1 buffer
(containing ethanol and guanidium chloride, supplied
by kit). The liquid in the collection tube was then
discarded. Clean and new collection tube was
attached to the column and 500 μL AW2 buffer
(containing ethanol, provided by the kit) was run
through the column followed by a 3-minute
centrifugation at 14,000 rpm. Collection tube was
discarded and the column was transferred to 1.5 mL
autoclaved and UV irradiated microcentrifuge tube.
The template was eluted once from the column with
40 µL AE buffer (containing 10 mM Tris-Cl and 0.5
mM EDTA, pH 9, supplied by the kit) whereas, for S.
aureus DNA extraction, the template was eluted once
from the column with 60 μL AE buffer. The column
was then incubated at room temperature for 1 minute
before centrifugation at 8,000 rpm for 1 minute
3 RESULTS AND DISCUSSION
3.1 Oligonucleotides Design and
Selections
It is necessary to target for than a single gene when
using PCR-based methods such as real-time PCR.
This is because single gene detections be limiting due
to the varying occurrence of genes in different strains
of the same bacterial species. For example, the nhe
and cerA genes used for B. cereus detection, are
present in 65-75% and 90-95% of B. cereus strains
respectively (De Santis et al., 2008; Martínez-Blanch
et al., 2009). While, nuc gene that encodes the S.
aureus specific thermostable nuclease enzyme which
has been used to confirm the presence of S. aureus in
culture-based detection
3
were found in only 75-78%
of phenotypically positive S. aureus strains in milk
and porcine products (Salem-Bekhit et al., 2010;
Velasco et al., 2018). While htrA gene is consistently
found in all S. aureus strains (Chiang et al., 2007;
Cremonsi et al., 2014), the study on htrA gene
prevalence in S. aureus strains is very limited in
comparison to the more commonly used nuc gene.
Targeting more than a single gene is necessary in
comprehensively identifying S. aureus and B. cereus
with real-time PCR.
The primers sequences designed in Table 2 for S.
aureus’ nuc and htrA genes and B. cereus’ cerA and
nhe genes were analyzed against the sequences in the
Genbank database. They were found to be specific to
only the target bacteria strains.
Figure 1: Melting curve analysis
of singleplex
amplifications of (A) B. cereus’ nhe and cerA genes and (B)
S. aureus’ nuc and htrA genes.
The suitability of the assays were further analyzed
in a singplex reactions to ensure that they would
produce distinctive T
m
s in the melting curve analysis
for the multiplex reactions. The amplicons (for
positive controls) produced single distinguishable
melting peaks for both B. cereus’s nhe and cerA at
74.2 ± 0.151
o
C and 80.0 ± 0.153
o
C respectively
(Figure 1A) and S. aureus’s htrA and nuc genes at
78.2 ± 0.096
o
C and 81.3 ± 0.154
o
C respectively
(Figure 1B), which proved the suitability of dsDNA
binding EvaGreen
®
dye as the detection chemistry for
duplex reactions targeting dual genes of both B.
cereus and S. aureus.
16th AFC 2019 - ASEAN Food Conference
200
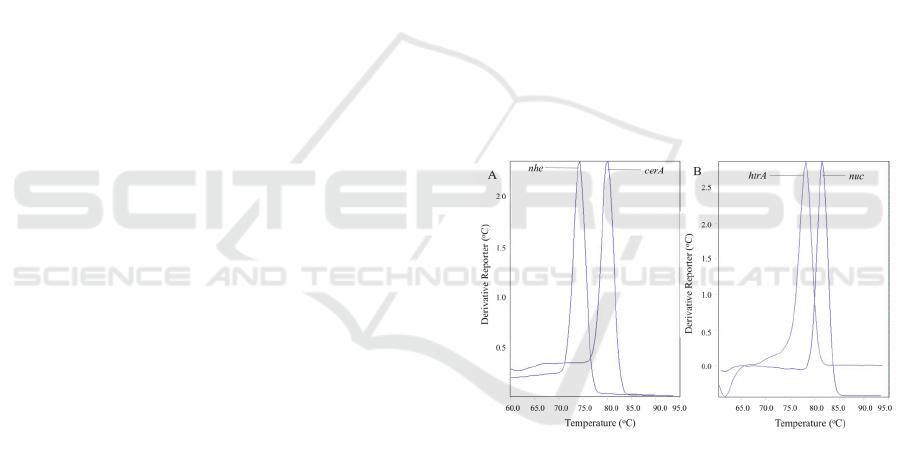
3.2 Development of Duplex Real-time
PCR Assays
Since, dsDNA binding EvaGreen
®
dye was used,
post-PCR melting curve analysis were performed to
ensure that each target in the duplex S. aureus and B.
cereus reactions are distinguishable. Both of the
duplex reactions showed that B. cereus’ nhe (T
m
=
78.7 ± 0.136
o
C) and cerA (T
m
= 83.8 ± 0.0783
o
C) and
S. aureus’ nuc (T
m
= 78.1 ± 0.151
o
C) and htrA (T
m
=
83.8 ± 0.153
o
C) amplification produce distinct and
easily identifiable melting peaks (Figure 2). Each
amplifications produced primer-dimers for the
negative controls of both S. aureus and B. cereus
duplex real-time PCR reactions. However, the
primer-dimer T
m
peaks were lower for both the target
genes and were easily differentiated from the target
amplicons’ T
m
peaks.
So they are suitable for the multiplex reaction with
the EvaGreen
®
dye. The variation in T
m
s
of the
amplicons are dependent on base compositions and to
some extent the length of the amplicons
(Nitsche,
2007). This study found that despite the relatively
same amplicon lengths of both the targets for S.
aureus and B. cereus duplex detections - the
experimented amplicons’ T
m
were distinctively
different. Since the lengths are relatively similar, the
guanine and cytosine nucleobases content of the
amplicons that contributes more to their T
m
difference
(Haynie, 2001). As shown in Table 2, the amplicons
with higher GC content (B. cereus’ BCcerA and S.
aureus’ SAnuc) have higher T
m
values in comparison
to amplicons with lower GC content (B. cereus’
BCnhe and S. aureus’ SAhtrA). The reason for this
correlation is that nucleobases guanine and cytosine
pairs form three hydrogen bonds, which stabilizes the
DNA double-helix structure more than the two
hydrogen bonds formed by nucleobases adenine and
thymine pairs (Marmur and Doty, 1962; Tropp,
2008). Thus, DNA with higher GC content requires
more energy to break the triple hydrogen bonds and
thus have higher T
m
(Marmur and Doty, 1962).
However, the addition of post-PCR melting curve
analysis will add to the detection time. Therefore to
compensate, the proposed duplex assays (S. aureus
duplex and B. cereus duplex) were specifically design
to amplify with the same fast protocol amplification
condition, which takes about approximately 30
minutes before post-PCR melting curve analysis.
Thus, both duplex assays can run at the same time to
reduce the detection time and to streamline the
process for detection of both S. aureus and B. cereus
in a single run.
Figure 2: Melting curve analysis for the assay for fast cycle
amplification of (A) B.cereus, (B) S. aureus.
3.3 Sensitivity and Cross-reactivity
Analysis
The sensitivity of the dual targets individual
detections of S. aureus and B. cereus were analyzed
for fast amplification protocol. The results of the
sensitivity tests are listed in Table 3 for both B. cereus
and S. aureus detection.
Table 3: Sensitivity analysis results for individual duplex
detections of B. cereus and S. aureus.
Bacterial
tar
g
et
Gene LOD
(
f
g
/reaction
)
Probability
(
%
)
B. cereus nhe 6.0×10
1
50.0
cerA 6.0 100.0
S. aureus nuc 3.0×10
2
83.3
htrA 3.0×10
1
50.0
The cross-reactivity of the duplex assays for fast
cycle amplifications were tested against the other
bacterial species listed in Table 1. The post-PCR
melting curve analysis did not show any specific T
m
peaks for bacterial species other than the peaks for
positive controls (B. cereus ATCC 14579 and S.
aureus ATCC 24923). The overall table view of the
cross-reactivity results is listed in Table 1 for B.
cereus and S. aureus multiplex reactions.
Therefore, even though combining the fast
amplification cycle and multiplexing decrease the
sensitivity of the assay, relatively high sensitivities
were still obtained for both S. aureus and B. cereus’
duplex assays. The B. cereus duplex’s limits of
detections (LODs) were 1 cell/reaction and 10
cell/reaction for cerA and nhe genes, respectively.
Whereas for S. aureus duplex the LODs of 10
cell/reaction and 100 cell/reaction for nuc and htrA
genes, respectively. The sensitivity obtained for both
Fast EvaGreen Real-time Duplex PCR for the Individual Detection of Staphylococcus aureus and Bacillus cereus using a Uniform
Amplification Strategy
201

duplex assays are comparable to the sensitivity
obtained for the previous singleplex B. cereus and S.
aureus detection (Salihah et al., 2018; Salihah et al.,
2019).
Furthermore, both the duplex assays are highly
specific to B. cereus and S. aureus, free from cross-
reactivity with other bacterial species in-silico and in-
vitro (Table 1). Thus both duplex is highly specific
and fairly sensitive.
3.4 Qualitative Detection in Milk
The suitability of the proposed dual gene targets
detection of both S. aureus and B. cereus was further
evaluated with simulated milk samples under the fast
cycle amplification condition for simultaneous
detection of S. aureus and B. cereus in a single run. S.
aureus and B. cereus DNAs were directly extracted
from milk samples and were then amplified and
detected by individual duplex assays in separate PCR
tubes and were analyzed together under a single
amplification condition. This allowed simultaneous
detection of dual gene targets of S. aureus and B.
cereus.
The assay detected S. aureus in milk samples
having at least 10 cells/reaction while nuc gene was
targeted while at least 100 cells/reaction was required
when htrA gene was targeted. The sensitivity
obtained for nuc and htrA gene were comparable to
those observed in the sensitivity analysis with pure
genomic DNA dilutions (Table 3). Hence, the
proposed assay claims the capability of detecting S.
aureus with as low as 500 cells/mL of simulated milk
sample. On the other hand, for B. cereus detection
sensitivity of 1 cell/reaction was observed for cerA
gene and 10 cells/reaction for nhe gene. This was
equivalent to 25 cells of B. cereus in 1 mL of
artificially inoculated milk.
This indicated the suitability of the assays to
detect target bacterial pathogens against background
microflora in complex food products such as milk.
The suitability were tested practically by using the no
pre-enrichment and no-expensive enzymes lysis
method of an adapted Qiagen DNeasy blood and
tissue kit previously developed by Salihah et al.
(2019). This further reduced detection time (direct
detection without the need of the additional pre-
enrichment step) and cost (no need to use expensive
enzyme lysis).
4 CONCLUSION
Therefore, we developed a real-time PCR dual gene
B. cereus and S. aureus detections system using a
single set of amplification condition to run two
individual duplex assays with EvaGreen
®
dye. The
assays demonstrated a highly specific and sensitive
detection of both gene targets of each species and
showed highly specific amplification against a large
set of background microflora. Further analysis is
needed to assess the applicability of the proposed
assay against at least five different strains of S. aureus
and B. cereus to validate both assays according to ISO
16140. Since, analysis of the T
m
of the amplicons was
a part of the detection method - the reproducibility
(i.e. inter- and intra-assay) of the amplicons’ T
m
needed to be calculated from a range of B. cereus and
S. aureus strains, as shown by Wehrle and colleagues
(2010). In addition, it might need to demonstrate the
capability of the multiplexed assays in detecting a
wide range of strains, and in case of B. cereus
multiplex assay, an inclusion of other enterotoxigenic
B. cereus strains might be tested. Furthermore, an
inclusion of other target genes for both B. cereus and
S. aureus in the assay might help to find more genetic
indicators of the bacterial pathogens. Primer pairs
could have been designed to target the hbl, cytk1 and
ces genes (Wehrle et al., 2009; Wehrle et al., 2010) to
measure the enteropathogenic potential of B. cereus
strains. For S. aureus multiplex detection, the
inclusion of primer pairs targeting the enterotoxin
gene cluster (egc), enterotoxin genes sea, seb, sec,
sed, see, entC as well as S. aureus specific femA gene
(Tamarapu et al., 2001; Pelisser et al., 2009; Fusco et
al., 2011) could be considered.
Overall, the use of dsDNA binding dyes like
EvaGreen
®
dye in this study provides an advantage
over probe-based chemistry as it is not only easier to
design and cheaper but also free from the limitation
of unavailability of compatible probe-dyes for current
real-time PCR instruments
(Agindotan et al., 2007).
In conclusion, the study claims to develop a highly
specific and sensitive multiplex assay to detect two
target genes of both B. cereus and S. aureus. This
multiplex assay was cost-effective as it used
EvaGreen
®
dyes chemistry and as both multiplex
reactions were run under a single amplification
condition which gave the benefit of streamlining the
detection of B. cereus and S. aureus.
16th AFC 2019 - ASEAN Food Conference
202

ACKNOWLEDGEMENTS
Nur Thaqifah Salihah would like to thank the
Ministry of Education, Brunei Darussalam for the
opportunity given to undertake a Ph.D programme at
Universiti Brunei Darussalam (UBD).
REFERENCES
Agindotan, B.O., Shiel, P.J., Berger, P.H., 2007. Simultaneous
detection of potato viruses PLRV, PVA, PVX and PVY
from dormant potato tubers by Taqman
®
real-time RT-
PCR. Journal of Virological Methods, 142, 1-9.
Ahmed, M.U., Nahar, S., Safavieh, M., Zourob M., 2013. Real-
time electrochemical detection of DNA using electrostatic
interaction of redox probe. Analyst, 138 (3), 907-91.
Applied Biosystem, N.D. Applied Biosystem 7500 Fast and
7500 Real-time systems: Specification Sheet.
Bennett, R.W., Lancette, G.A., 2012. ‘Staphylococcus aureus’
in Lampel, K.A., Al-Khaldi, S., Cahill, S.M. (eds).
Bacteriological Analytical Manual. 8
th
edition. Available
at: https://www.fda.gov/food/laboratory-methods-
food/bam-staphylococcus-aureus . (Accessed date: 10
June 2019).
Bennett, R.W., Monday, S.R., 2003. ‘Staphylococcus aureus’
in Miliotis, M.D., Bier, J.W. (eds). International handbook
of foodborne pathogens, Marcel Dekker. New York.
Cremonesi, P., Pisani, L.F., Lecchi, C., Ceciliani, F., Martino,
P., Bonastre, A.S., Karus, A., Balzaretti, C., Castiglioni, B.,
2014. Development of 23 individual TaqMan
®
real-time
PCR assays for identifying common foodborne pathogens
using single set of amplification conditions. Food
Microbiology, 43, 35-40.
Chiang, Y., Fan, C., Liao, W., Lin, C., Tsen, H., 2007. Real-
time PCR detection of Staphylococcus aureus in milk and
meat using new primers designed from the heat shock
protein gene htrA sequence. Journal of Food Protection,
70(12), 2855-2859.
De Santis, E.P.I., Foddai, A., Virdis, S., Marongiu, P., Pilo,
A.L., Scarano, C., 2008. Toxin gene pattern in Bacillus
cereus group strains isolated from sheep ricotta cheese.
Veterinary Research Communications,32, S323-S326.
Eischeid, A.C., 2011. SYTO dyes and EvaGreen outperform
SYBR Green in real-time PCR. BMC Res Notes, 4, 263.
Fusco, V., Quero, G.M., Morea, M., Blaiotta, G., Visconti, A.,
2011. Rapid and reliable identification of Staphylococcus
aureus harbouring the enterotoxin gene cluster (egc) and
quantitative detection in raw milk by real-time PCR.
International Journal of Food Microbiology, 144(3), 528-
537.
Giglio, S., Monis, P.T., Saint, C.P., 2003. Demonstration of
preferential binding of SYBR Green I to specific DNA
fragments in real-time multiplex PCR. Nucleic Acids
Research, 31(22), e136.
Guinebretiére, M.H., Velge, P., Couvert, O., Carlin, F.,
Debuyser, M.L., Nguyen-The, C., 2010. Ability of
Bacillus cereus group strains to cause food poisoning
varies according to phylogenetic affiliation (group I to VII)
rather than species affiliation. Journal of Clinical
Microbiology, 48(9), 3388-3391.
Gunasekera, T.S., Sørensen, A., Attfield, P.V., Sørensen, S.J.,
Veal, D.A., 2002. Inducible gene expression by
nonculturable bacteria in milk after pasteurization. Applied
and Environmental Microbiology, 68, 1988-1993.
Haynie, D.T., 2001. Biological thermodynamics, Cambridge
University Press, Cambridge.
Hu, Z., Zhu, C., Chang, H., Guo, W., Liu, D., Xiang, W.,
Wang, X., 2014. Development of a single-tube duplex
EvaGreen real-time PCR for the detection and
identification of EHV-1 and EHV-4. Applied Microbiology
and Biotechnology, 98(9), 4179-4186.
Klaassen, C.H.W., de Valk, H.A., Horrevorts, A., 2003.
Clinical Staphylococcus aureus isolate negative for Sa442
fragment. Journal of Clinical Microbiology, 41(9), 4493.
Martínez-Blanch, J.F., Sánchez, G., Garay, E., Aznar, R., 2009.
Development of a real-time PCR assay for detection and
quantification of enterotoxigenic members of Bacillus
cereus group in food samples. International Journal of
Food Microbiology, 135, 15-21.
Marmur, J., Doty, P., 1962. Determination of the base
composition of deoxyribonucleic acid from its thermal
denaturation temperature. Journal of Molecular Biology,
5, 109-118.
Nitsche, A., 2007. ‘Oligonucleotide design for in-house real-
time PCR applications in microbiology’ in Mackay, I.M.
(ed). Real-time PCR in microbiology: From diagnostic to
characterization, Caister Academic Press. Norfolk.
Pelisser, M.R., Klein, C.S., Ascoli, K.R., Zotti, T.R., Arisi,
A.C.M., 2009. Occurrence of Staphylococcus aureus and
multiplex PCR detection of classic enterotoxin genes in
cheese and meat products. Brazilian Journal of
Microbiology, 40(1), 145-148.
Postollec, F., Falentin, H., Pavan, S., Combrisson, J., Sohier,
D., 2011. Recent advances in quantitative PCR (qPCR)
applications in food microbiology. Food Microbiology,
28(5), 848-861.
Rajkowski, K.T., Bennett, R.W., 2003. ‘Bacillus cereus’ in
Miliotis M.D., Bier J.W. (eds). International handbook of
foodborne pathogens, Marcel Dekker. New York.
Safavieh, M., Ahmed, M.U., Sokullu, E., Ng, A., Braescu, L.,
Zourob, M., 2014. A Simple Cassette as Point-of-Care
Diagnostic Device for Colorimetric Bacteria Detection.
Analyst, 139, 482-487.
Safavieh, M., Ahmed, M.U., Zourob, M., 2012.
Electrochemical assay in microfluidic for rapid detection
and quantification of Escherichia coli. Biosensors and
Bioelectronics, 31(1), 523-528.
Safavieh, M., Ahmed, M.U., Zourob, M., 2013. High
throughput low cost electrochemical device for S. aureus
bacteria detection. IEEE Sensors,1-4.
Safavieh, M., Ahmed, M.U., Zourob, M., 2014. High
throughput Real time Electrochemical monitoring of
LAMP Using a Redox for Pathogenic Bacteria Detection.
Biosensors and Bioelectronics, 15(58), 101-106.
Safdar, M., Junejo, Y., 2015. Development and validation of
fast duplex real-time PCR assays based on SYBR Green
Fast EvaGreen Real-time Duplex PCR for the Individual Detection of Staphylococcus aureus and Bacillus cereus using a Uniform
Amplification Strategy
203

florescence for detection of bovine and poultry origins in
feedstuffs. Food Chemistry, 173, 660-664.
Salem-Bekhit, M.M., Muharram, M.M., Alhosiny, I.M.,
Hashim, M.E.S.Y., 2010. Molecular detection of genes
encoding virulence determinants in Staphylococcus aureus
strains isolated from bovine mastitis. Journal of Applied
Science Research, 6(2), 121-128.
Salihah, N.T., Hossain, M.M., Abdul Hamid, M.R.W., Ahmed,
M.U., 2018. A comparison of ZEN double-quenched
probe and SYBR GreeER chemistries in the real-time PCR
based quantitative detection of enterotoxigenic Bacillus
cereus in milk. Malaysian Journal of Microbiology, 14(1),
34-40.
Salihah, N.T., Hossain, M.M., Abdul Hamid, M.R.W., Ahmed,
M.U., 2019. A novel, rapid, and sensitive real-time PCR
assay for cost-effective detection and quantification of
Staphylococcus aureus in food samples with ZEN™
double-quenched probe chemistry. International Foor
Research Journal, 26(1), 193-201.
Salihah, N.T., Hossain, M.M., Lubis, H., Ahmed, M.U., 2016.
Trends and Advances in Food Analysis Using Real Time
Polymerase Chain Reaction. Journal of Food Science and
Technology, 53(5), 2196–2209.
Stenfors Arnesen, L.P., Fagerlund, A., Granum, P.E., 2008.
From soil to gut: Bacillus cereus and its food poisoning
toxins. FEMS Microbiology Reviews, 32(4), 579-606.
Tallent, S.M., Rhodehamel, E.J., Harmon, S.M., Bennett,
R.W., ‘Bacillus cereus’ in Lampel, K.A., Al-Khaldi, S.,
Cahill, S.M. (eds). Bacteriological Analytical Manual. 8
th
edition. Available at:
https://www.fda.gov/food/laboratory-methods-food/bam-
staphylococcus-aureus . (Accessed date: 10 June 2019).
Tamarapu, S., McKillip, J.L., Drake, M., 2001. Development
of a Multiplex Polymerase Chain Reaction assay for
detection and differentiation of Staphylococcus aureus in
dairy products. Journal of Food Protection, 64(5), 664-
668.
Tlili, C., Sokullu, E., Safavieh, M., Tolba, M., Ahmed, M.U.,
Zourob, M., 2013. Bacteria Screening, Viability, And
Confirmation Assays Using Bacteriophage-
Impedimetric/Loop-Mediated Isothermal Amplification
Dual-Response Biosensors. Analytical Chemistry, 85 (10),
4893–4901.
Tolba, M., Ahmed, M.U., Tlili, C., Eichenseher, F., Loessner,
M.J., Zourob, M., 2012. Bacteriophage endolysin-based
electrochemical impedance biosensor for the rapid
detection of Listeria cells. Analys, 137, 5749-5756.
Tropp, B.E., 2008. Molecular biology: Genes to proteins,
Jones and Bartlett publisher. Massachusetts.
Velasco, V., Vergara, J.L., Bonilla, A.M., Muñoz, J., Mallea,
A., Vallejos, D., Quezada-Aguiluz, M., Campos, J., Rojas-
García, P., 2018. Prevalence and Characterization of
Stapylococcuss aureus Strains in the Pork Chain Supply in
Chile. Foodborne Pathogens Diseases, 15(5), 262-268.
Wehrle, E., Didier, A., Moravek, M., Dietrich, R., Märtlbauer,
E., 2010. Detection of Bacillus cereus with
enteropathogenic potential by multiplex real-time PCR
based on SYBR Green I. Molecular Cell Probes, 24(3),
124-130.
Wehrle, E., Moravek, M., Dietrich, R., Bürk, C., Didier, A.,
Märtlbauer, E., 2009. Comparison of multiplex PCR,
enzyme immunoassay and cell culture methods for the
detection of enterotoxigenic Bacillus cereus. Journal
Microbiological Methods, 78(3), 265-270.
16th AFC 2019 - ASEAN Food Conference
204
