
Multi-dataset Training for Medical Image Segmentation as a Service
Javier Civit-Masot
1
, Francisco Luna-Perejón
2
Lourdes Duran-Lopez
2
, J. P. Domínguez-Morales
2
,
Saturnino Vicente-Díaz
2
, Alejandro Linares-Barranco
2
and Anton Civit
2
1
COBER S.L., Avenida Reina Mercedes, s/n, 41012, Seville, Spain
2
School of Computer Engineering, Avenida Reina Mercedes, s/n, 41012, Seville, Spain
Keywords: Deep Learning, Segmentation as a Service, U-Net, Optic Disc and Cup, Glaucoma.
Abstract: Deep Learning tools are widely used for medical image segmentation. The results produced by these
techniques depend to a great extent on the data sets used to train the used network. Nowadays many cloud
service providers offer the required resources to train networks and deploy deep learning networks. This
makes the idea of segmentation as a cloud-based service attractive. In this paper we study the possibility of
training, a generalized configurable, Keras U-Net to test the feasibility of training with images acquired, with
specific instruments, to perform predictions on data from other instruments. We use, as our application
example, the segmentation of Optic Disc and Cup which can be applied to glaucoma detection. We use two
publicly available data sets (RIM-One V3 and DRISHTI) to train either independently or combining their
data.
1 INTRODUCTION
1.1 Cloud based Segmentation
Segmentation is the process of automatic detection of
limits within an image. In medical images we find a
high variability both in the data sources and capture
technologies used (X-ray, CT, MRI, PET, SPECT,
endoscopy, etc.). Human anatomy also shows very
significant variations.
Deep Learning methods are being increasingly
used to process medical images (Litjens et al., 2017).
The effectiveness of these systems is conditioned by
the number and variety of the training images. If we
want to implement cloud-based services, they will
have to be trained with new data set samples
periodically. These images will most probably come
from different sources and, thus, we need to answer
some significant questions: Should we train the
networks specifically for images acquired with each
of the available instruments? Is it possible to train a
network with data from one instrument and make
predictions for other different instruments? What
happens if we train with combined data? It would be
very difficult to implement a reliable image
segmentation service without knowing the answer to
these questions.
Several segmentation researchers (Sevastopolsky,
2017) (Al-Bander et al., 2018) have used several
different data sets for their works, however, they
always train and test with each of these data sets
independently. In this paper we propose to compare
this traditional method with a new approach where we
preprocess and mix the data from several datasets and
use it to create independent data sets for training and
validation.
In this work we will use a generalized U-Net
architecture as our training network, and study, as our
example problem, the detection of the optical disc and
cup in fundus images. However, the same techniques
can be applied almost directly to the segmentation of
2D images in industrial applications, automatic
driving, detection of people, etc.
1.2 Convolution / Deconvolution
Networks
We will use a generalized U-Net (Ronneberger,
Fischer, & Brox, 2015) as our example network as it
is one of the most commonly used fully convolutional
network (FCN) families for the segmentation of
biomedical images.
The basic architecture of our network is shown in
Fig. 1. The network consists of descending layers
formed by two convolution layers with RELU
542
Civit-Masot, J., Luna-Perejón, F., Duran-Lopez, L., Domínguez-Morales, J., Vicente-Díaz, S., Linares-Barranco, A. and Civit, A.
Multi-dataset Training for Medical Image Segmentation as a Service.
DOI: 10.5220/0008541905420547
In Proceedings of the 11th International Joint Conference on Computational Intelligence (IJCCI 2019), pages 542-547
ISBN: 978-989-758-384-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
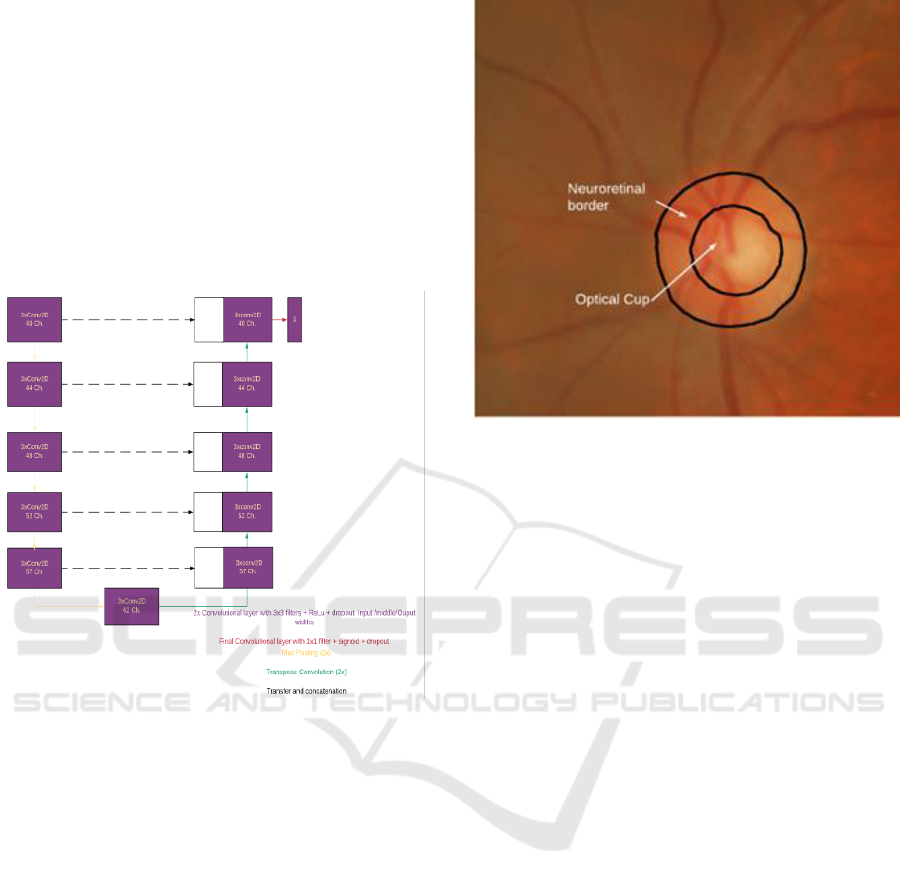
activation and dropout. The result of each layer is
sub-sampled using a 2x2 max pool layer and used as
input to the next layer. The 6th layer corresponds to
the lowest level of the network and has a structure like
the other descending layers. From this layer the data
is oversampled by transposed convolution, merged
with the output data of the corresponding downwards
layer and applied to a block similar to those used in
the descending layers. The last layer of the network is
a convolution layer with a width equal to the number
of classes to be segmented, which is just one in our
case.
Figure 1: Proposed generalized U-Net Architecture.
We choose this specific architecture for our test
as it has a moderate number of training parameters
(near 1M), which allows us to train it using free GPU
resources in the cloud and, when training with a
single data set, produces results that are very similar
to those obtained by other researchers.
1.3 Optical Disc and Cup
Glaucoma is a set of diseases that cause damage to the
optic nerve in the back of the eye and can cause loss
of vision. Glaucoma is one of the main causes of
blindness and is estimated that it will affect around 80
million people worldwide by 2020.
Only when the disease progresses, with a
significant loss of peripheral vision, the symptoms
that may lead to total blindness begin to be noticed.
Early detection is, thus, essential.
Many risk factors are associated with glaucoma
but intraocular hypertension (IH) is the most widely
accepted.
Figure 2: Neuroretinal border and Cup.
IH can cause irreversible damage to the optic
nerve or optic disc (OD). The OD is the beginning of
the optic nerve and is the point where the axons of
retinal ganglion cells come together. It is also the
entry point for the major blood vessels that supply the
retina and it corresponds to a small blind spot in the
retina. The optic disc can be visualized by various
techniques such as color fundus photography. The
OD is divided into two regions as shown in Fig. 3: a
peripheral zone called the neuroretinal border and a
white central region called the optic cup (OC).
Glaucoma produces pathological cupping of the
optic disc. As glaucoma advances, the cup enlarges
until it occupies most of the disc area. The ratio of the
diameter of OC to OD is known as CDR and is a well-
established indicator for the diagnosis of glaucoma
[9]. Therefore, the correct determination of this
diameters is key to the correct calculation of the CDR.
Human segmentation of OD and OC is a slow and
error prone process. Thus, automated segmentation is
attractive as, in many cases, it can be more objective
and faster than humans.
Several approaches have been proposed for
fundus image OD/OC segmentation. The existing
methods for automated OD and OC segmentation in
background images can be classified into three main
categories (Thakur & Juneja, 2018): templates based
on form matching and traditional machine learning
based on random forests, support vector machines, K-
means, etc. (e.g. (Kim, Cho, & Oh, 2017)), active
contours and deformable models (e.g. (Mary et al.,
2015)), and more recently, deep learning-based
methods (e.g. (Zilly, Buhmann, & Mahapatra,
2017),(Al-Bander et al., 2018)).
Multi-dataset Training for Medical Image Segmentation as a Service
543
The aim of this paper is to study the influence of
the dataset selection on the results. We will use a
segmentation approach based on (Sevastopolsky,
2017) but with significant modifications to make it
flexible and suitable for cloud-based implementation.
2 MATERIALS AND METHODS
For this work we used the Google Collaboratory
iPython development environment. The environment
has very good support for Keras for implementing
and training networks on GPUs in Google cloud. Our
network is based on (Sevastopolsky, 2017) but with
very significant modifications:
- We use a different dual image generator and use
it for both training and testing.
Figure 3: Disc images from RIM and DRISHTI datasets.
- We use a parameterizable recursive U-net model
which allows us to easily change many parameters
necessary to compare different implementations of U-
Net.
- We use 120 image batches for both training and
testing and train for 15 epochs using 150 training
steps and 30 testing steps per epoch. We use an Adam
optimizer algorithm in most cases with a 0.00075
learning rate. These values have proven suitable for
training in U-Net architectures and provide good
results with reasonable training times.
- We have tested several generalized U-Net
configurations and finally decided to use the
lightweight configuration shown in Fig.1
Regarding the datasets we use publicly available
RIM-ONE v3 and DRISHTI datasets. RIM ONE-v3
(Fumero, Alayón, Sanchez, Sigut, & Gonzalez-
Hernandez, 2011), from the MIAG group of the
University of La Laguna (Spain), consists of 159
fundus images which have been labelled by expert
ophthalmologists for both disc and cup. DRISHTI-
GS (Sivaswamy, Krishnadas, Joshi, Jain, & Tabish,
2014), from Aravind Eye hospital, Madurai, India
consists of 101 fundus images also labelled for disc
and cup.
The code we use for both OD and OC
segmentation is the same and the only difference is
the loading and pre-processing of images and masks.
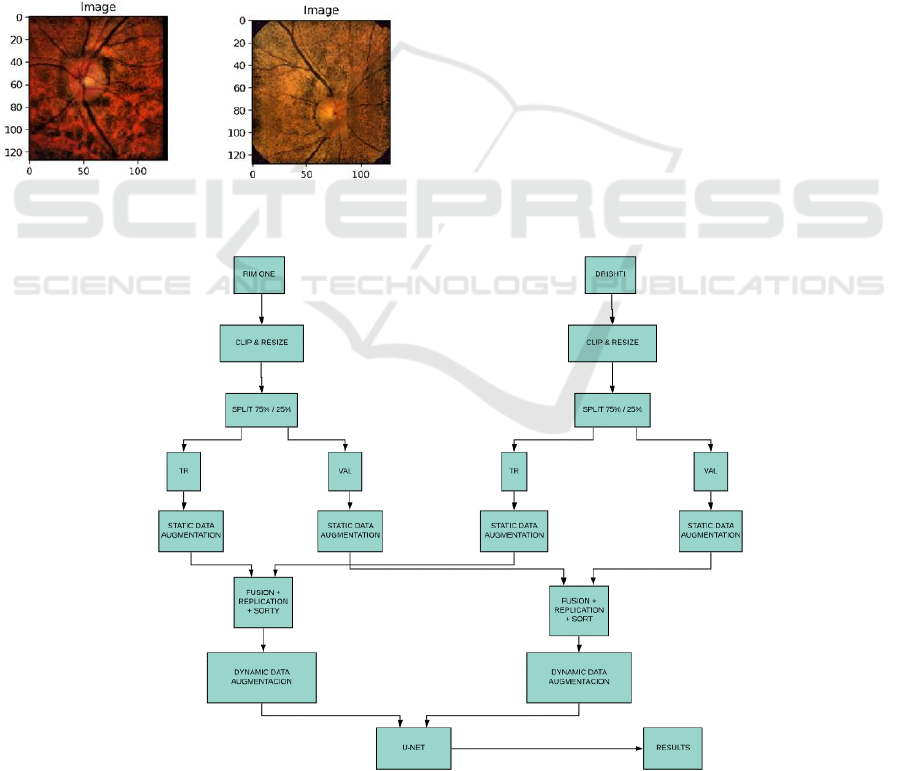
Figure 4: Multi-dataset-based training approach. For single dataset fusion step is not needed.
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
544
As already mentioned, our final objective is to
perform disc and cup detection as a service in the
cloud and, for this purpose, it is necessary that we are
independent, as much as possible, from the specific
characteristics of the captured image. As an example,
in Fig. 3 we can see that images coming from the three
different datasets have very different characteristics.
Our approaches for disc and cup segmentation are
very similar. Fig. 5 shows the methodology used for
cup segmentation when using a mixed dataset for
training and validation (Zoph et al., 2019). When we
train with either RIM-ONE or DRISHTI we use the
same approach without the fusion step.
Originally, we start by clipping and resizing the
original images in the datasets. When we segment the
disc, we remove a 10% border in all the edges of the
image to reduce black borders in the images. When
we segment the cup, we select the area that contains
the disc plus an additional 10% from the original
images. After clipping we resize the images to
128x128 pixels and perform a clip limited contrast
equalization.
After the equalization we do data set splitting. For
each dataset we use 75% of the images for training
and 25% for validation. It is essential to split the
datasets before performing any data augmentation to
ensure that the training and validation sets are
completely independent from each other. After
splitting we perform, for each used dataset, static data
augmentation by creating images with modified
brightness and different adaptive contrast parameters.
When we train with a mixed dataset after the static
data augmentation, we do the fusion of the data from
the different datasets. This process is done
independently for the training and validation dataset.
In the fusion process we perform data replication and
shuffling so that we provide longer vectors as input
for our dynamic image generators. The image
generators do data augmentation by performing
random rotations, shifting, zooming and flipping on
the extended fused dataset images.
As one of the main glaucoma indicators is the
CDR, i.e. the relation between the OD and the OC
diameters we introduce a new parameter RRP -Radii
Ratio parameter- which is the relation between the
radius of the predicted segmented disc and the radius
of the correct disc. We estimate the radii as the square
root of the segmented area divided by pi.
Apart from the mean Dice coefficient over the
validation data set we use an additional quality
parameter that is the percentage of the images where
the estimated radius error is less than 10%.
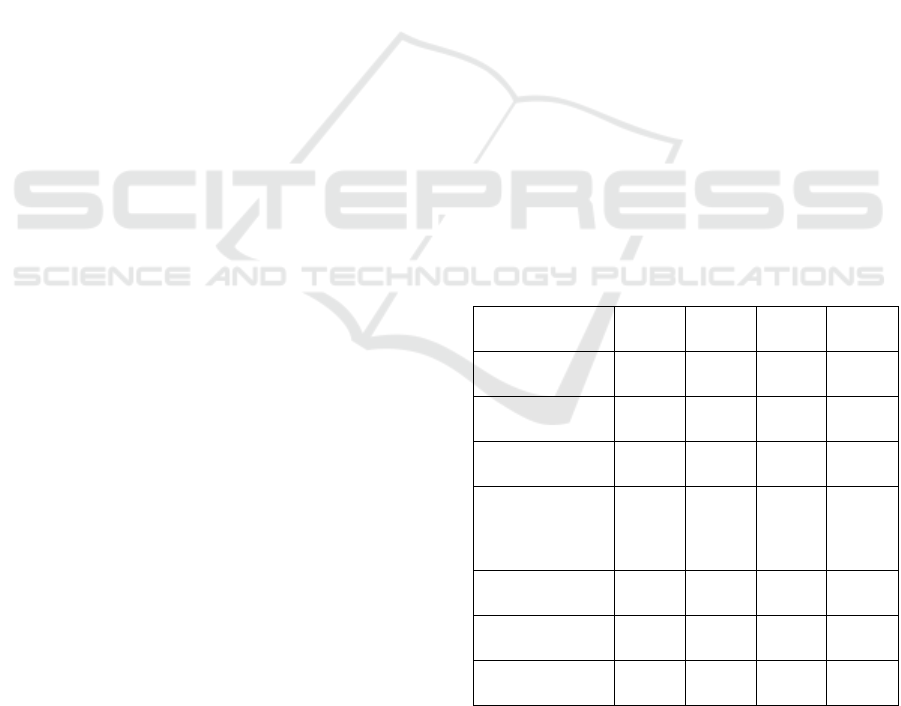
3 RESULTS
In Table I we show the Dice coefficients for the Disc
and Cup segmentation for three different training
scenarios:
- We train using 75% of the DRISHTI dataset and we
validate with the remaining DRISHTI and with the
RIM ONE validation data set.
- We train using 75% of RIM ONE the dataset and we
validate with the remaining RIM ONE and with the
DRISHTI validation data set.
- We train with 75% of a combined data set and
validate with the rest of the combined data set.
We can see in the table that when training with
DRISHTI we get very reasonable results when testing
with images from the same dataset with a Dice
coefficient above 0.98 for both OD and OC
segmentation. However, if we validate this network
with the RIM ONE data set result fall below 0.50.
A very similar situation happens if we train with
the RIM ONE data sets. If we validate with the RIM
ONE test data, we get Dice coefficients that are above
0.96 but this value falls below 0.66 when we test with
DRISHTI data.
If we train with a combined data set, we get results
that are more stable when testing with both datasets.
For OD segmentation we get a 0.96 Dice value for
DRISHTI and a 0.87 for RIM ONE. In the case of OC
segmentation these values fall to 0.94 and 0.82.
Table 1: Dice coefficient for OC and OD.
Author
Disc
DRI
Disc
RIM
Cup
DRI
Cup
RIM
(Zilly et al., 2017)
0.97
-
0.87
-
(Zilly et al., 2017)
0.95
0.90
0.83
0.69
(Sevastopolsky,
2017)
-
0.94
-
0.82
(Shankaranarayan
a, Ram, Mitra, &
Sivaprakasam,
2017)
-
0.98
-
0.94
Drishti Trained
0.98
0.50
0.98
0.42
RIM Trained
0.66
0.97
0.61
0.96
Multi-dataset
0.96
0.87
0.94
0.82
In table I we have also included results from other
papers that have studied the OD/OC segmentation
problem using Deep Learning based approaches and
training with, at least, one of the datasets that we use.
Multi-dataset Training for Medical Image Segmentation as a Service
545

In all these cases the researchers have trained and
tested independently with the different datasets.
Even though our network is very light when we
train with a single dataset, we get similar results to
those obtained by other researchers. For DRISHTI
dataset training we obtained a Dice coefficient of 0.98
for both OD and OC segmentation. This compares
favourably with 0.97 and 0.87 (Zilly et al., 2017).
When training with RIM ONE we obtain 0.97 for OD
and 0.96 for OC. This also compares well with 0.98
and 0.94 (Shankaranarayana et al., 2017).
The most important result from table I comes
from the data that is not available in other studies, i.e.,
when we train with a dataset and use the network with
data captured with another source, we get poor
prediction result.
Table I also shows that when we train with a
combined dataset the network performs well doing
predictions from both datasets.
In Table II we show the percentage of the
predictions that estimate the radius with an error
below 10%. This data is clinically very relevant as the
ratio between the cup and disc radii, i.e., the CDR, is
directly related to glaucoma.
When we train with a specific dataset, almost all
the radii for the testing data from the same dataset are
predicted with less than 10% error. However, the radii
prediction for the other dataset are much worse and,
in some case, we never get errors below 10%. As can
be seem in the table this situation improves very
significatively when we train with a mixed dataset.
Table 2: Images with less than 10% radius error.
Disc
DRI
Disc
RIM
Cup DRI
Cup
RIM
Drishti
Trained
100
38
100
0
RIM
ONE
Trained
62
100
0
95
Multi-
dataset
100
82
100
54
4 CONCLUSIONS AND FUTURE
WORK
We have been able to show that by using data from
different data sets, doing adequate image pre-
processing and performing very significant data
augmentation, both statically and dynamically, we
have been able to perform cup and disc segmentation
getting results with a performance that is equivalent
to that obtained by other authors using a single dataset
for evaluation and testing. This is, at least, a first
approach at the possibility of running this type of
segmentations as a service on the cloud.
We have also introduced a new clinically
significant parameter (Radii Ratio parameter- RRP)
that is very useful to estimate the accuracy of the
CDR.
We have shown that a very deep lightweight U-
Net derivative can perform as well as other heavier
less deeper alternatives for OD/OC segmentation.
This work has shown the advantages of using a
dataset that combines data from different sources
using aggressive data augmentation. Much work is
necessary to improve the commercial viability of this
type of service. In this work we have trained with a
mixed dataset but, in real life, we would have to start
training with the available data and do retraining as
more and more image data from different sources
becomes available. It would be necessary to
adequately study the behaviour of this type of trained
network with existing and new datasets.
ACKNOWLEDGEMENTS
This work was partially supported by the NPP project
funded by SAIT (2015-2018) and by the Spanish
government grant (with support from the European
Regional Development Fund) COFNET (TEC2016-77785-
P). Development in Cloud environment was supported by
Google Cloud platform research credit program
REFERENCES
Al-Bander, B., Williams, B., Al-Nuaimy, W., Al-Taee, M.,
Pratt, H., & Zheng, Y. (2018). Dense fully
convolutional segmentation of the optic disc and cup in
colour fundus for glaucoma diagnosis. Symmetry,
10(4), 87.
Fumero, F., Alayón, S., Sanchez, J. L., Sigut, J., &
Gonzalez-Hernandez, M. (2011). RIM-ONE: An open
retinal image database for optic nerve evaluation.
Paper presented at the 2011 24th international
symposium on computer-based medical systems
(CBMS).
Kim, S. J., Cho, K. J., & Oh, S. (2017). Development of
machine learning models for diagnosis of glaucoma.
PLoS One, 12(5), e0177726.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., . . . Sánchez, C. I. (2017).
A survey on deep learning in medical image analysis.
Medical image analysis, 42, 60-88.
Mary, M. C. V. S., Rajsingh, E. B., Jacob, J. K. K.,
Anandhi, D., Amato, U., & Selvan, S. E. (2015). An
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
546

empirical study on optic disc segmentation using an
active contour model. Biomedical Signal Processing
and Control, 18, 19-29.
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-net:
Convolutional networks for biomedical image
segmentation. Paper presented at the International
Conference on Medical image computing and
computer-assisted intervention.
Sevastopolsky, A. (2017). Optic disc and cup segmentation
methods for glaucoma detection with modification of
U-Net convolutional neural network. Pattern
Recognition and Image Analysis, 27(3), 618-624.
Shankaranarayana, S. M., Ram, K., Mitra, K., &
Sivaprakasam, M. (2017). Joint Optic Disc and Cup
Segmentation Using Fully Convolutional and
Adversarial Networks, Cham.
Sivaswamy, J., Krishnadas, S., Joshi, G. D., Jain, M., &
Tabish, A. U. S. (2014). Drishti-gs: Retinal image
dataset for optic nerve head (onh) segmentation. Paper
presented at the 2014 IEEE 11th international
symposium on biomedical imaging (ISBI).
Thakur, N., & Juneja, M. (2018). Survey on segmentation
and classification approaches of optic cup and optic disc
for diagnosis of glaucoma. Biomedical Signal
Processing and Control, 42, 162-189.
Zilly, J., Buhmann, J. M., & Mahapatra, D. (2017).
Glaucoma detection using entropy sampling and
ensemble learning for automatic optic cup and disc
segmentation. Computerized Medical Imaging and
Graphics, 55, 28-41.
Zoph, B., Cubuk, E. D., Ghiasi, G., Lin, T.-Y., Shlens, J.,
& Le, Q. V. (2019). Learning Data Augmentation
Strategies for Object Detection. arXiv preprint
arXiv:1906.11172.
Multi-dataset Training for Medical Image Segmentation as a Service
547
