
Topological Approach for Finding Nearest Neighbor Sequence in Time
Series
Paolo Avogadro
a
and Matteo Alessandro Dominoni
b
Universit
`
a Degli Studi di Milano-Bicocca, Viale Sarca 336/14, 20126, Milano, Italy
Keywords:
Time Series, Anomaly, Discord, Nearest Neighbor Distance.
Abstract:
The aim of this work is to obtain a good quality approximation of the nearest neighbor distance (nnd) pro-
file among sequences of a time series. The knowledge of the nearest neighbor distance of all the sequences
provides useful information regarding, for example, anomalies and clusters of a time series, however the com-
plexity of this task grows quadratically with the number of sequences, thus limiting its possible application.
We propose here an approximate method which allows one to obtain good quality nnd profiles faster (1-2
orders of magnitude) than the brute force approach and which exploits the interdependence of three different
topologies of a time series, one induced by the SAX clustering procedure, one induced by the position in time
of each sequence and one by the Euclidean distance. The quality of the approximation has been evaluated
with real life time series, where more than 98% of the nnd values obtained with our approach are exact and
the average relative error for the approximated ones is usually below 10%.
1 INTRODUCTION AND
RELATED WORKS
The large amount of data produced by sensors im-
plies that human analysis of time series needs to be
supported by machine learning techniques. One of
the first problems encountered at the time of compar-
ing sequences within a time series is that their length
can span few hundreds of points, and for this rea-
son some form of dimensionality reduction becomes
propaedeutic for further investigations. From this
point of view the symbolic aggregate approximation
(SAX) algorithm (Lin et al., 2003) has proven to be
very effective, as it scales linearly with the size of
the time series, and provides efficient clustering. For
these reasons it has been used as the basis of a large
number of works on the filed (Keogh, 2019). During
the analysis of a time series one often looks for se-
quences which carry particular significance. For ex-
ample, anomaly search in time series is a particularly
active research field (Chandola et al., 2009), among
the many anomaly concepts, the idea of discords and
a pioneering method for finding them was introduced
by (Keogh et al., 2005). One of the limitations of dis-
cord search is that the length of the sequences is an
a
https://orcid.org/0000-0001-5170-4479
b
https://orcid.org/0000-0001-8481-6311
input parameter, however a priori a researcher does
not know the length of an anomaly. In order to over-
come this problem it has been proposed to use the
Kolmogorov complexity of the symbolic sequences
obtained with the SAX procedure to define a new con-
cept of anomaly called RRA which produces similar
results compared to discord search (Senin et al., 2014)
(Senin et al., 2018) and it also has the advantage of
being much faster than HOT SAX. At the other side
of the spectrum one might be interested in finding re-
peated patterns in a time series, for example in the
form of motifs (Chiu et al., 2003) (Patel et al., 2002)
(Lin et al., 2002).
Many of the indicators which allow to character-
ize a time series are obtained by calculating the Eu-
clidean distance between the sequences of a time se-
ries, and in fact the number of calls to the distance
function is often employed for assessing the speed
of an algorithm (Senin et al., 2018). In this respect
a complete knowledge of the distances of all the se-
quences introduces a wealth of information which can
be subsequently used for different primitives. Follow-
ing this perspective the articles of the Matrix Profile
series (Yeh et al., 2016) (Zhu et al., 2016) (and fol-
lowing papers) proposed very fast algorithms which
allow to determine the distance between all the se-
quences of a time series. In this article we thus pro-
vide an approach which allows one to obtain an ap-
Avogadro, P. and Dominoni, M.
Topological Approach for Finding Nearest Neighbor Sequence in Time Series.
DOI: 10.5220/0008493302330244
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 233-244
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
233

proximate nearest neighbor profile (formally defined
in Sec. 2.1.1) for all the sequences of a time series
which, at variance to the whole matrix profile, is just
the set of all the distances between a sequence and its
closest neighbor. With a good quality nnd profile, it
is possible to make quantitative statistical assessments
regarding the properties of the time series and obtain
new forms of indicators such as the one defined in
(Avogadro et al., ).
2 THREE DIFFERENT
TOPOLOGIES
In this section we detail three topologies of a time se-
ries (intended as notions of neighborhood):
• The Euclidean topology: which determines the
values of nnd
• The SAX topology: which is based on dimen-
sionality reduction and provides a quick grouping
within symbolic sequences
• The time topology: which is naturally determined
by the position of the points of a time series
2.1 Topology Induced by the Euclidean
Metric
The Euclidean distance between two sequences pro-
vides useful insights regarding their similarity. In par-
ticular, if one considers a sequence of s points as a
s−dimensional vector, it becomes straightforward to
use the Euclidean distance to define clusters of se-
quences, and as a result, to find anomalies or recurrent
patterns. We denote with S
k
the sequence of length s,
where the first point of the sequence is at time k. The
n
th
point of the sequence S
k
is denoted as s
k
n
. Accord-
ing to this notation, the Euclidean distance between
two sequences (S
k
and S
j
) is obtained with:
d(S
k
, S
j
) =
s
s
∑
n=1
s
k
n
− s
j
n
2
(1)
It is useful to remind that the order of neighbors (e.g.
the nearest neighbor, the second nearest neighbor, ...)
does not change by applying a monotone function
to this quantity (only the nearest neighbor distance
changes). This in turn implies that the same nearest
neighbor is obtained by using the Euclidean distance
or, for example, the d
2
distance (where no square root
is performed):
d
2
(S
k
, S
j
) =
∑
s
n=1
s
k
n
− s
j
n
2
.
2.1.1 Terminology
• The nearest neighbor distance of a sequence S
i
is
defined as:
nnd(S
i
) = min
j:|i− j|≥s
d(S
i
, S
j
), (2)
where the index j runs on all of the possible val-
ues within the search space U, as long as they ex-
clude self matches (|i − j| ≥ s). The concept of
non self match (Keogh et al., 2005) is necessary
to avoid “spurious” low values of nnd because of
partly overlapping sequences.
• The nnd profile is the set of all the possible nnds of
a given search space. An example of nnd profile
is displayed in Fig. 5 (left).
• The nnd density is obtained by dividing in bins
the range of all the possible nnd values and count-
ing the number of sequences which belong to each
bin. As an example, the discord belongs to the
righter-most bin, since, by definition, it has the
highest nnd value.
It should be noted that the concept of discord is
strictly linked to the search space U (the set of all the
sequences on which the minimization takes place). In
general, since the procedure for obtaining the nnd of
a sequence S is a minimization process, an increase
of the search space can only lead to a decrease of the
nnd. In detail, given two search spaces such that U
1
⊂
U
2
it is true that:
nnd
U
1
(S) ≥ nnd
U
2
(S). (3)
2.2 SAX Topology
The SAX algorithm (Lin et al., 2003) allows to pro-
duce a quick clusterization of a time series. Two
sequences can, in fact, be considered as neighbors
according to the SAX topology, if they belong to
the same cluster (a.k.a. symbolic sequence or s-
sequence). Notice that this topology is different from
the one induced by the Euclidean distance, since two
sequences can belong to two different SAX clusters
(e.g. they are not close according to SAX), but they
can be closest neighbors according to the Euclidean
metric (and vice versa). In order to understand this, it
might be useful to summarize the SAX procedure:
• Each sequence is fragmented in sub-sequences of
a given length (i.e. a 56 points sequence can be
divided in 7 consecutive sub-sequences, each of 8
points.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
234

• From each of these sub-sequences, the algebraic
average value is extracted and collected (piece-
wise aggregate approximation or PAA). This tech-
nique allows to pass from a sequence to a re-
duced sequence (r-sequence) of points, where this
r-sequence is smaller than the original one. Each
point of the r-sequence is an average of the points
of the original sequence.
• All the possible values of the averages are further
grouped in intervals (the number of intervals is the
dimension of the alphabet associated to the SAX
procedure) and each interval is assigned to a letter.
For example, let’s consider a sequence composed
of 20 points; we decide to group them in 5 sub-
sequences (each of 4 points). The correspond-
ing r-sequence contains only 5 points which are
then converted into a 3 letters alphabet, where
the letters are obtained with the following vocab-
ulary: [0, 3) → a, [3, 5) → b, [5, 8] → c (consid-
ering that all the points of the r-sequences lay
in the interval [0, 8]). As a result the sequence
34433013225661872103 turns into the symbolic
sequence babca:
sequence 3, 4, 4, 3, 3, 0, 1, 3, 2, 2, 5, 6, 6, 1, 8, 7, 2, 1, 0, 3
r-sequence 14/4, 7/4, 15/4, 22/4, 6/4
r-sequence 3.5, 1.75, 3.75, 5.5, 1.5
s-sequence b a b c a
Thanks to SAX, sequences giving rise to the same
the symbolic sequence are naturally grouped together.
The s-sequences are thus natural clusters for the se-
quences.
2.2.1 Curse of Dimensionality
Since the intervals defining the letters are “sharp”, the
nearest neighbor of a r-sequence close to the borders
of its s-sequence (cluster) might be in a neighboring
cluster. A letter, in fact, does not bring the informa-
tion regarding the fact that the average of points is
close to the center or to the borders (where other let-
ters begin).
A SAX cluster (symbolic sequence) of n letters
can be approximately seen as a hypercube (Fig. 1),
where each side corresponds to the interval of values
defining a letter. Actually, the object which correctly
represents a SAX cluster is not a hyper-cube since the
intervals don’t need to have the same length. In re-
ality this object is a n-dimensional parallelepiped or
parallelotope (where n is the number of letters of the
sequence). However, since the reasoning related to a
hypercube does not modify the results but it simplifies
the notation, in the following, we will keep on think-
ing in these terms. For high dimensional spaces, most
of the volume of a hypercube is close to its surface,
i.e. the probability to find a randomly placed point
close to the borders of the hypercube approaches one
as the number of sides (n) increases to infinity. In or-
der to better understand this fact it is possible to divide
the volume of the hypercube in two concentric parts:
an internal hypercube and an external shell (which is
just the difference between the whole hypercube and
the inner one). It is easy to calculate the inner volume
(where no point has coordinates within a small quan-
tity, ε l, from one of the faces) since it is an hy-
percube whose side is l − 2ε. The shell between the
inner hypercube and the full one represents the vol-
ume where the points are close to the surface, while
the inner hypercube is the region where the points are
far form the surface. In a scenario of a random dis-
tribution of the points within the symbolic sequence,
the volume of the shell is a good approximation of the
probability of finding a sequence close to the surface
of the symbolic sequences; vice-versa the volume of
the inner hypercube represent the probability of find-
ing a randomly placed point far from the surface. The
ratio of the volume of the inner hypercube and the
full one decreases geometrically with the dimension
of the clusters:
inner volume
volume
=
l − 2ε
l
n
n→∞
−−−→ 0
This implies that, as the dimensionality grows, most
of the volume is located in the external shell of the
hypercube. A point belonging to this region must be
close to at least one of its faces, and thus it is close to a
neighbor hypercube. From this perspective, the SAX
topology becomes less and less likely to approximate
the Euclidean topology as the length of the sequences
increases, since most of the sequences will be close
to the border of the SAX cluster and thus their clos-
est neighbor might be in a neighboring SAX cluster.
Nonetheless it is natural to think that SAX neighbors
are also likely to be Euclidean neighbors (this is in
fact the idea at the basis of HOT SAX (Keogh et al.,
2005)).
2.3 Time Topology
By time topology we simply refer to how distant
in time are the beginnings (or the ends) of two se-
quences, for example the nearest time-neighbors of S
i
are: S
i−1
and S
i+1
. In general the time distance be-
tween two sequences is simply:
d
t
(S
k
, S
j
) = |k − j| (4)
Topological Approach for Finding Nearest Neighbor Sequence in Time Series
235
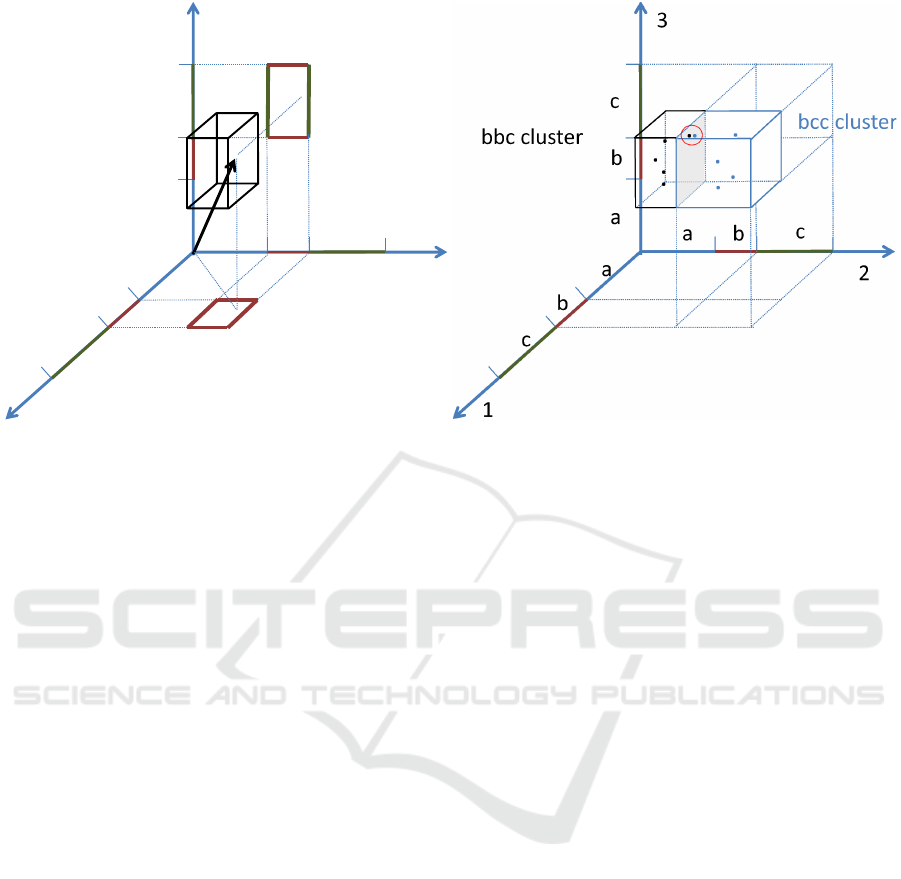
a
b
c
a
b
c
a
b
c
bbc cluster
1
2
3
.
Figure 1: (Left) A 3-dimensional parallelepiped corresponding to the symbolic sequence bbc, and a point whose coordinates
are averages of the subsequences. (Right) The nearest neighbor of one point of the bbc symbolic sequence is in the next
cluster, bcc, this suggests that the sequences giving rise to those points might be closest Euclidean neighbors but belonging to
different SAX clusters.
3 TOPOLOGY AS A ROAD-MAP
TO FIND THE APPROXIMATE
nnd PROFILE
The idea of this research is to go beyond a brute
force calculation, where one has to scan among all
the Euclidean distances in order to find the nearest
neighbor of a sequence. This can be achieved with
a clever selection method which allows to reduce the
total search space and thus the calculation time. This
selection procedure is guided by the different topolo-
gies present in the time series, allowing to aim more
precisely and reducing the problems related with the
curse of dimensionality.
Here we introduce the prescription to find the ap-
proximate nnd profile for a quick implementation, in
the rest of the section we will detail the reasons of the
steps. For each sequence S:
1. Perform an extensive nnd search within its own
SAX cluster (Sec. 3.1).
2. Perform an extensive nnd search within its own
SAX cluster, where the size of the alphabet has
been increased from n to n + 1, in respect to step
1 (Sec. 3.2).
3. Execute a search based on time topology (Sec.
3.3). If the nnd up to this point is lower than the
current min(nnd): analyse the next sequence (go
to point 1 with the next sequence).
4. If nnd(S) is still higher than the current min(nnd):
scan the other clusters from the smallest to the
biggest, until the nnd drops below the current min.
When these steps are applied to all the sequences of
the time series, the result is a good approximation of
the nnd profile. Since this strategy runs on an ex-
tended search space compared with the one used by
HOT SAX, it is assured that the discord of the time
series is going to be found. For improving the quality
of the approximate nnd profile, this procedure should
be repeated a few times (10 in the case of the results
of Sec. 4, thus ensuring to find the first 10 discords).
In order to avoid useless calculations (Bu et al., ), if a
sequence cannot be the discord (because its nnd value
is too low), we skip points 1, 2 when calculating the
2
nd
discord or above.
The quality of the resulting approximated nearest
neighbor profile will be analysed in Sec. 4. As a refer-
ence, we will now consider the HOT SAX algorithm,
and we will detail how to modify it in order to follow
the procedure just outlined and the reason for these
steps.
HOT SAX.
Let’s consider the approximate nnd profile obtained
with an application of HOT SAX (Keogh et al., 2005).
HOT SAX is a successful algorithm which allows to
find anomalies in time series. The idea of this algo-
rithm is to find those sequences which are particularly
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
236

different (according to the Euclidean distance) from
all the others: the discords. The discord is defined as
the sequence of the time series which has the maxi-
mum value of nnd. A brute-force discord search re-
quires two nested loops (and thus it is quadratic in
time with the number of sequences). The outer loop
runs on all the sequences. For each sequence S
i
of the
outer loop, the inner loop runs on all the sequences S
j
(| j − i| ≥ s), in order to find the nnd(S
i
). The inner
loop is the minimization process needed to find the
nearest neighbor. At the opposite, the external loop is
a maximization process aimed at finding the sequence
for which the nnd is the highest.
The discord sequence is the one with the highest
value of nnd, while the k
th
discord is defined as the
sequence with the highest nnd in a restricted search
space, which excludes all the sequences which (par-
tially) overlap with any of the previous (k − 1)
th
dis-
cords. The core idea of the HOT SAX algorithm is to
provide a smart method to exit from the inner loop, in
order to dramatically reduce the execution time. This
is done by re-ordering both the inner and the outer
loops.
At the beginning a SAX clusterization is per-
formed. The outer loop is re-ordered by positioning
at first small (SAX) clusters and at the end the biggest
ones. For each sequence of the outer loop, the mini-
mization procedure (for finding the nearest neighbor
distance of the sequence under observation) begins
by scanning those sequences which are in the same
SAX cluster, and for this reason they are good close
neighbor candidates. If a cluster contains only few se-
quences, it can be seen as an almost empty space and
it is a likely region where to find isolated sequences.
This reordering implies that, after a very few steps of
the outer loop the system will have found a good dis-
cord candidate, i.e. a sequence with a high value of
nnd. At this point as soon as the present nnd value of
a sequence (calculated in the inner loop) drops below
the actual highest nnd value, we are sure that that se-
quence cannot be a discord and it is possible to skip
the rest of the minimization process.
The rationale of these choices is that small (SAX)
clusters (in the limit containing only one sequence)
are good places where to find anomalies. The nnd
of a sequence of a small cluster will likely be high
(and it is better to search for it at the beginning in
order to have high nnd to confront with); while, on
the contrary, big clusters of sequences will contain
sequences with small nnd. Once the first sequences
have been calculated, since they are likely to be good
discord candidates, for the remaining ones, it will be
very likely that the inner loop quickly returns approx-
imate nnd values lower than the actual best discord
candidate. At this point the rest of the inner loop can
be skipped since we are sure that the sequence under
investigation cannot be the discord. These smart or-
derings, in practice, allow to skip most of the inner
loops reducing greatly the complexity of the calcu-
lation. Clearly the execution speed depends on the
time series under consideration, however this algo-
rithm has proven to be extremely efficient in many
practical cases. HOT SAX is thus essentially based
on comparing two different kinds of topologies, the
one induced by the Euclidean metric and the one of
the SAX procedure.
At the end of a HOT SAX calculation each se-
quence has an approximate neighbor (the one at the
time of exiting the inner loop) which determines an
approximate nnd. An approximate profile obtained in
this way as in Fig. 3 (left), however, is very differ-
ent from the exact one of Fig. 5 (left). This is not a
surprise, since the purpose of the algorithm is exactly
to exit from the minimization procedure in order to
avoid useless calculations.
3.1 Extensive nnd Search within the
SAX Cluster of Origin
Let’s introduce here the first modification of HOT
SAX which improves the nnd profile. Since the code
exits from inner loop as soon as the nnd of a se-
quence is below the current maximum, following the
SAX-Euclidean topology connection, in order to ob-
tain lower values of nnd, it is rather straightforward to
force the calculation to continue for all the sequences
of the same SAX cluster. If the connection between
SAX and Euclidean topology was perfect, this modi-
fication would assure to find the exact nearest neigh-
bor of each sequence. This in practice cannot hap-
pen. The Euclidean distance does induce a notion of
“closeness” however it does not define automatically
a clusterization.
The rationale of scanning the whole cluster where
the sequence belongs is that, if the sequence under
investigation is close to the center of the SAX cluster,
the Euclidean neighbor has a high chance of being
within the cluster itself. This procedure, is likely to
return the exact nearest neighbor for sequences which
are common within the time series (and thus belong
to big clusters).
3.2 Modifying the Size of the SAX
Alphabet
Because of the curse of dimensionality (Sec. 2.2.1),
for r-sequences near the border of their cluster there is
Topological Approach for Finding Nearest Neighbor Sequence in Time Series
237

a high chance that the closest neighbors might belong
to a different s-sequence (see Fig. 1).
There is a first easy cure for this problem. It is pos-
sible, in fact, to apply the SAX procedure two times
in order to produce different symbolic sequences. For
example the second time increasing by 1 the num-
ber of letters of the alphabet for the symbolic repre-
sentation. This prescription, in fact, moves the bor-
ders among the letters, so r-sequences close to the
borders have a high probability of being relocated
in other clusters (maybe containing their true neigh-
bors). Let’s consider a context where the alphabet
contains three letters associated to the following inter-
vals: [0, 3) −→ a; [3, 5) −→ b; [5, 8] −→ c. If the sec-
ond coordinates of two r-sequences are respectively
2.99 and 3.01 the corresponding letters would be a
and b (although these coordinates are very close). For
example:
3.95
2.99
7.21
→
b
a
c
;
3.82
3.01
7.38
→
b
b
c
(5)
By using an alphabet of 4 letters, however, the
intervals associated to each letter would move and
the two points would likely be associated to the
same letter, thus increasing the probability to find the
Euclidean nearest neighbor within the same cluster:
[0, 2) −→ a; [2, 4) −→ b; [4, 6) −→ c; [6, 8] −→ d.
The new symbolic sequences associated to the two r-
sequences are:
3.95
2.99
7.21
→
b
b
d
;
3.82
3.01
7.38
→
b
b
d
(6)
Notice that changing the size of the alphabet used
for the symbolic representation does not change the
sequence under investigation, in this way each se-
quence belongs to two (or more if applied many
times) different SAX clusters, where the probability
to find the closest Euclidean neighbor increases. This
procedure increases the size of the search space where
we can apply the minimization for finding the nnd
of each sequence, thus increasing the probability to
find a better approximation of the exact value. The
drawback of this procedure is to further slow down
the search (since it increases the search space for each
sequence). Since the two clusters might contain over-
lapping sequences it is useful to keep track of the ones
already checked in the first part of the algorithm and
avoid doing the same calculations two times.
3.3 Time for a More Accurate Search
By applying the steps of Sec. 3.1 and Sec. 3.2 it is
possible to obtain a nnd profile which becomes closer
to the exact one, however it is possible to notice that
there is still quite a big number of “suspicious” spikes,
for example in Fig. 4 (left). Up to this point we have
been exploiting the connection between SAX and Eu-
clidean topology. For a further improvement of the
nnd profile, it could be tempting to perform an ex-
tensive search also on the neighboring SAX clusters.
This approach however has two main problems.
• The curse of dimensionality implies that the num-
ber of neighboring clusters grows geometrically,
and there is no simple technique to understand if
any of them is better than the others.
• The amount of sequences to be searched becomes
very big thus rendering the approximate search
not valuable.
As a solution we propose to exploit the time topol-
ogy, which, at this point, can provide useful sugges-
tions regarding the position of close neighbors for
each sequence. Let’s consider the nearest neighbor
distance as a function of the index of the sequence i,
nnd(S
i
), where i runs over all sequences of the time
series. If the time series shows a certain degree of
regularity, we can expect that also nnd(S
i
) should be
pseudo-smooth (but for the points where there are true
anomalies). By pseudo-smooth we mean that, it is
possible to obtain upper bounds for nnd(S
i+1
) as a
function of nnd(S
i
) (and vice-versa). The reason is
rather simple, and in order to show it we will make
use of the d
2
distance instead of the Euclidean one,
since it allows to get rid of the square root (but the
order of the distances does not change). We will de-
note with p
i
, where i is the time, the single points of
the time series. With this notation, if the length of
the sequences is s = 10, the last point of sequence
S
43
is p
52
= s
43
10
. Let’s consider a sequence, S
i
, and
its Euclidean nearest neighbor located, for example,
at time i + k (where k is higher than the length of
the sequence s, to prevent a self-match condition), in
this case nnd(S
i
) = d
2
(S
i
, S
i+k
). The nearest neighbor
distance of the next sequence, S
i+1
, is related with
nnd(S
i
), by Eq 7.
nnd(S
i+1
) ≤ d
2
(S
i+1
, S
i+k+1
) = nnd(S
i
) + (p
i+s
− p
i+s+k
)
2
− (p
i
− p
i+k
)
2
,
(7)
• The first inequality of Eq. 7 holds because, by def-
inition, nnd(S
i+1
) is the minimum among all the
distances between S
i+1
and all the other sequences
of the time series.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
238
• the second part of Eq. 7 is true because of the defi-
nition of the d
2
distance function, which is a sum-
mation of squares: d
2
(S
i
, S
i+k
) =
∑
j=1,s
(p
i+ j
−
p
i+k+ j
)
2
If one compares the two distances:
d
2
(S
i
, S
i+k
) = (p
i
− p
i+k
)
2
+ (p
i+1
− p
i+k+1
)
2
+ ...
+(p
i+s−1
− p
i+k+s−1
)
2
d
2
(S
i+1
, S
i+k+1
) = (p
i+1
− p
i+k+1
)
2
+ ...
+(p
i+s−1
− p
i+k+s−1
)
2
+(p
i+s
− p
i+k+s
)
2
,
• It is easy to notice that the first and the last ad-
dend are the only differences between the two dis-
tances, and by hypothesis nnd(S
i
) = d
2
(S
i
, S
i+k
).
In detail the value of (p
i+s
− p
i+k+s
)
2
− (p
i
− p
i+k
)
2
in Eq. 7 determines whether the inequality implies a
strict limit or not. Since, by definition, the sequences
beginning at time i and at time i + k are the closest
neighbors, it seems reasonable to expect that the parts
of the time series which follow these two sequences
might resemble each other and thus be close in terms
of Euclidean distance. The information provided by
Eq. 7 is not restricted to the case in which one knows
the exact nnd(S
i
), but it can be used also in the case
in which the nnd(S
i
) is approximated, simply provid-
ing a looser upper bound on the value of nnd(S
i+1
).
Moreover this is true not only from one sequence to
the next one, but a similar relation exists also for the
previous sequence. At this point we have noticed
that time topology can provide good hints on where
to search for nearest neighbors, once an approximate
nnd profile is already present.
This reasoning follows the same basic idea of
the HOT SAX procedure applied to a different
topology, i.e. to provide a hint regarding where to
find Euclidean neighbor of a sequence. With a motto,
a sequence could “say”:
The Euclidean-neighbor of my time-neighbor,
is likely to be a time-neighbor of my Euclidean
neighbor.
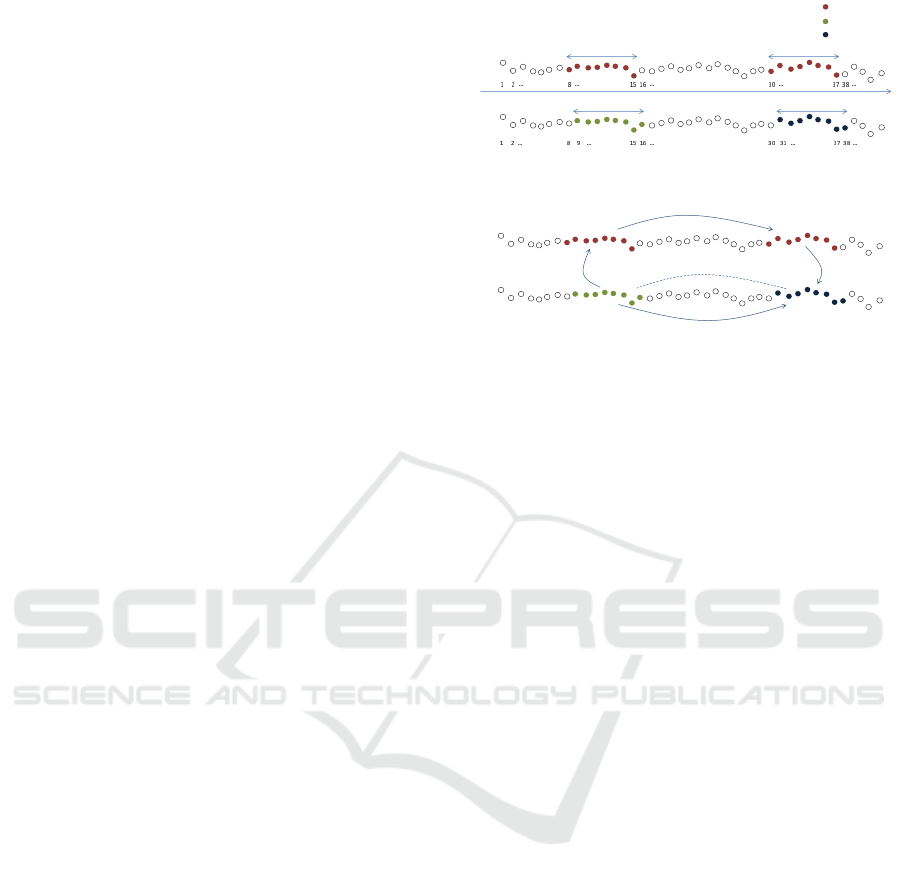
An example of the utilization of the procedure is
shown in Fig. 2, and here synthesized:
• S
8
is a sequence of length 8. Its Euclidean neigh-
bor has been found within those sequences which
share the same same SAX cluster, in particular is
S
8+22
.
• The (approximate) nnd of sequence S
9
(which is
the time-neighbor of S
8
) among its SAX neigh-
bors, is much higher than nnd(S
8
), not following
the constraints of Eq. 7. This is suspicious, since
we expect the nnd profile to be rather smooth (but
for the anomalies).
S
8+22
S
8
S
9
S
9+22
SAX
time
time
Euclidean
not SAX
cluster A
cluster B
cluster C
Clusters
Exploiting different topologies:
Figure 2: Different kinds of neighborhood: (top figure) Se-
quence S
8
and S
9
are time-neighbors (but they belong to
two different SAX clusters, the red and the green one).
Sequence S
8
and S
8+22
belong to the same SAX cluster
(the red one), moreover this latter sequence is the clos-
est Euclidean neighbor of the former. Sequence S
9
(be-
cause of the curse of dimensionality) does not belong to the
same cluster as its closest Euclidean neighbor, S
9+22
, and
there would be no reason to check the distance of the two,
however (bottom figure) thanks to the following passages:
S
9
time
−−→ S
9−1
SAX
−−−→ S
9−1+22
time
−−→ S
9−1+22+1
we can guess
that S
9
and S
9+22
are likely Euclidean neighbors.
• S
9+22
does not belong to the same SAX cluster
as S
9
(it belongs to a neighbor cluster, due to the
curse of dimensionality). SAX clustering, in this
case, does not provide good suggestions on where
to search for a good Euclidean neighbor of se-
quence S
9
.
• However, thanks of the time topology it is possi-
ble to say that S
9−1+22+1
is a good candidate for
being a close Euclidean neighbor of S
9
as shown
in Fig. 2 (the summation of the index of the pos-
sible neighbor sequence has not been carried out
in order to emphasize the three passages to obtain
it: time, SAX, and time again).
Clearly it is possible to check on both time direc-
tions in order to improve the quality of the results.
It is also possible to do more loose checks where one
takes into account, not just the first time neighbors,
but also more time-distant sequences (up to time dis-
tance= 10 in the present article). This implies that,
for sequence S
i
, we first check the nearest neighbor of
S
i+ j
( j ≤ 10), denoted with S
l
. Later, in order to im-
prove nnd(S
i
), we calculate d(S
i
, S
l− j
). Actually in
the present implementation we select the sequences to
be checked with a simple heuristic, namely only if the
nnd calculated up to that point presents a non-smooth
behavior (i.e. Eq. 7 is not respected).
Topological Approach for Finding Nearest Neighbor Sequence in Time Series
239
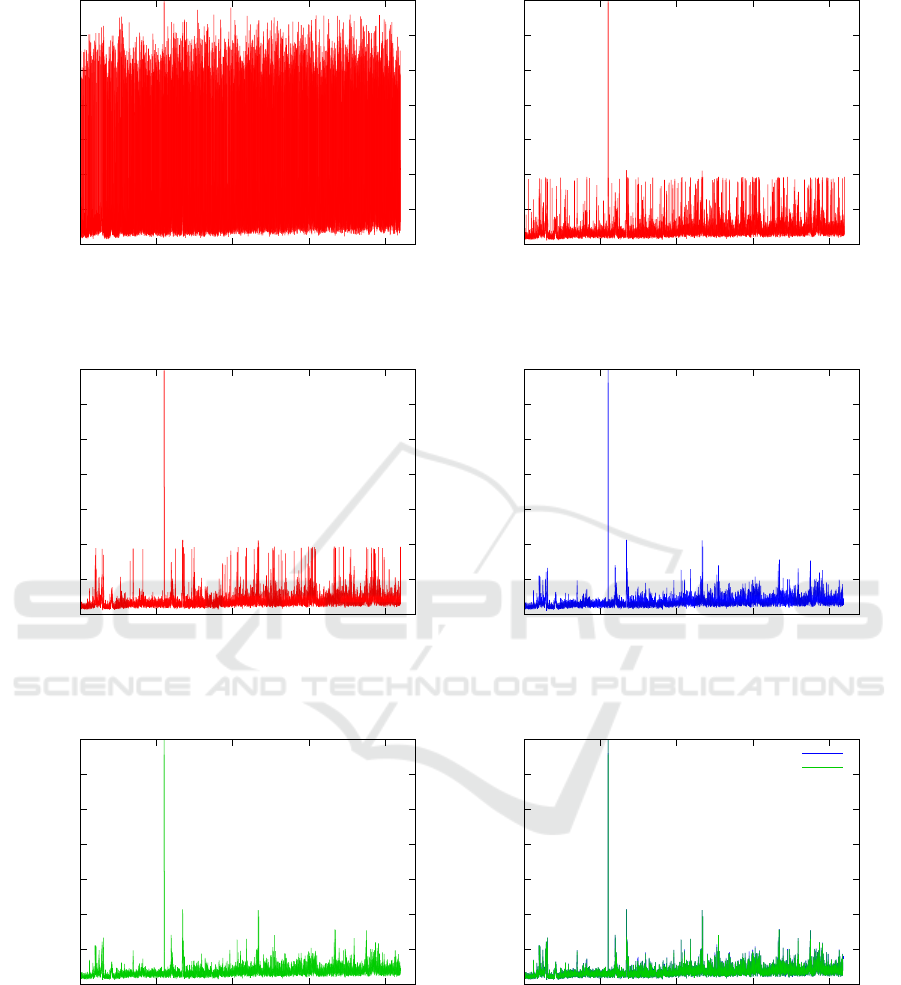
0
200
400
600
800
1000
1200
1400
0 50000 100000 150000 200000
nnd
sequence time index
0
200
400
600
800
1000
1200
1400
0 50000 100000 150000 200000
nnd
sequence time index
Figure 3: (Left) Approximate nnd profile for all the sequences, obtained with HOT SAX. (Right) The nnd profile obtained by
extending the search space in order to include the whole cluster where each sequence belongs.
0
200
400
600
800
1000
1200
1400
0 50000 100000 150000 200000
nnd
sequence time index
0
200
400
600
800
1000
1200
1400
0 50000 100000 150000 200000
nnd
sequence time index
Figure 4: (Left) Approximate nnd profile, obtained with the extended search space including two different kinds of SAX
clusters (with 3 and 4 letters alphabets respectively). (Right) The nnd profile calculated including also the time-topology.
0
200
400
600
800
1000
1200
1400
0 50000 100000 150000 200000
nnd
sequence time index
0
200
400
600
800
1000
1200
1400
0 50000 100000 150000 200000
nnd
sequence time index
Approximated nnd
Exact nnd
Figure 5: (Left) Brute force calculation of the exact nnds. (Right) Both the exact (green line) and time-topology approximated
nnd profiles (blue line). Since the approximation obtained with the help of time topology is very close to the exact calculations,
the two curves are essentially identical; only a closer look, as in Fig. 7 (right), can allow to distinguish the differences.
4 VALIDATION
In this section we evaluate to which extent the ex-
act nnd profile can be matched with the topologically
approximated one, and what is the gain in terms of
speed. In detail, let’s consider the nnd profile ob-
tained in five different cases, where each search space
includes the previous one:
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
240
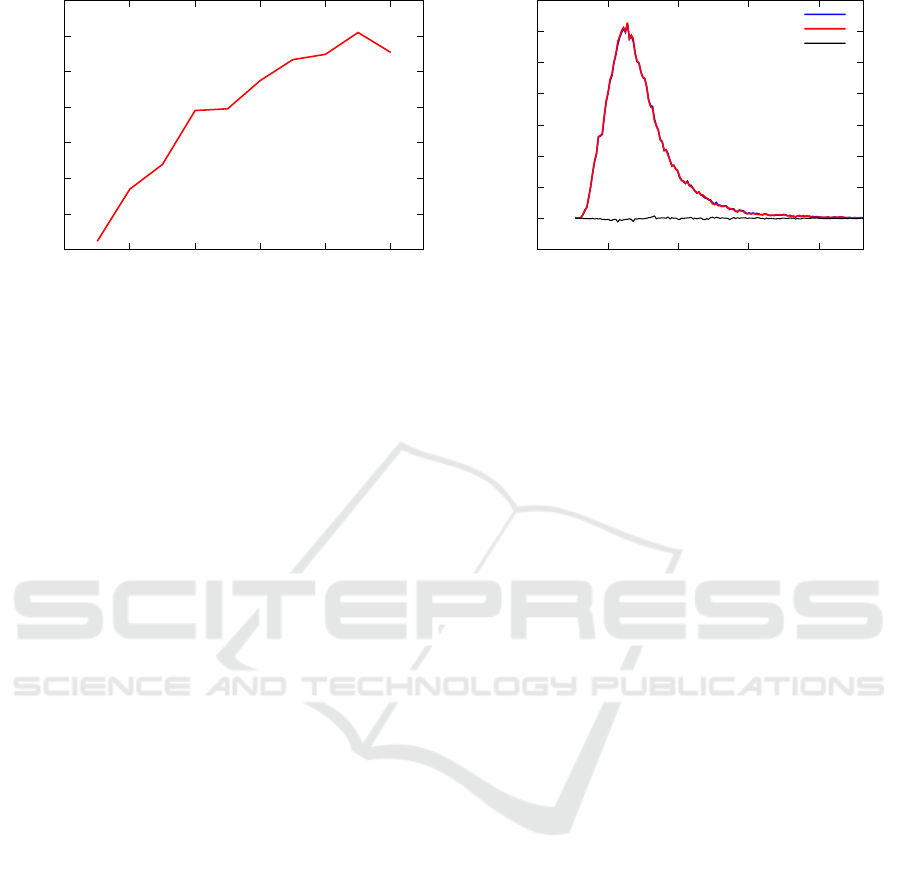
20
30
40
50
60
70
80
90
0 40000 80000 120000 160000 200000
Brute-Force/approximate time
Number of sequences
-0.005
0
0.005
0.01
0.015
0.02
0.025
0.03
0.035
0 50 100 150 200
fraction
nnd
topologically approx. nnd distribution
exact nnd distribution
difference
Figure 6: (Left) Ratio of the execution speed of the brute force algorithm and the topological approach for calculating the
nnd profile as a function of the size of the time series to be analysed. In particular we analysed chunks of increasing size
of ECG300. (Right) The topologically approximated nnd densities and the exact one are very similar, and shown here along
with their difference. The interval of nnd values spans from about 0 to about 1400, in order to improve readability, in this
picture, we show values of nnd up to 231.3 since they exhaust 99.5% of the cumulative distribution function, and they allow
to see the main differences between the two densities.
1. The approximate nnd values resulting from the
application of the HOT SAX algorithm is shown
in Fig. 3 (left).
2. Extending the nnd search to all the sequences of
the same cluster produces the approximate nnd of
Fig. 3 (right).
3. Extending the nnd search to all the sequences of
the cluster obtained by using one more letter in the
alphabet returns the nnd profile of Fig. 4 (left).
4. Using also the time topology for the nnd search
provides a clear improvement as of Fig. 4 (right).
5. The results of the exact calculation associated to
the brute force algorithm is in Fig. 5 (left).
In Fig. 5 (right) we show both the exact and the topo-
logically approximated nnd profiles for a direct vi-
sual comparison. The time series under consideration,
ECG300, belongs to the MIT-BIH ST change database
available at Physionet (Goldberger et al., e 13). It is
the ECG of a person, sampled 536976 times. In the
pictures, for a better readability of the images we limit
the series to 210000 points (however when taking into
account the accuracy of the approximation we con-
sider all the points). The parameters of the HOT SAX
search are:
• Sequence length: 56 points
• Alphabet size: 3 letters
• 8 points are used to form one letter (which implies
that each sequence/cluster of 56 points contains 7
letters).
It is clear that the approximate nnd profile re-
turned by HOT SAX (Fig. 3 on the left) is useless
for obtaining statistics. This is normal, since the ap-
proach of HOT SAX is to skip all the calculations
which are not necessary for finding discords, and not
to try to produce good quality nnds. A clear improve-
ment appears if we modify HOT SAX, by forcing it
to run over all the sequences of the cluster associated
to a sequence, as in Fig. 3 (right). In this case, the
algorithm does not exit the loop as soon as the cur-
rent value of the nnd drops below the actual best dis-
cord candidate (as it would do with a normal applica-
tion of HOT SAX), but it keeps on updating the nnd
associated to the sequence. Unfortunately, because
of the curse of dimensionality we can also expect
that many sequences with small Euclidean distance
should be present in neighbor SAX clusters, but the
number of such clusters grows exponentially with the
length of the r-sequences. An easy remedy is to per-
form two times the SAX procedure with two differ-
ent alphabets, i.e in the present example we explored
SAX clusters obtained with 3 letters and with 4 let-
ters. With this approach each sequence belongs to two
different clusters (s-sequences), moreover, since the
range where the letters are chosen passes from even
to odd (or vice versa), it seems reasonable that some
of the neighbors of the r-sequence (which were just
outside of the borders of the first SAX cluster) might
fall inside the second SAX cluster. It is worth remem-
bering that the clusters are based on the r-sequences,
and this induces further uncertainty regarding the fact
that two sequences belonging to the same SAX clus-
ter are also Euclidean neighbors. In Fig. 4 (left) there
is the result of this extension which shows a clear im-
provement of the nnds (we remind that the approx-
imate nnds are always greater or equal to the exact
Topological Approach for Finding Nearest Neighbor Sequence in Time Series
241
0
50
100
150
200
250
300
350
400
450
66000 66200 66400 66600 66800 67000 67200 67400 67600 67800
Two kinds of clusters
Three topologies
0
50
100
150
200
250
300
350
400
450
66000 66200 66400 66600 66800 67000 67200 67400 67600 67800
Three topologies
Exact calculation
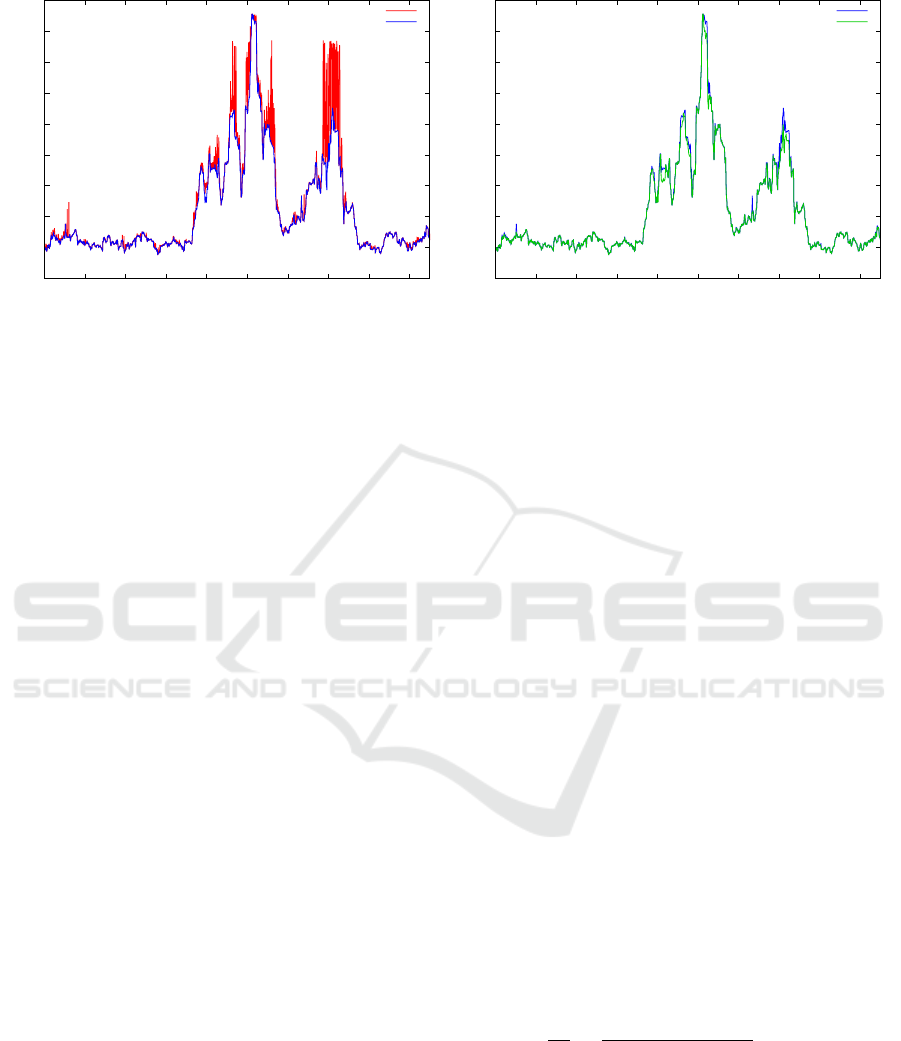
Figure 7: (Left) Detail of the nnds obtained after applying the three topology search (blue) and using two SAX clusters (red).
(Right) Detail of the nnds obtained with the exact calculation (gray) and after applying the three topology search (blue).
Table 1: Comparison of the results of the application of the
topologically approximated nearest neighbor distance pro-
file to real time series (Goldberger et al., e 13) (Laguna
et al., 1997) and a brute force calculation. The files sel con-
tain 225000 points, while the bidmc15 series 60000. The
subscripts (
2
,
3
,
4
and
5
) refer to the column of the files
under investigation, second, third, etc. The speedup is ob-
tained as the ratio of the number of calls to the distance
function of the brute force algorithm and the number of calls
of our approximate algorithm.
file name % of exact nnds Err speedup
sel0606
2
99.6 0.05 83
sel0606
3
99.5 0.05 116
sel102
2
98.8 0.05 40
sel102
3
99.7 0.06 26
sel123
2
98.8 0.05 32
sel123
3
99.2 0.04 14
bidmc15
2
99.6 0.15 5
bidmc15
3
99.8 0.09 30
bidmc15
4
98.4 0.05 40
bidmc15
5
98.9 0.06 58
ones, since the only difference among the two is re-
lated to the search space where the minimization takes
place, which is restricted in the case of the approxi-
mate values). At this point one has obtained a good
approximation of the nnd profile, however with a de-
tailed comparison of the approximate nnds and the
exact ones, Fig. 7 (left), it is possible to notice that
the former profile presents many spikes, while the ex-
act profile is more “smooth”. This is an indication
that the curse of dimensionality is still creating prob-
lems and another search mechanism needs to be intro-
duced. The three topology idea fits perfectly this ap-
proach and, by extending the search as per Sec. 3.3,
one obtains the nnd profile of Fig. 4 (right) which
is very close to Fig. 5 (left). At the end, in figure
5 (right) we compare the exact and the topologically
approximated nnd profile.
In order to better understand the relation between
the the time approximated nnd density and the exact
one, we show them in Fig. 6. Also in this case the dif-
ferences are so small to be almost unnoticeable. We
obtained the nnd densities by dividing the values of
the possible nnds in 1000 bins and for each bin we
counted the number of sequences with a nnd falling
within its limits. The difference among the two den-
sities is also shown for an improved readability. The
topologically approximated nnd density is so close to
the exact one that it can replace it for practical pur-
poses, and in particular when searching for statistical
properties of the nnd profile (Avogadro et al., ).
Fig. 7 (left) shows a detail of the nnd profile calcu-
lated with and without the time topology. It is clearly
visible a high number of spikes which correspond to
poor quality values of the nnd. When passing from
the topologically approximated nnd to the exact one
as in Fig. 7 (right), it is particularly visible that the
differences are more limited both in terms of quantity
and magnitude.
For a quantitative evaluation of the topologically
approximated nnd profile we counted the amount of
exact nnds calculated over the total. In the case of
ECG300 the exact nnd value has been obtained for
98% of the sequences. The average fractional error
of the approximate nnds has been obtained as:
Err =
1
N
a
N
∑
i=1
nnd
a
(S
i
) − nnd(S
i
)
nnd(S
i
)
= 3.1 · 10
−2
(8)
Where nnd
a
(S
i
) is the approximated nnd value asso-
ciated to the sequence S
i
, and N
a
is the amount of se-
quences for which only an approximate nnd has been
found (2% of the total number of sequences). Notice
that there is no reason to use the absolute value, since
nnd
a
(S
i
) ≥ nnd(S
i
) ∀i, the approximated distances are
in fact always greater or equal than the exact ones
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
242

(and in the present case there are no sequences for
which the nnd is exactly 0). In most of the cases (usu-
ally around 99% of the sequences), the numerator of
Eq. 8 is zero. In Table 1 we show the results of the ap-
plication of the topologically approximated nnd pro-
file for other time series.
5 CONCLUSIONS AND FUTURE
WORKS
When analysing time series, an nnd profile can be
useful in order to characterize the properties of the
sequences, for example to find out possible anoma-
lies in the form of discords or recurrent sequences.
Unfortunately a full nnd profile requires calculations
which scale quadratically with the number of points.
In the present article we propose a procedure which
can speed up the process greatly. The idea followed
in this article is to exploit different kinds of neigh-
borhoods for each sequence in order to constrain the
calculations to search spaces where the probability to
find the exact neighbor is very high. The three topolo-
gies are: the one introduced by SAX, the time topol-
ogy and the Euclidean topology. The reduced search
space thus obtained allows one to skip most of the cal-
culations while retaining a good accuracy for the nnds
of each sequence.
This is a heuristic selection procedure, and so it is
not possible to provide exact bounds in term of com-
putational complexity. However, the experimental re-
sults we obtained on real data time series are very
interesting since the speed-ups in respect to a brute
force algorithm are between 1 and 2 orders of mag-
nitude. There is also a clear trend indicating that the
ratio between the brute force computational time and
the computational time obtained with the time topol-
ogy increases with the size of the time series under
observation and for this reason our approach becomes
particularly appealing with large time series.
In terms of accuracy, the time-approximated nnd
profiles are very close to the exact ones, in the cases
under observation for more than 98% of the sequences
the exact nnd has been found, while for those se-
quences for which just an approximate nnd has been
found, the values are close (≈ 10%) to the correct
ones. It should be emphasized that, due to the nature
of the time-approximated nnd profile (which essen-
tially extends the search space of the HOT SAX algo-
rithm) the results automatically include highest nnds
(corresponding to the discords of the time series).
An interesting result implied by exploiting the
time topology is Eq. 7, which expresses an upper
bound for the nnd of a sequence, once the nnd of a
time neighbor of that sequence is known. In practice
this implies that, once an approximate nnd profile has
been obtained, it is very easy (linear with the size of
the time series) to check if some of the approximate
nnds are particularly distant from their correct value.
In summary, this approach allows to diminish sig-
nificantly the amount of calculations needed to obtain
the nnd profile of a time series at a reasonable loss of
precision. In the present literature the state of the art
is represented by the algorithms of the Matrix Profile
series (Yeh et al., 2016), however they scale quadrat-
ically with the length of the time series and they also
provide information which might be difficult to uti-
lize (they allow to obtain the distance from all the se-
quences while often times only the nearest neighbors
play an important role in determining the properties
of a sequence).
Future works include the application of MASS al-
gorithm which exploits the Fast Fourier Transform
to speed up the calculation of the distances (Mueen
et al., 2017) which is at the basis of (Yeh et al., 2016),
and it is known to greatly speed up the calculation of
distances between sequences.
It is possible to exploit, with minor modifications,
the procedure provided by this work in order to obtain
approximations of the second, third,..., k-th neighbors
of a sequence and we are in the process of implement-
ing and testing them for an even more complete pro-
file of the main properties of each sequence.
We are also implementing the time topology to
speed up the calculation of discords.
REFERENCES
Avogadro, P., Palonca, L., and Dominoni, M. A. Online
anomaly search in time series: significant online dis-
cords. under review.
Bu, Y., Leung, T.-W., Fu, A. W.-C., Keogh, E., Pei, J., and
Meshkin, S. WAT: Finding Top-K Discords in Time
Series Database, pages 449–454.
Chandola, V., Banerjee, A., and Kumar, V. (2009).
Anomaly detection: A survey. ACM Comput. Surv.,
41(3):15:1–15:58.
Chiu, B., Keogh, E., and Lonardi, S. (2003). Probabilis-
tic discovery of time series motifs. In Proceedings
of the Ninth ACM SIGKDD International Conference
on Knowledge Discovery and Data Mining, KDD ’03,
pages 493–498, New York, NY, USA. ACM.
Goldberger, A. L., Amaral, L. A. N., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E.,
Moody, G. B., Peng, C.-K., and Stanley, H. E.
(2000 (June 13)). PhysioBank, PhysioToolkit, and
PhysioNet: Components of a new research resource
for complex physiologic signals. Circulation,
101(23):e215–e220. Circulation Electronic Pages:
Topological Approach for Finding Nearest Neighbor Sequence in Time Series
243

http://circ.ahajournals.org/content/101/23/e215.full
PMID:1085218; doi: 10.1161/01.CIR.101.23.e215.
Keogh, E. (Accessed 11 July 2019). Welcome to the sax.
https://www.cs.ucr.edu/ eamonn/SAX.htm.
Keogh, E., Lin, J., and Fu, A. (2005). Hot sax: efficiently
finding the most unusual time series subsequence. In
Proceedings of the Fifth IEEE International Confer-
ence on Data Mining (ICDM’05), pages 226–233.
Laguna, P., Mark, R. G., Goldberger, A., and Moody, G. B.
(1997). A database for evaluation of algorithms for
measurement of qt and other waveform intervals in the
ecg. Computers in Cardiology, pages 24:673–676.
Lin, J., Keogh, E., Lonardi, S., and Chiu, B. (2003). A sym-
bolic representation of time series, with implications
for streaming algorithms. In Proceedings of the 8th
ACM SIGMOD Workshop on Research Issues in Data
Mining and Knowledge Discovery, DMKD ’03, pages
2–11, New York, NY, USA. ACM.
Lin, J., Keogh, E., Patel, P., and Lonardi, S. (2002). Find-
ing motifs in time series. In Proceedings of the The
2nd Workshop on Temporal Data Mining, the 8th ACM
Int’l Conference on KDD.
Mueen, A., Zhu, Y., Yeh, M., Kamgar, K., Viswanathan,
K., Gupta, C., and Keogh, E. (2017). The fastest sim-
ilarity search algorithm for time series subsequences
under euclidean distance. http://www.cs.unm.edu/
mueen/FastestSimilaritySearch.html.
Patel, P., Keogh, E., Lin, J., and Lonardi, S. (2002). Mining
motifs in massive time series databases. In 2002 IEEE
International Conference on Data Mining, 2002. Pro-
ceedings., pages 370–377.
Senin, P., Lin, J., Wang, X., Oates, T., Gandhi, S., Boedi-
hardjo, A. P., Chen, C., and Frankenstein, S. (2018).
Grammarviz 3.0: Interactive discovery of variable-
length time series patterns. ACM Trans. Knowl. Dis-
cov. Data, 12(1):10:1–10:28.
Senin, P., Lin, J., Wang, X., Oates, T., Gandhi, S., Boedi-
hardjo, A. P., Chen, C., Frankenstein, S., and Lerner,
M. (2014). Grammarviz 2.0: A tool for grammar-
based pattern discovery in time series. In Calders,
T., Esposito, F., H
¨
ullermeier, E., and Meo, R., ed-
itors, Machine Learning and Knowledge Discovery
in Databases, pages 468–472, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Yeh, C. M., Zhu, Y., Ulanova, L., Begum, N., Ding, Y., Dau,
H. A., Silva, D. F., Mueen, A., and Keogh, E. (2016).
Matrix profile i: All pairs similarity joins for time se-
ries: A unifying view that includes motifs, discords
and shapelets. In 2016 IEEE 16th International Con-
ference on Data Mining (ICDM), pages 1317–1322.
Zhu, Y., Zimmerman, Z., Senobari, N. S., Yeh, C. M., Fun-
ning, G., Mueen, A., Brisk, P., and Keogh, E. (2016).
Matrix profile ii: Exploiting a novel algorithm and
gpus to break the one hundred million barrier for time
series motifs and joins. In 2016 IEEE 16th Inter-
national Conference on Data Mining (ICDM), pages
739–748.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
244
