
Ontology Learning from Clinical Practice Guidelines
Samia Sbissi
1 a
, Mariem Mahfoudh
2,3 b
and Said Gattoufi
1 c
1
SMART Laboratory, Tunis University, Tunis, Tunisia
2
MIRACL Laboratory, University of Sfax, Sfax, Tunisia
3
ISIGK, University of Kairouan, Kairouan, Tunisia
Keywords:
Ontology Learning, Ontology Enrichment, SWRL, Word2Vec.
Abstract:
In order to assist professionals and doctors to make decisions about appropriate health care for patients who are
at risk of cardiovascular disease, we propose a decision support system based on OWL (Ontology Language
Web) ontology with SWRL (semantic web rule language) rules. The idea consists to parse clinical practice
guidelines (i.e. documents that contain recommendations and medical knowledges) to enrich and exploit
existing cardiovascular domain ontology. The enrichment process is conducted by ontology learning task. We
first pre-process the text and extract the relevant concepts. Then, we enrich the ontology not only by OWL DL
axioms, but also SWRL rules. To identify the similarity between terms texts and ontology concepts, we have
used a combination of methods as levenshtein similarity and Word2Vec.
1 INTRODUCTION
The clinical guidelines (CG) contain a set of recom-
mendations and knowledge used to guide health pro-
fessionals in making appropriate decisions and im-
proving the quality of care. Although health profes-
sionals are familiar with these guides, text-based ver-
sions have several limitations (Cabana et al., 1999;
Francke et al., 2008). The text of the recommenda-
tions may contain undefined terms in well-accepted
terminology, and unclear sentences which may lead
to an ambiguous interpretation of the content. The
size/complexity of the recommendations may be an
obstacle also in the sense that it may hide relevants
informations or discourage specialists from reading
all the document (Bonacin et al., 2013). In order to
deal with these problems, some tools were proposed
to code the clinical guidelines and to create computer-
interpretable guidelines (CIG) and there is an increas-
ing demand to convert this unstructured information
into structured information.
Ontology plays a key role in representing the
knowledge hidden in texts and makes it human and
computer understandable. An ontology is a formal
and structural way of representing the concepts and
a
https://orcid.org/0000-0002-5301-5156
b
https://orcid.org/0000-0001-7860-8604
c
https://orcid.org/0000-0001-7914-6165
relations of a shared conceptualization. More pre-
cisely, it can be defined as concepts, relations, at-
tributes and hierarchies present in the domain. On-
tologies can be created by extracting relevant in-
stances of information from text using a process
called ontology population. However, handcrafting
such big ontologies is a difficult task, and it is im-
possible to build ontologies for all available domains
(Asim et al., 2018). Therefore, instead of handcraft-
ing ontologies, research trend is now shifting toward
automatic extract ontology from the text that is de-
fined as an ontology learning process (Maedche and
Staab, 2001).
The process of ontology learning begins with the
extraction of terms and their synonyms from the
text. The corresponding terms and synonyms are con-
verted to the form of concepts. Then, taxonomic and
non-taxonomic relations between these concepts are
found. Finally, axiom schemata are instantiated and
general axioms are extracted from unstructured text.
This whole process is known as ontology learning
layer cake (Gardent and Mahfoudh, 2016).
In our work, we address the problem of ontology
learning that could be applied to the analyzed medical
text, in order to enrich an existing ontology. The
enriched ontology aims to infer and produce a recom-
mendation task. To this end, we collaborate with the
hospital of the "Rabta" (Tunis) to make an assistance
system that helps doctors to make decisions about
312
Sbissi, S., Mahfoudh, M. and Gattoufi, S.
Ontology Learning from Clinical Practice Guidelines.
DOI: 10.5220/0008169903120319
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 312-319
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

patients who are at risk of cardiovascular disease,
especially aortic dissection. The aortic dissection
is a partial disruption of the wall of the aorta that
may at any time evolve towards a complete rupture,
with consequent death (Criado, 2011). One of the
ontologies we found close to our domain is the
CVDO ontology
1
. It is an owl ontology, designed
to describe the entities related to cardiovascular
diseases. This ontology will be learnt and enriched
from a text called clinical practice guidelines (Erbel
et al., 2014). It is an evolving reference document
that contains a set of recommendations which
aim to assist professionals to master a medical
domain. Recommendation example: In patients
with an abdominal aortic diameter of
25-29 mm, new ultrasound imaging should
be considered 4 years later). Our goal is to
transform these recommendations into logical forms,
more precisely into Semantic Web Rule Language
(SWRL) rules. SWRL (Horrocks et al., 2004) is a
semantic web language, which is integrated directly
within OWL (Ontology web language) ontologies.
It allows defining rules in the form of logical impli-
cations between conditions and conclusions. This
transformation will be conducted by the ontology
learning process. We think that the ontology learning
process will help increase the number of transformed
rules from text to ontology (Sbissi et al., 2019) due
to the lack of concepts and relations in the initial
ontology.
The remainder of this paper is organized as fol-
lows. Section two gives a summary of related work.
Section three presents our approach. Section four
presents the implementation and results. Finally, we
conclude and we give some future work.
2 RELATED WORK
Studies done by (Séroussi et al., 2010) highlight
physicians low adherence to the text-based guide-
lines (27.2%), and high adherence to the evaluated
electronic version of the guidelines (86.1%). They
proposed a system called ASTI-GM that has been
designed to be computer-based thinking support on
how to decide, is a demand guideline-based CDSS
where the user interactively characterizes her patient
by browsing the system knowledge base to obtain
the recommended treatment. The translation from
text-based guidelines to computer-interpret-able one
requires a well-defined description language. Sev-
eral formalization techniques and methodologies have
1
http: //purl.bioontology.org/ontology/CVDO
been proposed in the literature. One of the most used
is the ontology. An important benefit of ontology is
the ability to specify axioms for reasoning.
The mapping process from text is called ontol-
ogy learning and it can be of three types : man-
ual, semi-automatic and automatic. In the manual
method, an ontology is constructed from the scratch
by domain experts and knowledge engineers using
the most painstaking procedures (Maedche, 2013). In
the semi-automatic method, the domain experts and
trained users use semi-automatic prototype. For ex-
ample, (Dramé et al., 2014) build a semi automatic-
multilingual domain ontology using UMLS Metathe-
saurus and parallel corpus. However, these meth-
ods are time-consuming and require domain experts.
Consequently, the automatic method of ontology
learning is becoming a major trending. Several sys-
tems are proposed : Text-to-Onto (Cimiano et al.,
2009), OntoGain (Drymonas et al., 2010), etc.
There are three major approaches for ontology
learning that are often used: statistical methods (e.g.
C/CN value, T F − IDF, word2vec etc.), machine
learning methods, and linguistic approaches (e.g.
POS patterns, parsing, WordNet, discourage anal-
ysis, etc.). Authors in (Wohlgenannt, 2015) have
built an ontological learning system by collecting ev-
idence from heterogeneous sources in a statistical ap-
proach. The candidate concepts were extracted and
the "is a" type of relations was constructed by us-
ing chi − square" co-occurrence significance score.
(Doing-Harris et al., 2015) made use of cosine sim-
ilarity, T F −IDF, also called C −value statistic, and
POS to extract the candidate collocates for construct-
ing an ontology.
In (Jiang and Tan, 2010; Wong et al., 2012),
authors used statistical methods to extract concepts
by computing the relevance of document words
based on term frequency-inverse document frequency
(TD/IDF) and similar measures. Often these methods
are combined.
In the medical field, there are a lot of non-
taxonomic relationships, such as symptoms and eti-
ologies of diseases, indications of medicines, aliases
of diseases or medicines, etc. Among them, ontol-
ogy learning from this type of text plays a big role
in using statistical and linguistic methods. (Mikolov
et al., 2013) propose a skip gram model implemented
in the word2vec system. The key idea is that words
with similar contexts should have similar meanings.
For example, if we see the two sentences "the pa-
tient complained of aortic dissection symptoms" and
"the patient reported aortic dissection symptoms", we
might infer that "complained" means the same thing
as "reported". As a result, these two words should be
Ontology Learning from Clinical Practice Guidelines
313
close in the representation space. (Minarro-Giménez
et al., 2014) learn to embed from unstructured med-
ical corpora crawled from PubMed, Merck Manuals,
Medscape and Wikipedia.
In this paper, we take this line of work further
by showing how to learn medical concept and rela-
tion from medical recommendation text. Specifically,
we show how to use a claims ontology consisting of
cardiovascular disease and recommendation text. We
show that with simple algorithmic adjustments, it is
possible to use the word2vec algorithm to learn em-
beddings on this type of longitudinal. In addition of
linguistic approach, mostly the extraction of concepts
was done together with the concept hierarchy extrac-
tion by looking for specific patterns in the texts. These
patterns included the Hearst patterns and other lexico-
syntactic patterns conducted by Pos tagging techni-
cal from NLP (Liu et al., 2011; Biemann, 2005).
We learn in this process new concepts and relations
that will be added to the ontology. The process of
adding new elements in ontology is called ontology
enrichment. All the previous work are interested to
extract new concepts and relations (taxonomic or se-
mantic) in order to enrich the ontology. However, in
our knowledge, very little works take into account to
enrich ontology by SWRL rules constructed after the
results obtained by ontology learning.
3 PROPOSED APPROACH
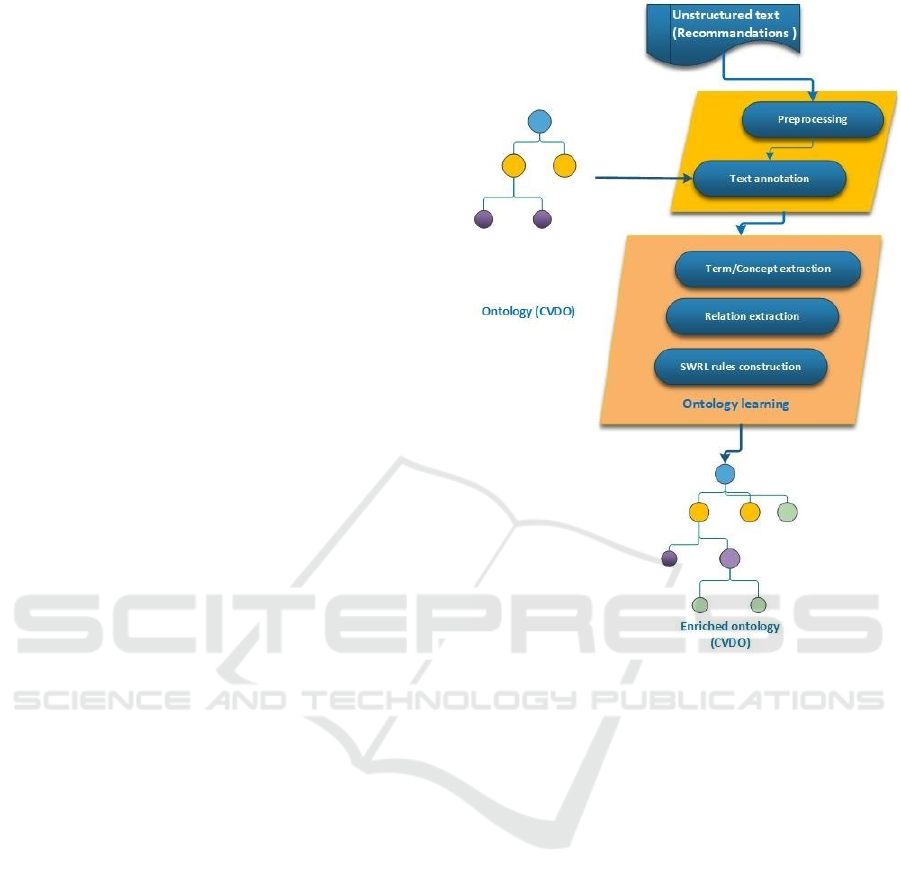
Our approach is described in fig.1 and described be-
low.
3.1 Ontology Learning
Different sub-tasks are included in the ontology learn-
ing: relevant terminology extraction, synonym terms
identification, concepts construction, concepts hierar-
chy organization, learning relations, relations hierar-
chy organization and axioms extraction (Asim et al.,
2018; Gyawali et al., 2017). To accomplish these sub-
tasks, we begin by the indispensable pre-processing
step.
Pre-processing. Our corpus is composed of a set of
medical recommendations defined by the European
Society of Cardiology ESC. An example of a recom-
mendation is presented below.
"If the anatomy is favourable and the expertise
available, endovascular repair (TEVAR) should be
preferred over open surgery."
Figure 1: Approche overview.
To pre-process our corpus, we have used linguis-
tic technical as part of speech tagging, sentence pars-
ing and lemmatization which are linguistic-based pre-
processing techniques used in almost every ontology
learning methodology. More details can be finding in
our previous work presented in (Sbissi et al., 2019).
Term/Concept Extraction. Several approaches use
linguistic method to extract terms and concepts (Is-
mail et al., 2015; Panchenko et al., 2016; Atapattu
et al., 2017). The text is tagged with parts of speech
to extract syntactic structures in a sentence such as
noun phrases and verb phrases.
In (Sbissi et al., 2019), we have used Levenshtein
measure and WordNet ontology to search similarity
between text and the ontology. In order to amelio-
rate our results, we propose to integrate other linguis-
tic and syntactic methods. We also use a statistical
method, Word2vec. Word2vec (Mikolov et al., 2013)
computes continuous vector representations for large
text data-sets. It provides high performance for mea-
suring syntactic and semantic similarities.
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
314
Taxonomic Relations. Subsumption relations (also
known as "is-a" or inclusion relations) provide a tree
view of the ontology and determine inheritance be-
tween concepts.
3.2 Ontology Enrichment
Ontology enrichment consists of adding or modify-
ing the existing ontology by performing one or sev-
eral ontology learning tasks (Mahfoudh et al., 2013).
We are not interested in only simple concepts and tax-
onomic relations, but also in SWRL rules extracted
automatically from medical guidelines. The enrich-
ment process attempts to facilitate text understand-
ing and automatic processing of textual resources,
moving from words to concepts and relationships. It
starts by extracting concepts/relationships from plain
text using linguistic processing such as part-of-speech
(POS) tagging and phrase chunking. The extracted
concepts and relationships are then arranged in the
initial ontology, using syntactic and semantic analy-
sis techniques. The text contains a set of recommen-
dations. The following example presents one recom-
mendation and how it is treated.
"In all patients with AD, medical therapy including
pain relief and blood pressure control is
recommended."
⇓
[(’In’, ’IN’), (’all’, ’DT’), (’patients’, ’NNS’),
(’with’, ’IN’), (’AD’, ’NNP’), (’,’, ’,’), (’medical’,
’JJ’), (’therapy’, ’NN’), (’including’, ’VBG’),
(’pain’, ’NN’), (’relief’, ’NN’), (’and’, ’CC’),
(’blood’, ’NN’), (’pressure’, ’NN’), (’control’,
’NN’), (’is’, ’VBZ’), (’recommended’, ’VBN’)]
Let (NN) be a noun, (VB) be a verb, etc. The
evolution of our ontology CVDO is conducted not
only by some changes of the addition of elements like
(concepts, object properties and data properties) re-
sulting from the ontology learning process but also
by adding the swrl rules extracted from the text of
the recommendations. The following example is an
SWRL extracted from a recommendation.
In patients with abdominal aortic diameter of 25
to 29 mm, new ultrasound imaging should be
considered.
⇓
Patient(?p) ∧hasAbdominalDiametre(?p, ?d) ∧
swrlb : greaterT han(?d, 25) ∧swrlb :
lessT han(?d, 29)− >
recommendedDiagnosis(?p, ”ultrasounImaging”)
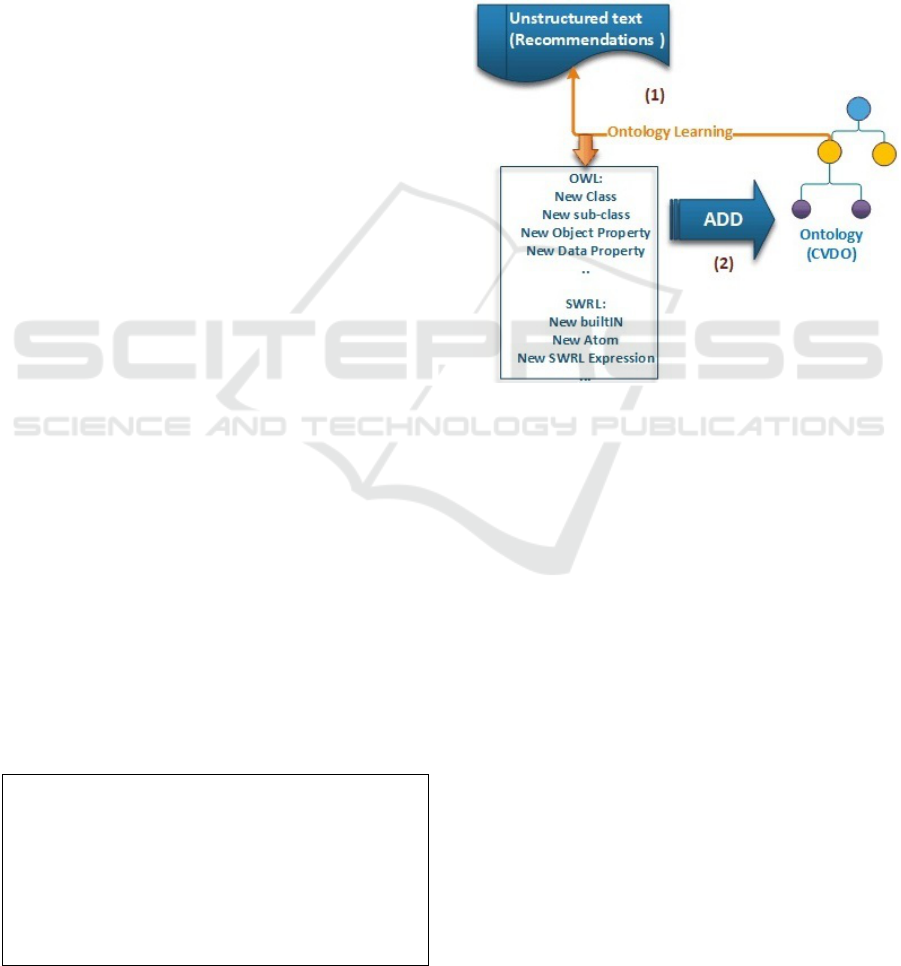
In our case, two big changes are replicated to the
ontology as it is illustrated in figure 2.
• OWL Changes: AddClass, AddSub-
Class, AddDataProperty, AddObjectProp-
erty,AddDataPropertyAssertion, AddObjectProp-
ertyAssertion, AddIndividual, etc.
• SWRL Changes: AddAtom, AddClassAtom,
SWRLBuiltIn, SWRLExpression, etc.
Figure 2: Changes replicated to ontology.
We can benefit from the syntactic relations of the
terms extracted from the text to determine the type of
change that will be added to the ontology. Example
a concept should be a noun, object property and data
property are a verb. After parsing the text, the extrac-
tion of syntactic relations between the terms as well
as the part of speech tag is used. We focus on whether
each extracted term will be a candidate to enrich our
initial ontology or not. We use a similarity measure to
compare each extracted term to the content of the ini-
tial ontology. The most populate change of ontology
enrichment is the aid of a new concept in the ontology.
The following example explains the different steps of
this process.
Ontology Learning from Clinical Practice Guidelines
315
AddClass(Cc,CVDO):
Input:
• Cc: candidate concept to be added (Cc is a
noun(NN)).
• CVDO: our existing ontology.
• Recommandation.txt: the text of recommenda-
tion analysed and parsed.
Case 1:
If Cc does not exist in CVDO Then:
Search similarity between Cc and concepts of
CVDO: Levenshtein measure is used.
• If we obtain a similarity: sim(Cc]=Ci Then
: add semantic relation IsSimilar(Cc)=Ci or
AddLabel.
Case 2:
If Cc no exist in CVDO and no similarity obtained
with concepts of the ontology Then:
AddClass(Cc,CVDO).
An other changes conducted by constructing
SWRL is add a class of atom as represented by the
following steps:
AddClassAtom(C,var):
C should be a class existing in CVDO
• If var is a variable representing an OWL indi-
vidual Then
write C(?var).
• If var is a name of individual Then
write C(var).
3.3 Evaluation of Ontology Evolution
During the process of ontology enrichment guided by
ontology learning, we search to maintain the consis-
tency of the ontology. The managing and the evalua-
tion of the evolution of ontologies can be at different
layers (Petasis et al., 2011):
• Lexical, vocabulary or data layer. We focus on
which concepts and instances have been included
in the ontology and the vocabulary used to iden-
tify them.
• Relational layer: the relations between the con-
cepts of the ontology:
– Hierarchy, taxonomy: an ontology almost al-
ways includes hierarchical inclusion relations
between its concepts.
– Semantic relations: it concerns other relations
besides inclusion and can be evaluated sepa-
rately.
We propose in our approach to preserving consis-
tency: each transformation is defined by a set of
negative application conditions (NAC) and derived
changes (DCH) (Mahfoudh et al., 2015). The ontol-
ogy inconsistencies treated by our work are:
• Data redundancy that can be generated following
and add or rename operation. This type of incon-
sistency is corrected by the NACs.
• Axioms contradiction, the addition of a new ax-
iom should not be accepted if it contradicts an ax-
iom already defined in the ontology. Many cases
are considered: (1) two classes cannot be disjoint
and equivalent at the same time, (2) two classes
that share a subsumption relation cannot be dis-
joints, etc.
In some cases, if we apply a change to one ontology
entity, it can depend on other ontology elements. Re-
ferring to previous work in (Mahfoudh et al., 2015),
the Table 1 presents some changes and the ontol-
ogy concepts which are related. We used the follow-
ing vocabulary: (Class (C), Property (P), ObjectProp-
erty(OP), DataProperty(DP), Individual (I), DataType
(DT)).
Table 1: Dependancy between changes and ontology enti-
ties.
C I OP DP
AddConcept
√
AddIndividual
√ √
AddDataproperty
√
AddObjectProperty
√ √
AddSubClasses
√ √
AddObjectPropertyAssertion
√ √
AddDataPropertyAssertion
√ √
A number of changes could affect the ontology
when it is requested to be reflected in the existing on-
tology. Add concept is the most common change in
any ontology. New concepts emerge and have to be
accommodated in the concept hierarchy (subclass).
The addition of subclasses requires certain conditions
and generates changes at the ontology level.
3.3.1 Add Subclass
The AddSubClass (C1;C2) is defined as follow (Mah-
foudh et al., 2015):
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
316

• Precondition: C1;C2, the classes should exist in
the ontology.
• Negative Changes(NCH):
1. C1 vC2: condition to avoid redundancy. If C1
already is a subclass of C2, we will not add it;
2. C2 v C1: the subsumption relation cannot be
symmetric;
3. C1 v ¬C2:classes which share a subsumption
relation cannot be disjoint;
4. ∃Ci ∈ C(O).(C1 v Ci) ∧(Ci v C2): if there is
a class Ci which is the subClassOf the class C2
and the superClass of C1, then, C1 is already a
subClass of C2;
5. ∃(Ci,C j) ∈ C(O).(Ci v C1) ∧ (C j v C2) ∧
(Ci v ¬C j): classes which share a subsump-
tion relation cannot have subClasses that are
disjoint;
• Results: C1 vC2 , the axiom will be added to the
ontology.
4 IMPLEMENTATION AND
RESULTS
We developed a Java-based implementation to test
our approach. Stanford CoreNLP is used for pre-
processing of text to determine Pos tagging and chun-
ked tree. In the process of ontology learning and en-
richment, we need to read from ontology and to write
or add elements in the ontology. For this task, we
used the Jena API. Our ontology CVDO contains ini-
tially 514 concepts and the recommendations text is
composed of 614 words.
4.1 Search Links
Search links between CVDO ontology and a pre-
processed text of clinical practice guideline refer to
the semantic annotation process. We used the con-
cepts names to produce an expanded list of equivalent
or related terms. Each term of the input text may be
associated with one or more entities from the ontol-
ogy. To find the similarities, we have used (Sbissi
et al., 2019) :
1. exact matching: identifies the identical entities
(String) in the text and in the domain ontology ;
2. morphological matching: identifies the entities
with a morphological correspondence;
3. syntactical similarities: using Levenshtein mea-
sure (Levenshtein, 1966);
4. semantic matching: identifies the synonyms rela-
tions with WordNet ontology.
We present in table 2 the result of links process.
Table 2: Search links.
number of links Links(%)
Initially 28 4.38%
With similarity 190 30%
Only 30% of similarities was extracted. The only
relation between text and ontology is "is-similar-to".
This type of relationship is sufficient to an enrichment
task. We pass to the step 2 that’s ontology learning.
4.2 Ontology Learning
Term Extraction. Linguistic and syntactic analysis is
employed head-modifier principal to identify and ex-
tract complex terms in which the head of the complex
term takes the role of hypernym. X is a hyponym of
Y if Y is a type of X. Example a dissection aortic is a
hyponym of dissection aortic type B.
we used linguistic features (POS, etc.) and word em-
bedding features (word2vec).
Table 3: Word2vec process.
Word2vec Word2vec
(unigram) (bigram)
Iteration1 71.8% 76.2%
IterationN 76.6% 81.8%
We keep the process for word2vec as simple as
possible. After word2vec model generation, we fix
and apply the built-in word2vec similarity function to
get terms related to the seed terms.
In table 3, we iterate the algorithm, In the first itera-
tion, the algorithm needs first user intervention to re-
move from the result file all words that are far from
the domain. After the third iteration, the algorithm
offers automatically correspondence.
On the plus side, the word2vec implementation
is extremely simple and provides a high-percentage
of relevant concept candidates. On the minus side,
candidates suggested by word2vec are (as expected)
sometimes even too strongly related to the seed terms,
for example, syntactic variations such as plural forms
or near-synonyms.
Word2vec with bigram produce a better result. To ex-
plain it, let the same example of the rule:
"In all patients with AD, medical therapy in-
cluding pain relief and blood pressure control is
recommended."
*bigram[0,10]:(’in,’all’), (’all’,patients’),
..(”pain’,’relief’),..,(’blood’,’pressure’).
Ontology Learning from Clinical Practice Guidelines
317
In our case bigram could have good result because
the majority of concepts are composed noun. Also,
the same case to object property.
4.3 Ontology Enrichment
Thanks to the ontology learning technique, we were
able to extract concepts and relations between con-
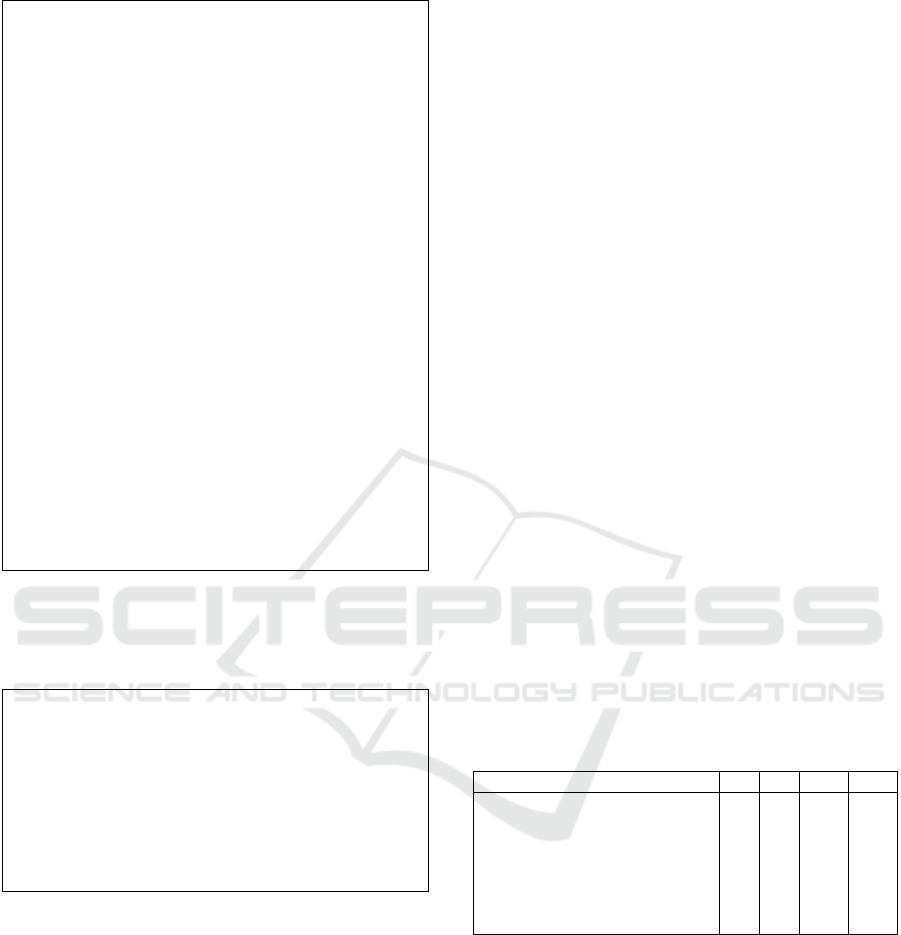
cepts that were missing in the ontology. The Figure
3 represents some concepts, object property and dat
property that we managed to extract.
Figure 3: New elements conducting the ontology enrich-
ment.
The ontology enriched with the rules is shown in
Figure 4. We illustrate in this figure some enriched
concepts with relations.
Figure 4: Enriched ontology.
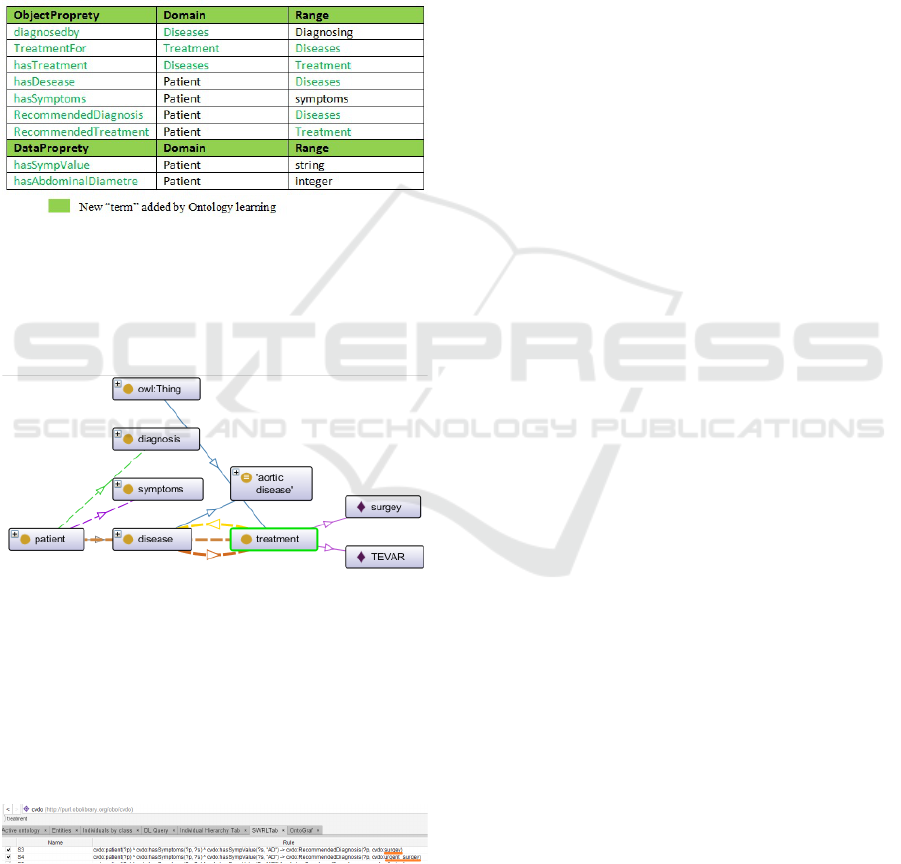
In the Figure 5, we can remark that we have two
recommendations in case of aortic dissection "AD".
Actually, these two rules belong to one recommenda-
tion. Our word2vec extraction process has considered
that surgery and urgent
s
urgey are distinct. So to im-
prove the results we want to use T F −IDF to measure
cosine similarity, T F −IDF, also called C −value.
Figure 5: Conlict rules.
5 CONCLUSION
The paper presented a method to automatically learn-
ing ontology from unstructured text, a clinical prac-
tice guidelines (CG) in order to enrich an existing on-
tology. CG presents a set of recommendations and
knowledge which aims to assist doctors to make de-
cisions about appropriate health care for patients who
are at risk of cardiovascular disease.
The ontology learning process starts with
analysing the text by the pre-processing technics
using the Stanford core NLP. Then, it passes to
relevant terminology extraction, synonym of terms
identification, concepts construction, concept hi-
erarchy organization, learning relations, relations
hierarchy organization and axioms extraction. To
extract term/concept, we used Levenshtein mea-
sure, WordNet ontology and the statistical method
word2vec. For other relations we used the chunking
tree and hearst pattern to search hierarchic relations.
Once these elements are extracted, we have updated
the ontology by adding them. The ontological
enrichment process do not treat only OWL concepts
and axioms but also integrates SWRL rules which
will be used to reasoning tasks.
As a future work, we aim to use enriched ontol-
ogy and the SWRL rules to build a medical decision
support system for the cardiologists.
REFERENCES
Asim, M. N., Wasim, M., Khan, M. U. G., Mahmood, W.,
and Abbasi, H. M. (2018). A survey of ontology learn-
ing techniques and applications. Database, 2018.
Atapattu, T., Falkner, K., and Falkner, N. (2017). A com-
prehensive text analysis of lecture slides to generate
concept maps. Computers & Education, 115:96–113.
Biemann, C. (2005). Ontology learning from text: A survey
of methods. In LDV forum, volume 20, pages 75–93.
Bonacin, R., Pruski, C., and Da Silveira, M. (2013). Ar-
chitecture and services for formalising and evaluating
care actions from computer-interpretable guidelines.
International Journal of Medical Engineering and In-
formatics, 5(3):253–268.
Cabana, M. D., Rand, C. S., Powe, N. R., Wu, A. W.,
Wilson, M. H., Abboud, P.-A. C., and Rubin, H. R.
(1999). Why don’t physicians follow clinical practice
guidelines?: A framework for improvement. Jama,
282(15):1458–1465.
Cimiano, P., Mädche, A., Staab, S., and Völker, J. (2009).
Ontology learning. pages 245–267.
Criado, F. J. (2011). Aortic dissection: a 250-year perspec-
tive. Texas Heart Institute Journal, 38(6):694.
Doing-Harris, K., Livnat, Y., and Meystre, S. (2015). Au-
tomated concept and relationship extraction for the
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
318

semi-automated ontology management (seam) sys-
tem. Journal of biomedical semantics, 6(1):15.
Dramé, K., Diallo, G., Delva, F., Dartigues, J. F., Mouil-
let, E., Salamon, R., and Mougin, F. (2014). Reuse
of termino-ontological resources and text corpora for
building a multilingual domain ontology: an applica-
tion to alzheimer’s disease. Journal of biomedical in-
formatics, 48:171–182.
Drymonas, E., Zervanou, K., and Petrakis, E. G. (2010).
Unsupervised ontology acquisition from plain texts:
the ontogain system. pages 277–287.
Erbel, R., Aboyans, V., Boileau, C., Bossone, E., Bar-
tolomeo, R. D., Eggebrecht, H., Evangelista, A., Falk,
V., Frank, H., et al. (2014). 2014 esc guidelines on the
diagnosis and treatment of aortic diseases. European
heart journal, 35(41):2873–2926.
Francke, A. L., Smit, M. C., de Veer, A. J., and Mistiaen,
P. (2008). Factors influencing the implementation of
clinical guidelines for health care professionals: a sys-
tematic meta-review. BMC medical informatics and
decision making, 8(1):38.
Gardent, C. and Mahfoudh, M. (2016). Overview and com-
parison of existing deep semantic parsers. Technical
report, ModelWriter Project, LORIA-CNRS.
Gyawali, B., Shimorina, A., Gardent, C., Cruz-Lara, S., and
Mahfoudh, M. (2017). Mapping natural language to
description logic. In European Semantic Web Confer-
ence, pages 273–288. Springer.
Horrocks, I., Patel-Schneider, P. F., Boley, H., Tabet, S.,
Grosof, B., Dean, M., et al. (2004). Swrl: A semantic
web rule language combining owl and ruleml. W3C
Member submission, 21:79.
Ismail, R., Bakar, Z. A., and Rahman, N. A. (2015). Extract-
ing knowledge from english translated quran using nlp
pattern. Jurnal Teknologi, 77(19).
Jiang, X. and Tan, A.-H. (2010). Crctol: A semantic-based
domain ontology learning system. Journal of the
American Society for Information Science and Tech-
nology, 61(1):150–168.
Levenshtein, V. I. (1966). Binary codes capable of cor-
recting deletions, insertions and reversals. In Soviet
physics doklady, volume 10, pages 707–710.
Liu, K., Hogan, W. R., and Crowley, R. S. (2011). Nat-
ural language processing methods and systems for
biomedical ontology learning. Journal of biomedical
informatics, 44(1):163–179.
Maedche, A. and Staab, S. (2001). Ontology learning for
the semantic web. IEEE Intelligent systems, 16(2):72–
79.
Mahfoudh, M., Forestier, G., Thiry, L., and Hassenforder,
M. (2013). Consistent ontologies evolution using
graph grammars. In International Conference on
Knowledge Science, Engineering and Management,
pages 64–75. Springer.
Mahfoudh, M., Forestier, G., Thiry, L., and Hassenforder,
M. (2015). Algebraic graph transformations for for-
malizing ontology changes and evolving ontologies.
Knowl.-Based Syst., 73:212–226.
Mikolov, T., Chen, K., Corrado, G., and Dean, J. (2013).
Efficient estimation of word representations in vector
space. arXiv preprint arXiv:1301.3781.
Minarro-Giménez, J. A., Marin-Alonso, O., and Samwald,
M. (2014). Exploring the application of deep learning
techniques on medical text corpora. Studies in health
technology and informatics, 205:584–588.
Panchenko, A., Faralli, S., Ruppert, E., Remus, S., Naets,
H., Fairon, C., Ponzetto, S. P., and Biemann, C.
(2016). Taxi at semeval-2016 task 13: a taxonomy
induction method based on lexico-syntactic patterns,
substrings and focused crawling. In Proceedings of
the 10th International Workshop on Semantic Evalua-
tion (SemEval-2016), pages 1320–1327.
Petasis, G., Karkaletsis, V., Paliouras, G., Krithara, A., and
Zavitsanos, E. (2011). Ontology population and en-
richment: State of the art. pages 134–166.
Sbissi, S., Mahfoudh, M., and Gattoufi, S. (2019). Map-
ping clinical practice guidelines to SWRL rules. pages
283–292.
Séroussi, B., Bouaud, J., Sauquet, D., Giral, P., Cornet,
P., Falcoff, H., and Julien, J. (2010). Why gps do
not follow computerized guidelines: an attempt of ex-
planation involving usability with asti guiding mode.
Studies in health technology and informatics, 160(Pt
2):1236–1240.
Wohlgenannt, G. (2015). Leveraging and balancing het-
erogeneous sources of evidence in ontology learning.
pages 54–68.
Wong, W., Liu, W., and Bennamoun, M. (2012). Ontology
learning from text: A look back and into the future.
ACM Computing Surveys (CSUR), 44(4):20.
Ontology Learning from Clinical Practice Guidelines
319
