
Stochastic Information Granules Extraction for Graph Embedding and
Classification
Luca Baldini
a
, Alessio Martino
b
and Antonello Rizzi
c
1
Department of Information Engineering, Electronics and Telecommunications, University of Rome "La Sapienza",
Via Eudossiana 18, 00184 Rome, Italy
Keywords:
Pattern Recognition, Supervised Learning, Granular Computing, Graph Embedding, Inexact Graph Matching.
Abstract:
Graphs are data structures able to efficiently describe real-world systems and, as such, have been extensively
used in recent years by many branches of science, including machine learning engineering. However, the
design of efficient graph-based pattern recognition systems is bottlenecked by the intrinsic problem of how
to properly match two graphs. In this paper, we investigate a granular computing approach for the design
of a general purpose graph-based classification system. The overall framework relies on the extraction of
meaningful pivotal substructures on the top of which an embedding space can be build and in which the
classification can be performed without limitations. Due to its importance, we address whether information
can be preserved by performing stochastic extraction on the training data instead of performing an exhaustive
extraction procedure which is likely to be unfeasible for large datasets. Tests on benchmark datasets show that
stochastic extraction can lead to a meaningful set of pivotal substructures with a much lower memory footprint
and overall computational burden, making the proposed strategies suitable also for dealing with big datasets.
1 INTRODUCTION
Graphs are powerful data structures able to capture
relationships between elements. This representative
power in describing patterns under a structural and
topological viewpoint makes graphs a flexible and ac-
curate abstraction especially when nodes and/or edges
can be equipped with labels (in this case, we refer to
as labelled graphs). Indeed, they have been widely
used to model a plethora of real-world phenomena,
including biological systems (Giuliani et al., 2014;
Krishnan et al., 2008; Di Paola and Giuliani, 2017),
functional magnetic resonance imaging (Richiardi
et al., 2013), computer vision (Bai, 2012) and online
handwriting (Del Vescovo and Rizzi, 2007b). On the
other hand, it is rather common in pattern recognition
to represent the input pattern as a feature vector ly-
ing in an n-dimensional vector space. This is mainly
due to the relatively simple underlying math whether
some properties are satisfied. In fact, the resulting
space can easily be equipped by an adequate metric
satisfying the properties of non-negativity, identity,
symmetry and triangle inequality (P˛ekalska and Duin,
a
https://orcid.org/0000-0003-4391-2598
b
https://orcid.org/0000-0003-1730-5436
c
https://orcid.org/0000-0001-8244-0015
2005; Martino et al., 2018a; Weinshall et al., 1999).
This can not be easily achieved in structured domains
and, for this reason, the main drawback when repre-
senting entities with graphs is the unpractical, non-
geometric space to whom they belong to. A rather
natural approach to tackle this problem when design-
ing a classification system, is to use an ad-hoc dissim-
ilarity measure working directly in the input space:
this allows to reuse some of the well-known pattern
recognition techniques for supervised learning, e.g.
the K-Nearest Neighbour (K-NN) algorithm (Cover
and Hart, 1967). Related to this approach, we con-
sidered Graph Edit Distances (GEDs) (Neuhaus and
Bunke, 2007) that operate directly in the structured
domain (i.e., graphs), measuring the dissimilarity be-
tween two graphs, say G
1
and G
2
, as the minimum
cost sequence of atomic operations (namely, substi-
tution, deletion and insertion of nodes and/or edges)
needed to transform G
1
into G
2
. A very interest-
ing strategy that has gained much attention relies on
Graph Kernels (Vishwanathan et al., 2010; Ghosh
et al., 2018): these methods exploit the so-called ker-
nel trick, that is the inner product between graphs in a
vector space induced by a (semi)definite positive ker-
nel function. The classification task can heavily rely
on well-known kernelized algorithms, the seminal ex-
Baldini, L., Martino, A. and Rizzi, A.
Stochastic Information Granules Extraction for Graph Embedding and Classification.
DOI: 10.5220/0008149403910402
In Proceedings of the 11th International Joint Conference on Computational Intelligence (IJCCI 2019), pages 391-402
ISBN: 978-989-758-384-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
391

ample being Support Vector Machines (Cortes and
Vapnik, 1995). The last method, that is closely related
to this work, is Graph Embedding. In this approach,
the input pattern from the structured graphs domain
G is mapped into an embedding space D. Clearly, the
designing of the mapping function φ : G → D with
D ⊆ R
m
is crucial in this procedure and some effort
must be ensured to fill the informative and seman-
tic gap between the two domains. For this purpose,
a Granular Computing (Bargiela and Pedrycz, 2008)
approach based on the extraction of information gran-
ules together with symbolic histograms (Del Vescovo
and Rizzi, 2007a) can be pursued in order to obtain
an efficient mapping function able to reflect the infor-
mation carried by the structured data into the vector
space. This method allows the use of common classi-
fication and data-driven systems and can achieve not
only performance similar to the state-of-art classifiers
(Bianchi et al., 2014a), but can also provide useful in-
formation through the extracted granules, as they are
human-interpretable. Unfortunately, an heavy com-
putational effort is necessary and often, as the dataset
size increases, the problem may become unfeasible,
especially under the memory footprint viewpoint.
In this paper, starting from the classification sys-
tem developed by (Bianchi et al., 2014a), we explore
an alternative approach for substructures extraction
that will be used to synthesize the alphabet, i.e. the set
of information granules on the top of which the em-
bedding space is built. In particular, a lighter stochas-
tic procedure has been developed and compared to
exhaustive method from (Bianchi et al., 2014a); this
procedure takes advantage of Breadth First Search
(BFS) and Depth First Search (DFS) algorithms for
graph traversing.
This paper is organized as follows: in Section 2
we give an overview of Granular Computing both as
an information processing paradigm and as a frame-
work in order to build data-driven classification sys-
tems for structured data; in Section 3 we introduce
GRALG, the graph-based classification system core
of this work, highlighting the improvements with re-
spect to its original implementation. Section 4 regards
computational results both in terms of performances
and computational burden with respect to the origi-
nal implementation and, finally, Section 5 draws some
conclusions and future directions.
2 EMBEDDING VIA DATA
GRANULATION
Granular Computing is often described as a human-
centered information processing paradigm (Howard
and Lieberman, 2014; Yao, 2016) based on formal
mathematical entities known as information gran-
ules (Han and Lin, 2010; Bargiela and Pedrycz,
2006). The human-centered computational concept
in soft computing and computational intelligence was
initially developed by Lofti Zadeh through fuzzy
sets (Zadeh, 1979) that exploits human-inspired ap-
proaches to deal with uncertainties and complexities
in data. The process of ’granulation’, intended as the
extraction of meaningful aggregated data, mimics the
human mechanism needed to organize complex data
from the surrounding environment in order to sup-
port decision making activities and describe the world
around (Pedrycz, 2016). For this reason, Granular
Computing can be defined as a framework for analyz-
ing data in complex systems aiming to provide human
interpretable results (Livi and Sadeghian, 2016).
The importance of information granules resides in
the ability to underline properties and relationships
between data aggregates. Specifically, their synthe-
sis can be achieved by following the indistinguisha-
bility rule, according to which elements that show
enough similarity, proximity or functionality shall be
grouped together (Zadeh, 1997). With this approach,
each granule is able to show homogenous semantic in-
formation from the problem at hand (Pedrycz, 2010).
Furthermore, data at hand can be represented using
different levels of ’granularity’ and thus different pe-
culiarities of the considered system can emerge (Yao,
2008; Pedrycz and Homenda, 2013; Yao and Zhao,
2012; Wang et al., 2017; Yang et al., 2018). When
analyzing a system with high level of detail, one shall
expect a huge number of very compact information
granules since, straightforwardly, finer details are of
interest. On the other hand, the level of abstraction
increases when decreasing the granularity level: as a
result, one shall expect a lower number of very pop-
ulated, yet less compact, information granules. De-
pending on this resolution, a problem may exhibit dif-
ferent properties and different atomic units that show
different representations of the system as a whole.
Clearly, an efficient and automatic procedure to se-
lect the most suitable level of abstraction according to
both the problem at hand and the data description is
of utmost importance.
A mainstream approach in order to synthesize a
possibly meaningful set of information granules can
be found in data clustering. Since its direct connec-
tion with the concept of ’granules-as-groups’, cluster
analysis has been widely explored in the context of
granular computing (Pedrycz, 2005; Pedrycz, 2013).
When designing a clustering method for information
granules synthesis, the parameters of the algorithm
must be tuned in an appropriate way in order to select
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
392

the relevant features at a suitable resolution (granu-
larity) for the problem at hand. According to (Ding
et al., 2015), typically three main factors can impact
the resulting data partitioning: (dis)similarity mea-
sure, threshold parameter and cluster representatives.
The threshold defines whether a given pattern belongs
or not to a specific cluster. In our point of view,
this threshold changes the granularity and therefore
the level of detail considered. A typical clustering
algorithm that endows a threshold in order to deter-
mine pattern-to-cluster assignments is the Basic Se-
quential Algorithmic Scheme (BSAS) (Theodoridis
and Koutroumbas, 2008) that performs a so-called
free clustering procedure, i.e. the number of clusters
shall not be defined a-priori as in other data clustering
paradigms, notably k-clustering (Martino et al., 2017;
Martino et al., 2018b; Martino et al., 2019). Varying
the threshold parameter impacts on how patterns will
be aggregated into clusters. A suitable (dis)similarity
function is in charge to measure the (dis)similarity
in order to aggregate data entities in a proper man-
ner. Since the clustering procedure is usually per-
formed in the input (structured) domain, not only the
(dis)similarity measure, but also the cluster represen-
tative shall be tailored accordingly. In order to repre-
sent clusters in structured domains, the medoid (also
called MinSOD) is usually employed (Del Vescovo
et al., 2014) mainly due to the following reason: its
evaluation relies only on pairwise dissimilarities be-
tween patterns belonging to the cluster itself, with-
out any algebraic structures that can not be defined
in non-geometric spaces (Martino et al., 2017). The
clusters representatives from the outcoming partition
can be considered as symbols belonging to an alpha-
bet A = {s
1
,...,s
m
}: these symbols are the pivotal
granules on the top of which the embedding space can
be built thanks to the symbolic histograms paradigm.
According to the latter, each pattern can be described
as an m-length integer-valued vector which counts in
position i the number of occurrences of the i
th
symbol
drawn from the alphabet. The embedding space can
finally be equipped with algebraic structures such as
the Euclidean distance or the dot product and standard
classification systems can be used without limitations.
3 THE GRALG CLASSIFICATION
SYSTEM
GRALG (GRanular computing Approach for La-
belled Graphs) is a general purpose classification sys-
tem suitable for dealing with graphs and based on
Granular Computing. GRALG has been originally
proposed in (Bianchi et al., 2014a) and lately suc-
cessfully applied in the context of image classifica-
tion (Bianchi et al., 2014b; Bianchi et al., 2016). In
this Section, the main blocks of the system are de-
scribed separately (Sections 3.1–3.4), along with the
way they cooperate in order to perform the training
(Section 3.5) and testing phases (Section 3.6).
3.1 Extractor
The goal of this block regards the extraction of sub-
structures from the input set S ⊂ G. In the origi-
nal GRALG implementation, this procedure used to
compute exhaustively the set of possible subgraphs
from any given graph G ∈ S. The maximum order o,
namely the maximum number of vertices for all sub-
graphs, is an input parameter which must be defined
by the end-user. Obviously, the complexity of the pro-
cedure strongly depends on this parameter: in fact,
the asymptotically combinatorial behaviour of an ex-
haustive extraction makes this method unfeasible for
large graphs and/or for high value of o, both in terms
of running time and memory usage. The procedure
used to expand each node of a given graph to a pos-
sible subgraph of order 2, caching in memory the re-
sulting substructures, and then expanding and storing
them iteratively until the desired maximum order o is
reached. At the end of the extraction procedure, the
resulting set of substructures S
g
is returned.
3.1.1 Random Subgraphs Extractor based on
BFS and DFS
The new procedure randomly draws a graph G ∈ S
and then selects a seed node v ∈ G for a traversal strat-
egy based on either BFS or DFS in order to extract a
subgraph. Both the extractions (graph G from S and
node v from G) are performed with uniformly random
distribution. Alongside o (maximum subgraph order),
a new parameter W determines the cardinality of S
g
.
Algorithm 1: Random Extractor.
procedure EXTRACTRND(Graph Set
S = {G
1
,.. .,G
n
} with G = {V ,E}, W max
size of subgraphs set, empty set of subgraphs S
g
, o
max order of extracted subgraph)
while |S
g
| ≤ W do
for order = 1 to o do
Random extract a graph G from S
Random extract a vertex v from V
g = EXTRACT(G,v,order)
S
g
= S
g
∪ g
return Subgraph Set S
g
with |S| = W
Stochastic Information Granules Extraction for Graph Embedding and Classification
393

Algorithm 1, which summarizes this procedure, relies
on a procedure called EXTRACT (separately described
in Algorithm 2) that performs a graph traverse using
one of the two well-known algorithms:
Breadth First Search: Starting from a node v, BFS
performs a traverse throughout the graph explor-
ing first the adjacent nodes of v, namely those with
unitary distance, and then moving farther only af-
ter the neighbourhood is totally discovered. A
First-In-First-Out policy is in charge to organize
the list of neighbours for the considered vertex, in
order to give priority to adjacent nodes. The algo-
rithm can be summerized as follow:
1. Select the starting vertex v.
2. Push v in a queue list Q.
3. Pop u the first element of the queue from Q.
4. For each neighbour s of u, push s to Q if s is not
mark as visited.
5. Mark u as a visited vertex.
6. Repeat 3-5 until Q is empty.
Depth First Search: In this strategy, a given graph
is traversed starting from a seed vertex v, but un-
like the BFS search, the visit follows a path with
increasingly distance from v and backtracks only
after all the vertices from the selected path are dis-
covered. A Last-In-First-Out policy is in charge to
organize the list of neighbours for the considered
vertex, in order to visit in-depth vertices first. The
steps of the algorithm are:
1. Select the starting vertex v.
2. Push v in a stack list S.
3. Pop u the last element from stack S.
4. For each neighbour s of u, push s in S if s is not
marked as visited.
5. Mark u as visited.
6. Repeat 3-5 until S is empty.
Algorithm 2: BFS/DFS graph extraction.
procedure EXTRACT(Graph G, Vertex v, order ac-
tual order of extracted subgraph)
graph g of vertex V
g
and E
g
initially empties
repeat
{V
g
,E
g
} ← BFS/DFSsearch with seed
node v
until |V
g
| = order
g = {V
g
,E
g
}
return g
In Algorithm 2, these methods are employed to pop-
ulate the set of vertices V
g
and edges E
g
for the sub-
graph g: a vertex is added to V
g
as soon as it is marked
as visited, whereas an edge is added to E
g
by consid-
ering the current and the last visited vertices.
3.2 Granulator
This module is in charge to compute the alphabet
symbols A starting from the subgraphs belonging to
the set S
g
, as returned by the Extractor defined in Al-
gorithm 1. The information granules are synthesized
by performing the BSAS clustering algorithm on S
g
.
The BSAS algorithm relies on two parameters Q and
θ, respectively the maximum number of allowed clus-
ters and a threshold dissimilarity below which a pat-
tern can be included in its nearest cluster
1
. Regarding
θ, it is worth noting that different values lead to dif-
ferent partitions and a binary search is deployed to
generate an ensemble of partitions, each of which is
obtained with a different value for θ. For every clus-
ter C in the resulting partitions, a cluster quality index
F(C) is defined as:
F(C) = η · Φ(C) +(1 − η) ·Θ(C) (1)
where the two terms Φ(C) and Θ(C) are defined re-
spectively as:
Φ(C) =
1
|C| − 1
∑
i
d(g
∗
,g
i
) (2)
Θ(C) = 1 −
|C|
|S
g
tr
|
(3)
where, in turn, g
∗
is the representative of cluster C
and g
i
the i
th
pattern in the cluster. In other words,
the quality index (1) sees a convex linear combination
between the compactness Φ(C) and the cardinality
Θ(C), weighted by a parameter η ∈ [0,1]. From Sec-
tion 2, g
∗
is the MinSOD of cluster C, defined as the
element that minimizes the sum of pairwise distances
with respect to all other patterns in the cluster. The
dissimilarity measure driving both Eq. (2) and the
overall clustering procedure is defined as a weighted
GED, described in details in Section 3.2.1. Eq. (1)
needs to be evaluated for all clusters in the partitions
(regardless of the corresponding θ), yet only repre-
sentatives belonging to clusters whose quality index
is above a threshold τ
F
are eligible to be included in
A: in this way, only well-formed clusters (i.e., com-
pact and populated) are considered.
3.2.1 Dissimilarity Measure and Inexact Graph
Matching
The core dissimilarity measure in GRALG is a
weighted GED, which is based on the same ratio-
1
If a pattern cannot be included in one of the available clus-
ters, it can be used to initialize a new cluster, provided that
the number of already-available clusters is below Q.
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
394

nale behind other well-known edit distances, such as
the Levenshtein distance between strings (Cinti et al.,
2019). Accordingly, it is possible to define some edit
operations on graphs: deletion, insertion, substitution
of both nodes and edges. Each of these operations
can be possibly associated to a weight in order to tune
the penalty induced by a particular transformation.
In GRALG, six weights for edit operations are taken
into account in order to establish the importance of
substitutions, deletions and insertions for vertices and
edges.
Formally speaking, the GED between G
1
and G
2
can be defined as a function d : G ×G → R, such that:
d(G
1
,G
2
) = min
(e
1
,...,e
k
)∈X (G
1
,G
2
)
k
∑
i=1
c(e
i
) (4)
where X (G
1
,G
2
) is the (possibly infinite) set of
prospective edit operations needed to transform the
two graphs into one another. Obviously, defining the
costs c(·) for edit operations is the crucial facet in
any GED. The optimal match described in Eq. (4)
is unpractical due to exponential complexity (Bunke
and Allermann, 1983; Bunke, 1997; Bunke, 2000;
Bunke, 2003), thus a suitable algorithm for a subopti-
mal search is mandatory (Tsai and Fu, 1979). In light
of these observations, let us now describe the dissim-
ilarity measure used in GRALG.
Let G
1
= (V
1
,E
1
,L
v
,L
e
), G
2
= (V
2
,E
2
,L
v
,L
e
)
be two fully labelled graphs with nodes and edges
labels set L
v
and L
e
and let o
1
= |V
1
|, o
2
= |V
2
|,
n
1
= |E
1
|, n
2
= |E
2
| be the number of nodes and edges
in the two graphs, respectively. For the sake of gen-
eralization, the two graphs are likely to have different
sizes, hence we suppose o
1
6= o
2
and n
1
6= n
2
. Further,
let us define suitable dissimilarity measures between
vertices and edges, respectively d
π
v
v
: L
v
×L
v
→ R and
d
π
e
e
: L
e
× L
e
→ R, possibly depending on some pa-
rameters π
v
and π
e
(Wang and Sun, 2015; Di Noia
et al., 2019). The strategy adopted in GRALG is
called node Best Match First (nBMF) (Bianchi et al.,
2016): by following a greedy strategy, nBMF matches
most similar nodes first and then matches edges in-
duced by those pairs. The procedure can be divided in
two consecutive routines called VERTEX NBMF and
EDGE NBMF, respectively.
Let us start from the former, technically described
in Algorithm 3. The first node from V
1
is selected
and matched with the most similar node from V
2
ac-
cording to d
π
v
v
. This pair is included in the set of node
matches M . Nodes involved in the pair are then re-
moved from their respective sets and the procedure
iterates until either V
1
or V
2
is empty. In terms of
edit operations, each match counts as a (node) sub-
stitution and the overall cost associated to nodes sub-
stitutions is given by the sum of their respective dis-
similarities. The overall cost for nodes insertions and
deletions is strictly related to the difference between
the two orders. Specifically, if o
1
> o
2
, then we con-
sider (o
1
−o
2
) node insertions. Conversely, if o
1
< o
2
,
then we consider (o
2
− o
1
) node deletions.
Algorithm 3: Node Best Match First Routine 1.
1: procedure VERTEX NBMF(G
1
,G
2
)
2: minDissimilary ← ∞
3: M ←
/
0
4: c
sub
node
= 0
5: repeat
6: Select a node v
a
∈ V
1
7: for all nodes v
b
∈ V
2
do
8: if d
π
v
v
(v
a
,v
b
) ≤ minDissimilarity then
9: minDissimilarity = d
π
v
v
(v
a
,v
b
)
10: V
1
= V
1
r v
a
and V
2
= V
2
r v
b
11: append (v
a
,v
b
) → M
12: c
sub
node
+= minDissimilarity
13: until V
1
=
/
0 ∨ V
2
=
/
0
14: if o
1
> o
2
then
15: c
ins
node
= (o
1
− o
2
)
16: else if o
1
< o
2
then
17: c
del
node
= (o
2
− o
1
)
18: return M , c
sub
node
, c
ins
node
, c
del
node
Now the procedure moves towards edges (Algorithm
4). For each pair of nodes in M , the procedure checks
whether an edge between the two nodes exists in both
E
1
and E
2
: if so, this counts as an edge substitu-
tion and its cost is given by the dissimilarity between
edges according to d
π
e
e
. Conversely, if the two nodes
are connected on G
1
only, this counts as an edge in-
sertion; if the two nodes are connected on G
2
only,
this counts as an edge deletion.
Algorithm 4: Node Best Match First Routine 2.
1: procedure EDGE NBMF(G
1
,G
2
)
2: for all (v
a
,v
b
) ∈ M from VERTEX NBMF do
3: if ∃e
a
∈ E
1
,e
b
∈ E
2
| e
a
= (v
a
,v
b
) ∧ e
b
=
(v
a
,v
b
) then
4: c
sub
edge
+= d
π
e
e
(e
a
,e
b
)
5: else if ∃e
a
∈ E
1
| e
a
= (v
a
,v
b
) then
6: c
ins
edge
+= 1
7: else if ∃e
b
∈ E
2
| e
b
= (v
a
,v
b
) then
8: c
del
edge
+= 1
9: return c
sub
edge
, c
ins
edge
, c
del
edge
By defining c
sub
edge
, c
ins
edge
, c
del
edge
, c
sub
node
, c
ins
node
, c
del
node
as the
overall edit costs on nodes and edges and by defining
Stochastic Information Granules Extraction for Graph Embedding and Classification
395

w
sub
node
, w
sub
edge
, w
ins
node
, w
ins
edge
, w
del
node
, w
del
edge
as the afore-
mentioned non-negative six weights which reflect the
importance of the three atomic operations (insertion,
deletion, substitutions) on nodes and edges, the to-
tal dissimilarity measures on vertices and edges of G
1
and G
2
, say d
V
(V
1
,V
2
) and d
E
(E
1
,E
2
), are respec-
tively computed as:
d
V
(V
1
,V
2
) = w
sub
node
· c
sub
node
+ w
ins
node
· c
ins
node
+ w
del
node
· c
del
node
d
E
(E
1
,E
2
) = w
sub
edge
· c
sub
edge
+ w
ins
edge
· c
ins
edge
+ w
del
edge
· c
del
edge
(5)
In order to avoid skewness due to the different sizes
between G
1
and G
2
, the latter can be normalized as
follows:
d
0
V
(V
1
,V
2
) =
d
V
(V
1
,V
2
)
max(o
1
,o
2
)
d
0
E
(E
1
,E
2
) =
d
E
(E
1
,E
2
)
1
2
(min(o
1
,o
2
) · (min(o
1
,o
2
) − 1))
(6)
And finally:
d(G
1
,G
2
) =
1
2
d
0
V
(V
1
,V
2
) + d
0
E
(E
1
,E
2
)
(7)
3.3 Embedder
This block aims at the definition of an embedding
function φ : G → D that maps the graphs space G into
an m-dimensional space D ⊆ R
m
.
The embedding relies on the symbolic his-
tograms paradigm (Del Vescovo and Rizzi, 2007a;
Del Vescovo and Rizzi, 2007b). After the alphabet
A = {s
1
,.. .,s
m
} has been computed by the Granu-
lator module, the embedding function φ
A
: G → R
m
consists in assigning an integer-valued vector h
(i)
(the
symbolic histogram) to each graph G
i
such that:
h
(i)
= φ
A
(G
i
) = [occ(s
1
),.. .,occ(s
m
)] (8)
where occ : A → N counts the occurrences of the sub-
graphs s
j
∈ A in the input graph G
i
. The counting
process of the symbols s
j
in G
i
is performed thanks to
the same GED described in Section 3.2.1 between s
j
and the subgraphs of G
i
. h
(i)
j
is increased only when
the dissimilarity between a subgraph of G
i
and the
symbol s
j
reach a symbol-dependent threshold value
τ
j
= Φ(C
j
)· ε, where ε is a user-defined tolerance pa-
rameter and C
j
is the cluster whose MinSOD is s
j
.
The resulting embedding space is defined as the space
spanned by the symbolic histograms of the form (8).
A not negligible issue of this procedure is the
computational burden related to the subgraphs extrac-
tion and comparison: the former exhaustive procedure
used to extract all subgraphs up to a desired order
from a given graph G
i
; then, for each subgraph, it used
to compute the GED with respect to all symbols in
A. In order to pursue the goal of avoiding an exhaus-
tive extraction, a lighter procedure has been deployed
and described in Algorithm 5. In this case, the algo-
rithm explores a graph by performing a traverse start-
ing from each node, which acts as seed node for the
BFS or DFS strategy
2
in order to extract subgraphs.
Furthermore, for limiting the number of subgraphs, if
a node v ∈ G already appears in one of the previously
extracted subgraphs, it will not be later considered as
a prospective seed node.
Algorithm 5: Extraction procedure for Embedder.
procedure EXTRACTEMBED(Graph G = {V ,E},
empty set S
ge
, o max order of extracted subgraph)
for all Vertices v in V do
g := empty graph
for order = 1 to o do
g = EXTRACT(G,v,order)
S
ge
= S
ge
∪ g
V = V r V
g
return Subgraph Set S
ge
3.4 Classifier
The classification module in GRALG relies on the K-
NN decision rule. In order to assign the class label to a
previously-unseen pattern, K-NN looks the K nearest
pattern and the classes they belong to and the test pat-
tern is classified according to the most frequent class
amongst the K nearest patterns.
The performance of the whole system is defined as
the accuracy on a given validation/test set, in turn de-
fined as the ratio of correctly classified patterns. It
is worth stressing that the classification procedure is
performed in a metric space due to the embedding
procedure, therefore the K-NN is equipped with a
plain Euclidean distance between vectors (i.e., sym-
bolic histograms).
3.5 Training Phase
The four blocks described in Sections 3.1–3.4 carry
out the atomic functions in GRALG and herein we
describe how they jointly work in order to synthesize
a classification model. Let S ⊂ G be a dataset of la-
belled graphs on nodes and/or edges and let S
tr
, S
vs
and S
ts
be three non-overlapping sets (training, vali-
dation and test set, respectively) drawn from S.
2
The Embedder must follow the same traverse strategy as
the Extractor: both of them shall use either DFS or BFS.
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
396

Table 1: Characteristic of IAM datasets used for testing: size of Training (tr), Validation (vl) and Test (ts) set, number of
classes (# classes), types of nodes and edges labels, average number of nodes and edges, whether the dataset is uniformly
distributed amongst classes or not (Balanced).
Database size (tr, vl, ts) # classes node labels edge labels Avg # nodes Avg # edges Balanced
Letter-L 750, 750, 750 15 R
2
none 4.7 3.1 Y
Letter-M 750, 750, 750 15 R
2
none 4.7 3.2 Y
Letter-H 750, 750, 750 15 R
2
none 4.7 4.5 Y
GREC 286, 286, 528 22 string + R
2
tuple 11.5 12.2 Y
AIDS 250, 250, 1500 2 string + integer + R
2
integer 15.7 16.2 N
The training procedure starts with the Extractor
(Section 3.1) that expands graphs in S
tr
using either
BFS or DFS in order to return the set of subgraphs S
g
tr
which are used as the main input for the Granulator
module.
3.5.1 Optimized Alphabet Synthesis via Genetic
Algorithm
The Granulator block (Section 3.2) depends on sev-
eral parameters whose suitable values are strictly
problem and data-dependent and are hardly known
a-priori. For this reason, a genetic algorithm is in
charge of automatically tune these parameters in or-
der to sythesize the alphabet A.
The genetic code is given by:
[Q τ
F
η Ω Π] (9)
where:
• Q is maximum number of allowed clusters for the
BSAS procedure
• τ
F
is the threshold that discards low quality clus-
ters in order to form the alphabet
• η is the trade-off parameter for weighting com-
pactness and cardinality in the cluster quality in-
dex (1)
• Ω = {w
sub
node
,w
sub
edge
,w
ins
node
,w
ins
edge
,w
del
node
,w
del
edge
} is
the set composed by the six weights for the GED
(see Section 3.2.1)
• Π = {π
v
, π
e
} is the set of parameters for the dis-
similarity measures between nodes d
π
v
v
and edges
d
π
e
e
, if applicable (see Section 3.2.1).
Each individual from the evolving population consid-
ers the set of subgraphs S
g
tr
extracted from S
tr
and runs
several BSAS procedures with different threshold
values θ where at most Q clusters can be discovered in
each run and where the dissimilarity between graphs
is evaluated using the nBMF procedure as in Section
3.2.1 by considering the six weights Ω and (possibly)
the parameters Π, if the vertices and/or nodes dissim-
ilarities are parametric themselves. At the end of the
clustering procedures, each cluster is evaluated thanks
to the quality index (1) using the parameter η for
weighting the convex linear combination and clusters
whose value is above τ
F
are discarded and their rep-
resentatives will not form the alphabet. Once the al-
phabet A is synthesized, the Embedder (Section 3.3)
extracts S
ge
tr
and S
ge
vs
from S
tr
and S
vs
and exploits A
in order to map both the training set and the validation
set towards a metric space (say D
tr
and D
vs
) using the
same GED previously used for BSAS, along with the
corresponding parameters Ω and Π. The classifier is
trained on D
tr
and its accuracy is evaluated on D
vs
.
The latter serves as the fitness function for the indi-
vidual itself. Standard genetic operators (mutation,
selection, crossover and elitism) take care of moving
from one generation to the next. At the end of the evo-
lution, the best individual is retained, especially the
portions of the genetic code Ω
?
and Π
?
, along with
the alphabet A
?
synthesized using its genetic code.
3.5.2 Feature Selection Phase
The Granulator may produce a large set of symbols in
A
?
, hence the dimension of the embedding space may
result large as well. In order to shrink the dimension-
ality of the embedding space (i.e., the set of mean-
ingful symbols), a feature selection procedure still
based on genetic optimization is in charge to discard
unpromising features, hence reducing the number of
symbols in A
?
, with a projection mask m ∈ {0,1}
|A
?
|
:
features corresponding to 1’s are retained, whereas
features corresponding to 0’s are discarded. The pro-
jection mask is the genetic code for this second ge-
netic optimization stage.
In this optimization step, each individual from the
evolving population projects D
tr
and D
vs
on the sub-
space marked by non-zero elements in m, say D
tr
and
D
vs
. The classifier is trained on D
tr
and its accuracy
is evaluated on D
vs
. The fitness function is defined
as a convex linear combination between the classifier
accuracy on D
vs
and the cost µ of the mask m defined
as:
µ = |m == 1| / |m| (10)
Stochastic Information Granules Extraction for Graph Embedding and Classification
397

weighted by a parameter α ∈ [0,1] which weights per-
formances and sparsity. At the end of the evolution
the best projection mask m
?
is retained and used in
order to consider the reduced alphabet A
?
.
3.6 Synthesized Classification Model
From the two genetic optimization procedures, Π
?
,
Ω
?
and A
?
are the main actors which completely char-
acterize the classification model, hence the key com-
ponents in order to classify previously unseen test
data. Specifically, given a set of test data S
ts
, the Em-
bedder evaluates S
ge
ts
and performs the symbolic his-
tograms embedding by matching symbols in A
?
using
the GED equipped with parameters Ω
?
and Π
?
(if ap-
plicable).
The K-NN classifier is trained on
D
?
tr
, namely the
training set projected using the best projection mask
m
?
and the final performance is evaluated on the em-
bedded test data.
4 TEST AND RESULTS
For addressing the proposed improvements over the
original GRALG implementation, different graph
datasets from the IAM repository (Riesen and Bunke,
2008) are considered (see Table 1 for list and descrip-
tion). Since labelled graphs on both nodes and edges
have been considered, suitable dissimilarity measures
have to be defined as well (cf. Section 3.2.1):
• Letter: Node labels are real-valued 2-dimensional
vectors v of x,y coordinates and therefore the dis-
similarity measure d
v
between two given nodes,
say v
(a)
and v
(b)
, is defined as the plain Euclidean
distance:
d
v
(v
(a)
,v
(b)
) = kv
(a)
− v
(b)
k
2
Conversely, edges are not labelled.
• AIDS: Node labels are composed by a string value
S
chem
(chemical symbol), an integer N
ch
(charge)
and a real-valued 2-dimensional vector v of x,y
coordinates. For any two given nodes, their dis-
similarity is evaluated as:
d
v
(v
(a)
,v
(b)
) = kv
(a)
− v
(b)
k
2
+ |N
(a)
ch
− N
(b)
ch
|+
+ d
s
(S
(a)
chem
,S
(b)
chem
)
where d
s
(S
(a)
chem
,S
(b)
chem
) = 1 if S
(a)
chem
6= S
(b)
chem
, and
0 otherwise. Conversely, the edge dissimilarity
is discarded since not useful for the classification
task.
• GREC: Node labels are composed by a string
(type) and a real-valued 2-dimensional vector v.
The dissimilarity measure d
v
between two differ-
ent nodes is then defined as:
d
v
(v
(a)
,v
(b)
) =
(
1 if type
(a)
= type
(b)
kv
(a)
− v
(b)
k
2
otherwise
Edge labels are defined by an integer value
f req (frequency) that defines the number of
(type,angle)-pairs where, in turn, type is a string
which may assume two values (namely, arc or
line) and angle is a real number. Given two edges,
say e
(a)
and e
(b)
their dissimilarity is defined as
follows:
1. If f req
(a)
= f req
(b)
= 1
d
e
(e
(a)
,e
(b)
) =
α · d
line
(angle
(a)
,angle
(b)
)
if type
(a)
= type
(b)
= line
β · d
arc
(angle
(a)
,angle
(b)
)
if type
(a)
= type
(b)
= arc
γ otherwise
2. If f req
(a)
= f req
(b)
= 2
d
e
(e
(a)
,e
(b)
) =
α
2
· d
line
(angle
(a)
1
,angle
(b)
1
)+
+
β
2
· d
arc
(angle
(a)
2
,angle
(b)
2
)
if type
(a)
= type
(b)
= line
α
2
· d
line
(angle
(a)
2
,angle
(b)
2
)+
+
β
2
· d
arc
(angle
(a)
1
,angle
(b)
1
)
if type
(a)
= type
(b)
= arc
γ otherwise
3. If f req
(a)
6= f req
(b)
d
e
(e
(a)
,e
(b)
) = δ
where d
line
and d
arc
are the module distance nor-
malized respectively in [−π,π] and [0, arc
max
].
α,β,γ, δ ∈ [0,1] is the set of parameters Π defined
in Section 3.5.1 which shall be optimized by the
genetic algorithm.
Table 2: Number of subgraphs extracted (o = 5) by the ex-
haustive procedure.
Dataset |S
g
tr
| |S
g
vl
| |S
g
ts
|
Letter-L 21165 20543 21435
Letter-M 8582 8489 8560
Letter-H 8193 7976 8111
GREC 27119 28581 50579
AIDS 35208 35692 220108
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
398

The implementation has been developed in C++, us-
ing the SPARE
3
(Livi et al., 2014) and Boost li-
braries
4
. Tests have been performed on a work-
station with Linux Ubuntu 18.04, 4-cores Intel i7-
3770K@3.50GHz equipped with 32GB of RAM.
For the sake of benchmarking, the number of sub-
graphs extracted from the training set, validation set
and test set by the former exhaustive procedure has
been reported in Table 2. In our tests, we followed
the random extraction procedure defined in Algorithm
1, setting up the maximum number of allowed sub-
graphs W equal to a given percentage of |S
g
tr
| (cf. Ta-
ble 2). The subgraphs for the embedding strategy are
extracted by following the procedure described in Al-
gorithm 5. Both of the traverse strategies (BFS and
DFS) have been considered for comparison and the
number of resulting subgraphs needed for the embed-
ding procedure are reported in Table 3.
The system parameters are defined as follow:
– W = 10%, 30%, 50% of |S
g
tr
|
– o = 5 the maximum order of the extracted sub-
graphs
– 20 individuals for the population of both genetic
algorithms
– 20 generations for the first genetic algorithm (al-
phabet optimization)
– 50 generations for the second genetic algorithm
(feature selection)
– α = 1 in the fitness function for the second genetic
algorithm (no weight to sparsity)
– K = 5 for the K-NN classifier
– ε = 1.1 as tolerance value for the symbolic his-
tograms evaluation.
Table 3: Number of subgraphs extracted for the embedding
block using Algorithm 5 with BFS and DFS.
Dataset Traverse |S
ge
tr
| |S
ge
vl
| |S
ge
ts
|
Letter-L
BFS 5451 5371 5428
DFS 4266 4192 4253
Letter-M
BFS 5311 5293 5243
DFS 4336 4234 4213
Letter-H
BFS 4513 4355 4305
DFS 4495 4391 4290
AIDS
BFS 6701 6833 41149
DFS 11776 11893 71294
GREC
BFS 5141 5119 9508
DFS 6076 6219 11223
3
https://sourceforge.net/projects/libspare/
4
http://www.boost.org/
In Figure 1, we compare the performances achieved
by the exhaustive procedure in terms of accuracy on
the test set (in percentage) and total wall-clock time
(in minutes) against the proposed subsampling proce-
dures. Due to the intrinsic randomness in the training
procedures, results herein presented have been aver-
aged across five runs. The random extraction proce-
dure has been tested with three different values of W ,
up to a maximum subgraph order o. It is notewor-
thy that aim of our analyses is to investigate on how
the subsampling rate impacts on accuracy, memory
footprint and running times: as such, all parameters
except W itself have been kept constant.
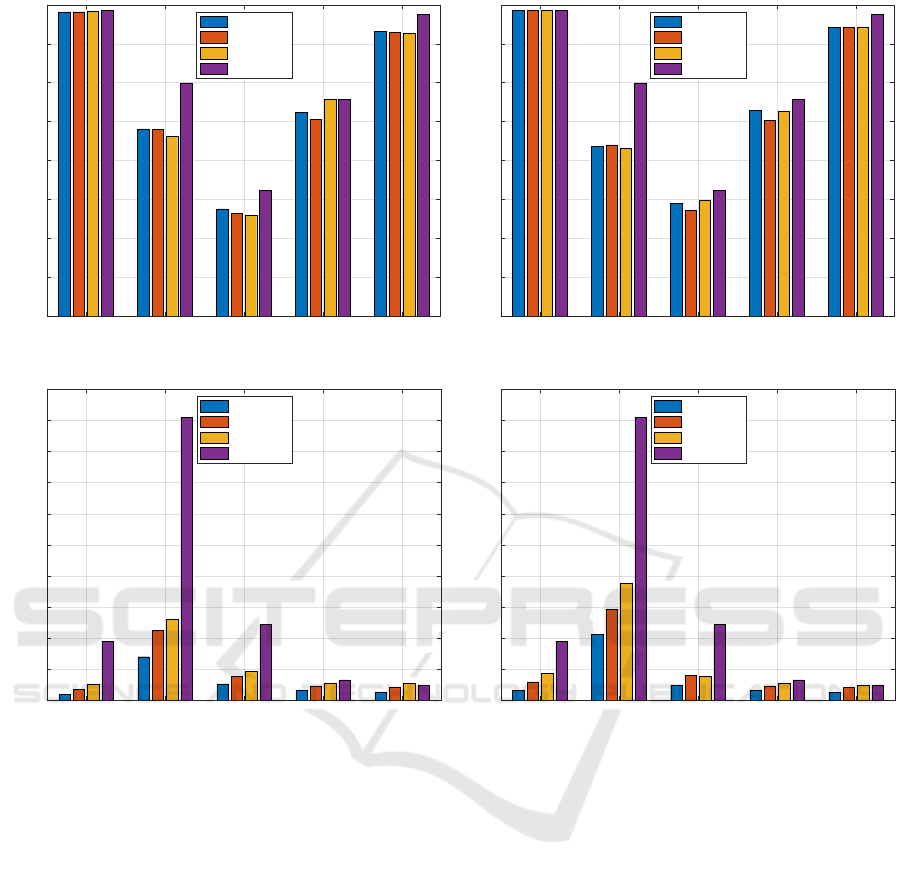
By matching Figures 1a and 1b, it is possible to
see that the novel strategies lead to comparable re-
sults (in terms of accuracy) with those obtained by
the exhaustive procedure for every value of W . The
only remarkable shift can be observed for GREC (ap-
proximately 5%). It is worth remarking that the per-
formances of the classification block are strongly in-
fluenced by the efficiency of the mapping function
in preserving the graph input space properties into
the R
m
space. This can be achieved only if the in-
formation granules extracted are indeed meaningful
representatives of the considered dataset(s). For all
datasets, clearly some properties emerge even by per-
forming a strong subsampling of the prospective sub-
graphs.
Other than comparable results in terms of accu-
racy, remarkable improvements in terms of running
time can be observed as well (Figures 1c and 1d).
This is due to the lower number of subgraphs returned
by the Extractor driving mainly the Granulator and
due to the traverse strategy adopted by the Embed-
der before the evaluation of the symbolic histograms.
Recalling Section 3.5.1, the genetic algorithm must
repeat several times the entire procedure of granula-
tion, embedding and classification in order to opti-
mize the parameters involved. This task involves the
GED computation many times, which can be very in-
tensive and time consuming. By matching Table 1
and Figures 1c–1d, clearly the advantages of subsam-
pling are more and more evident as the dataset size
increases and/or in presence of complex semantic in-
formation on nodes/edges, as their dissimilarity mea-
sures impact the overall GED computational burden.
5 CONCLUSIONS
In this paper, we addressed the possibility of design-
ing a Granular Computing-based classification system
for labelled graphs by performing stochastic extrac-
tion procedures on the training data in order to im-
Stochastic Information Granules Extraction for Graph Embedding and Classification
399
AIDS GREC Letter-H Letter-M Letter-L
60
65
70
75
80
85
90
95
100
Accuracy [%]
W = 10%
W = 30%
W = 50%
Exhaustive
(a) Accuracy on the Test Set (BFS)
AIDS GREC Letter-H Letter-M Letter-L
60
65
70
75
80
85
90
95
100
Accuracy [%]
W = 10%
W = 30%
W = 50%
Exhaustive
(b) Accuracy on the Test Set (DFS)
AIDS GREC Letter-H Letter-M Letter-L
0
20
40
60
80
100
120
140
160
180
200
Time [min]
W = 10%
W = 30%
W = 50%
Exhaustive
(c) Overall Running Time (BFS)
AIDS GREC Letter-H Letter-M Letter-L
0
20
40
60
80
100
120
140
160
180
200
Time [min]
W = 10%
W = 30%
W = 50%
Exhaustive
(d) Overall Running Time (DFS)
Figure 1: Comparison between the exhaustive procedure and the proposed stochastic sampling.
prove the information granulation procedure both in
terms of running time and memory footprint. The hy-
pothesis behind a stochastic granulation procedure is
that the information (regularities), whether present in
the dataset, can still be observed if subsamples of the
dataset itself are considered. In plainer words, mean-
ingful clusters are still visible.
In order to prove this concept, we equipped
GRALG with a different granulation procedure that
instead of finding information granules on the en-
tire set of possible subgraphs, such subgraphs are
extracted by performing stochastic extraction proce-
dures driven by well-known graph traversing algo-
rithms, namely DFS and BFS. These two strategies
are also considered when building the embedding
space, since the symbolic histograms paradigm relies
on counting how many times the symbols from the al-
phabet appear in the original graphs. Indeed, DFS and
BFS have been used to traverse the input graphs and
match the resulting subgraphs with the alphabet.
This lightweight procedure for extracting sub-
graphs both at granulation stage and at embedding
stage drastically outperforms the former exhaustive
procedure in terms of memory footprint and running
times and, at the same time, results in terms of accu-
racy on the test set are comparable with respect to the
former case. The achieved results somehow prove our
hypothesis, at least for the considered datasets, show-
ing that clustering techniques may be promising for
synthesizing information granules even with random
subsampling. This is particularly crucial in Big Data
scenarios, where the memory footprint is a delicate
issue and where redundancies and noisy patterns can
easily be found in massive datasets.
Nonetheless, the overall system keeps the pe-
culiar properties typical of information granulation-
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
400

based systems, namely the human-interpretability of
the synthesized model. Indeed, the resulting informa-
tion granules can give insights to field-experts about
the modelled system. This aspect is stressed by the
second genetic optimization, which is in charge of
shrinking the alphabet size, hence finding the subset
of information granules better related to the semantic
behind the classification problem at hand.
As already mentioned, subsampling procedures
are appealing especially in Big Data scenarios. As
such, future research avenues can consider the im-
plementation of the proposed alphabet synthesis tech-
niques in parallel and distributed frameworks (Dean
and Ghemawat, 2008; Zaharia et al., 2010), even-
tually following multi-agent paradigms (Cao et al.,
2009; Altilio et al., 2019), or by means of dedicated
hardware (Tran et al., 2016; Cinti et al., 2019) in or-
der to properly face massive datasets and/or datasets
with non-trivial semantic information on both nodes
and edges. Thanks to these paradigms, the dataset can
be shred across several computational units and, most
importantly, the GED evaluation can be performed in
parallel, being it the most computationally expensive
step in the synthesis procedure.
REFERENCES
Altilio, R., Di Lorenzo, P., and Panella, M. (2019). Dis-
tributed data clustering over networks. Pattern Recog-
nition, 93:603 – 620.
Bai, X. (2012). Graph-Based Methods in Computer Vision:
Developments and Applications: Developments and
Applications. IGI Global.
Bargiela, A. and Pedrycz, W. (2006). The roots of granular
computing. In 2006 IEEE International Conference
on Granular Computing, pages 806–809.
Bargiela, A. and Pedrycz, W. (2008). Toward a theory
of granular computing for human-centered informa-
tion processing. IEEE Transactions on Fuzzy Systems,
16(2):320–330.
Bianchi, F. M., Livi, L., Rizzi, A., and Sadeghian, A.
(2014a). A granular computing approach to the de-
sign of optimized graph classification systems. Soft
Computing, 18(2):393–412.
Bianchi, F. M., Scardapane, S., Livi, L., Uncini, A., and
Rizzi, A. (2014b). An interpretable graph-based im-
age classifier. In 2014 International Joint Conference
on Neural Networks (IJCNN), pages 2339–2346.
Bianchi, F. M., Scardapane, S., Rizzi, A., Uncini, A.,
and Sadeghian, A. (2016). Granular computing tech-
niques for classification and semantic characterization
of structured data. Cognitive Computation, 8(3):442–
461.
Bunke, H. (1997). On a relation between graph edit distance
and maximum common subgraph. Pattern Recogni-
tion Letters, 18(8):689 – 694.
Bunke, H. (2000). Graph matching: Theoretical founda-
tions, algorithms, and applications. In Proceedings of
Vision Interface, pages 82–88.
Bunke, H. (2003). Graph-based tools for data mining and
machine learning. In Perner, P. and Rosenfeld, A., ed-
itors, Machine Learning and Data Mining in Pattern
Recognition, pages 7–19, Berlin, Heidelberg. Springer
Berlin Heidelberg.
Bunke, H. and Allermann, G. (1983). Inexact graph match-
ing for structural pattern recognition. Pattern Recog-
nition Letters, 1(4):245 – 253.
Cao, L., Gorodetsky, V., and Mitkas, P. A. (2009). Agent
mining: The synergy of agents and data mining. IEEE
Intelligent Systems, 24(3):64–72.
Cinti, A., Bianchi, F. M., Martino, A., and Rizzi, A. (2019).
A novel algorithm for online inexact string matching
and its fpga implementation. Cognitive Computation.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
Machine learning, 20(3):273–297.
Cover, T. M. and Hart, P. E. (1967). Nearest neighbor pat-
tern classification. IEEE Transactions on Information
Theory, 13(1):21–27.
Dean, J. and Ghemawat, S. (2008). Mapreduce: simplified
data processing on large clusters. Communications of
the ACM, 51(1):107–113.
Del Vescovo, G., Livi, L., Frattale Mascioli, F. M., and
Rizzi, A. (2014). On the problem of modeling struc-
tured data with the minsod representative. Interna-
tional Journal of Computer Theory and Engineering,
6(1):9.
Del Vescovo, G. and Rizzi, A. (2007a). Automatic classi-
fication of graphs by symbolic histograms. In 2007
IEEE International Conference on Granular Comput-
ing (GRC 2007), pages 410–416. IEEE.
Del Vescovo, G. and Rizzi, A. (2007b). Online handwrit-
ing recognition by the symbolic histograms approach.
In 2007 IEEE International Conference on Granular
Computing (GRC 2007), pages 686–686. IEEE.
Di Noia, A., Martino, A., Montanari, P., and Rizzi, A.
(2019). Supervised machine learning techniques and
genetic optimization for occupational diseases risk
prediction. Soft Computing.
Di Paola, L. and Giuliani, A. (2017). Protein–Protein Inter-
actions: The Structural Foundation of Life Complex-
ity, pages 1–12. American Cancer Society.
Ding, S., Du, M., and Zhu, H. (2015). Survey on granularity
clustering. Cognitive neurodynamics, 9(6):561–572.
Ghosh, S., Das, N., Gonçalves, T., Quaresma, P., and
Kundu, M. (2018). The journey of graph kernels
through two decades. Computer Science Review,
27:88–111.
Giuliani, A., Filippi, S., and Bertolaso, M. (2014). Why
network approach can promote a new way of thinking
in biology. Frontiers in Genetics, 5:83.
Han, J. and Lin, T. Y. (2010). Granular computing: Models
and applications. International Journal of Intelligent
Systems, 25(2):111–117.
Howard, N. and Lieberman, H. (2014). Brainspace: Re-
lating neuroscience to knowledge about everyday life.
Cognitive Computation, 6(1):35–44.
Stochastic Information Granules Extraction for Graph Embedding and Classification
401

Krishnan, A., Zbilut, J. P., Tomita, M., and Giuliani, A.
(2008). Proteins as networks: usefulness of graph the-
ory in protein science. Current Protein and Peptide
Science, 9(1):28–38.
Livi, L., Del Vescovo, G., Rizzi, A., and Frattale Mas-
cioli, F. M. (2014). Building pattern recognition
applications with the spare library. arXiv preprint
arXiv:1410.5263.
Livi, L. and Sadeghian, A. (2016). Granular comput-
ing, computational intelligence, and the analysis of
non-geometric input spaces. Granular Computing,
1(1):13–20.
Martino, A., Giuliani, A., and Rizzi, A. (2018a). Gran-
ular computing techniques for bioinformatics pat-
tern recognition problems in non-metric spaces. In
Pedrycz, W. and Chen, S.-M., editors, Computational
Intelligence for Pattern Recognition, pages 53–81.
Springer International Publishing, Cham.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M. (2017).
Efficient approaches for solving the large-scale k-
medoids problem. In Proceedings of the 9th Inter-
national Joint Conference on Computational Intelli-
gence - Volume 1: IJCCI,, pages 338–347. INSTICC,
SciTePress.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M. (2018b).
Distance matrix pre-caching and distributed computa-
tion of internal validation indices in k-medoids clus-
tering. In 2018 International Joint Conference on
Neural Networks (IJCNN), pages 1–8.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M.
(2019). Efficient approaches for solving the large-
scale k-medoids problem: Towards structured data.
In Sabourin, C., Merelo, J. J., Madani, K., and War-
wick, K., editors, Computational Intelligence: 9th In-
ternational Joint Conference, IJCCI 2017 Funchal-
Madeira, Portugal, November 1-3, 2017 Revised Se-
lected Papers, pages 199–219. Springer International
Publishing, Cham.
Neuhaus, M. and Bunke, H. (2007). Bridging the gap be-
tween graph edit distance and kernel machines, vol-
ume 68. World Scientific.
Pedrycz, W. (2005). Knowledge-based clustering: from
data to information granules. John Wiley & Sons.
Pedrycz, W. (2010). Human centricity in computing with
fuzzy sets: an interpretability quest for higher order
granular constructs. Journal of Ambient Intelligence
and Humanized Computing, 1(1):65–74.
Pedrycz, W. (2013). Proximity-based clustering: a search
for structural consistency in data with semantic blocks
of features. IEEE Transactions on Fuzzy Systems,
21(5):978–982.
Pedrycz, W. (2016). Granular computing: analysis and de-
sign of intelligent systems. CRC press.
Pedrycz, W. and Homenda, W. (2013). Building the
fundamentals of granular computing: A principle
of justifiable granularity. Applied Soft Computing,
13(10):4209 – 4218.
P˛ekalska, E. and Duin, R. P. (2005). The dissimilarity rep-
resentation for pattern recognition: foundations and
applications.
Richiardi, J., Achard, S., Bunke, H., and Van De Ville, D.
(2013). Machine learning with brain graphs: predic-
tive modeling approaches for functional imaging in
systems neuroscience. IEEE Signal Processing Mag-
azine, 30(3):58–70.
Riesen, K. and Bunke, H. (2008). Iam graph database
repository for graph based pattern recognition and ma-
chine learning. In Joint IAPR International Work-
shops on Statistical Techniques in Pattern Recognition
(SPR) and Structural and Syntactic Pattern Recogni-
tion (SSPR), pages 287–297. Springer.
Theodoridis, S. and Koutroumbas, K. (2008). Pattern
Recognition. Academic Press, 4 edition.
Tran, H.-N., Cambria, E., and Hussain, A. (2016). Towards
gpu-based common-sense reasoning: Using fast sub-
graph matching. Cognitive Computation, 8(6):1074–
1086.
Tsai, W.-H. and Fu, K.-S. (1979). Error-correcting isomor-
phisms of attributed relational graphs for pattern anal-
ysis. IEEE Transactions on systems, man, and cyber-
netics, 9(12):757–768.
Vishwanathan, S. V. N., Schraudolph, N. N., Kondor, R.,
and Borgwardt, K. M. (2010). Graph kernels. Journal
of Machine Learning Research, 11(Apr):1201–1242.
Wang, F. and Sun, J. (2015). Survey on distance metric
learning and dimensionality reduction in data mining.
Data Mining and Knowledge Discovery, 29(2):534–
564.
Wang, G., Yang, J., and Xu, J. (2017). Granular
computing: from granularity optimization to multi-
granularity joint problem solving. Granular Comput-
ing, 2(3):105–120.
Weinshall, D., Jacobs, D. W., and Gdalyahu, Y. (1999).
Classification in non-metric spaces. In Kearns, M. J.,
Solla, S. A., and Cohn, D. A., editors, Advances
in Neural Information Processing Systems 11, pages
838–846. MIT Press.
Yang, J., Wang, G., and Zhang, Q. (2018). Knowledge
distance measure in multigranulation spaces of fuzzy
equivalence relations. Information Sciences, 448:18–
35.
Yao, Y. (2016). A triarchic theory of granular computing.
Granular Computing, 1(2):145–157.
Yao, Y. and Zhao, L. (2012). A measurement theory view
on the granularity of partitions. Information Sciences,
213:1–13.
Yao, Y.-Y. (2008). The rise of granular computing. Journal
of Chongqing University of Posts and Telecommuni-
cations (Natural Science Edition), 20(3):299–308.
Zadeh, L. A. (1979). Fuzzy sets and information granu-
larity. Advances in fuzzy set theory and applications,
11:3–18.
Zadeh, L. A. (1997). Toward a theory of fuzzy information
granulation and its centrality in human reasoning and
fuzzy logic. Fuzzy sets and systems, 90(2):111–127.
Zaharia, M., Chowdhury, M., Franklin, M. J., Shenker, S.,
and Stoica, I. (2010). Spark: Cluster computing with
working sets. In Proceedings of the 2nd USENIX
Conference on Hot Topics in Cloud Computing, Hot-
Cloud’10, pages 10–10. USENIX Association.
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
402
