
Development of a Gestational Diabetes Computer Interpretable
Guideline using Semantic Web Technologies
Garazi Artola
1
, Jordi Torres
1
, Nekane Larburu
1,2
, Roberto Álvarez
1,2
and Naiara Muro
1,2,3,4,5
1
Vicomtech Research Centre, Mikeletegi Pasalekua 57, 20009, San Sebastian, Spain
2
Biodonostia Health Research Institute, P. Doctor Begiristain s/n, 20014, San Sebastian, Spain
3
UPMC, Univ. Paris 06, Paris, France
4
INSERM, Université Paris 13, Paris, France
5
LIMICS, Sorbonne Paris Cité, UMR S 1142, Paris, France
Keywords: Computer Interpretable Guideline, Semantic Web Technologies, Ontology, Gestational Diabetes, Decision
Support System.
Abstract: The benefits of following Clinical Practice Guidelines (CPGs) in the daily practice of medicine have been
widely studied, being a powerful method for standardization and improvement of medical care quality.
However, applying these guidelines to promote evidence-based and up-to-date clinical practice is a known
challenge due to the lack of digitalization of clinical guidelines. In order to overcome this issue, the use of
Clinical Decision Support Systems (CDSS) has been promoted in clinical centres. Nevertheless, CPGs must
be formalized in a computer interpretable way to be implemented within CDSS. Moreover, these systems are
usually developed and implemented using local setups, and hence local terminologies, which causes lack of
semantic interoperability. In this context, the implementation of Semantic Web Technologies (SWTs) to
formalize the concepts used in guidelines promotes the interoperability and standardization of those systems.
In this paper, an architecture that allows the formalization of CPGs into Computer Interpretable Guidelines
(CIGs) supported by an ontology in the gestational diabetes domain is presented. This CIG has been
implemented within a CDSS and a mobile application has been developed for guiding patients based on up-
to-date evidence based clinical guidelines.
1 INTRODUCTION
Clinical practice is based on the latest and most
reliable clinical evidence to provide the best
healthcare to patients. During the last years, studies
have shown the benefits of following Clinical
Practice Guidelines (CPGs) in the daily practice of
medicine (Grimshaw and Russell, 1993), being a
powerful method for standardization and
improvement of the medical care quality. According
to the Institute of Medicine’s (IOM) definition, CPGs
are “systematically developed statements to assist
practitioner and patient decisions about appropriate
health care for specific clinical circumstances”
(Institute of Medicine, 1990).
However, applying these guidelines to promote
the best evidence-based and most up-to-date clinical
practice is a known challenge. There is a lack of
digitalization of the guidelines, which makes it
difficult to maintain them updated in a dynamic way
and implement them in computerized systems.
In order to overcome these issues, Clinical
Decision Support Systems (CDSS) that formalize
guidelines in a computer interpretable way (i.e. as
Computer Interpretable Guidelines or CIGs) are
promoted. In this context, the application of Semantic
Web Technologies (SWTs) to formalize the
guidelines’ concepts could be a key to promote the
interoperability and standardization of the clinical
knowledge, by giving the opportunity to pursue a
reuse of ontologies.
In this paper, an architecture that will allow the
formalization of CPGs into CIGs, supported by an
ontology to assure a semantic validity of all the
formalized information is presented. As a use case,
the implementation of a gestational diabetes CIG is
described. Moreover, a mobile based application is
presented as the front-end of the CDSS for a patient-
oriented guidance.
This paper is organized as follows. Section 2
describes the state of the art done about the different
concepts needed in this work. Section 3 introduces the
104
Artola, G., Torres, J., Larburu, N., Álvarez, R. and Muro, N.
Development of a Gestational Diabetes Computer Interpretable Guideline using Semantic Web Technologies.
DOI: 10.5220/0008068001040114
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 104-114
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

methodology used during the development of the
work. Section 4 explains a practical scenario, and
finally, Section 5 presents our conclusions and future
guidelines.
2 STATE OF THE ART
In this chapter, the state of the art in Semantic Web
Technologies (SWTs), Clinical Practice Guidelines
(CPGs) for Gestational Diabetes Mellitus (GDM)
domain, Computer Interpretable Guidelines (CIGs),
and Clinical Decision Support Systems (CDSS) is
described.
2.1 Semantic Web Technologies
(SWTs)
The technological breakthrough in biomedical
engineering and health informatics is producing a
huge amount of data coming from different sources,
which causes a limited interoperability of healthcare
systems (Kolias et al., 2014). The Semantic Web
1
is a
Web of data that wants to enable computers to
interpret and process information on the World Wide
Web. SWTs provide the tools to process the data in a
more effective way, create the framework for
interoperability between systems and integrate data
from various sources.
In this context, many researchers have made use
of these technologies to cope with problems related to
semantic interoperability of ontologies or clinical
datasets. As an example, the work presented by El-
Sappagh et al. (El-Sappagh et al., 2018) introduced
the Diabetes Mellitus Treatment Ontology (DMTO)
as a basis for shared-semantics, domain-specific,
standard, machine-readable, and interoperable
knowledge relevant to type 2 diabetes mellitus
(T2DM) treatment. However, to the best of our
knowledge, there is no available gestational diabetes-
centered ontology in the main open source ontology
repositories, such as BioPortal
2
.
As can be seen, ontologies are becoming an
important tool in the field of semantics for
interoperability. In this sense, there are different
ontology languages available for representing
information on the semantic web, such as RDF
Schema or Web Ontology Language (OWL). RDF
Schema allows to build a simple hierarchy of
concepts and properties, while OWL has the ability to
1
https://www.w3.org/standards/semanticweb/
2
http://bioportal.bioontology.org/
3
http://www.snomed.org/snomed-ct/five-step-briefing
specify far more about the properties and classes,
adding semantics to the schema (e.g. defining two
concepts as equivalent or inferring implicit facts).
One of the widely used ontology editors is Protégé
(Musen, 2015), which is fully compatible with the
latest OWL and RDF specifications. In Protégé,
terminologies are represented using classes, slots, and
facets (Noy and McGuinness, 2001), and it also
allows the use of annotation properties for adding
labels to the ontology classes and to link each concept
with its definition in validated and available standard
terminologies (e.g. SNOMED CT
3
, LOINC
4
, NCI
Thesaurus
5
, CIE-10-ES
6
). These permit the
representation of the biomedical concepts with stable
and unique codes, guaranteeing the interoperability of
the implemented knowledge.
2.2 Clinical Practice Guidelines
(CPGs)
CPGs are a set of criteria developed in a systematic
way to help professionals and patients in the decision-
making process, providing the latest evidence-based
diagnostic or therapeutic options when dealing with a
health problem or a specific clinical condition (Kredo
et al., 2016). Over the past years, these CPGs have
been widely used as part of CDSS by formalizing
them as CIGs. Such CIG-based CDSS have
demonstrated to be able to increase the chance of
impacting clinician behaviour compared to using only
narrative guidelines, as they provide updated patient
specific clinical data and advises at the point of care
(Latoszek-Berendsen et al., 2010).
Realizing that SWTs presented an increased
awareness when trying to cope with semantic
interoperability problems in ontologies, some other
researchers studied their usage to represent
computerized CPGs. For example, Hu et al. (Hu et al.,
2015) and Huang et al. (Huang et al., 2014) discuss
several use cases of semantic representation of
evidence-based medical guidelines, showing that they
are potentially useful for medical applications.
Due to our research interest, a state of the art in
Gestational Diabetes Mellitus (GDM) CPGs was
done. GDM is the most common metabolic disorder
of pregnancy, defined as a glucose intolerance
developed in the second or third trimester of
pregnancy (American Diabetes Association, 2016). It
confers an increased risk and complications during
pregnancy for both mother and child, including
4
https://loinc.org/
5
https://ncithesaurus-stage.nci.nih.gov/ncitbrowser/
6
https://eciemaps.mscbs.gob.es/
Development of a Gestational Diabetes Computer Interpretable Guideline using Semantic Web Technologies
105

cesarean delivery, shoulder dystocia, macrosomia,
and neonatal hypoglycemia (The HAPO Study
Cooperative Research Group, 2008). Furthermore,
women with GDM have a substantially increased risk
to develop type 2 diabetes and cardiovascular
diseases after pregnancy (Bellamy et al., 2009;
Sullivan et al., 2012). Therefore, strategies addressed
to optimize management of GDM including effective
prevention, and proper diagnosis and treatment are
mandatory (Chiefari et al., 2017).
There are several guidelines based on best
available and updated evidence for the GDM
management, such as the ones developed by the
National Institute for Health and Care Excellence
(NICE) or the World Health Organization (WHO).
These guidelines allow patient-centered decision
support by following several criteria: (i) the
measurement of clinical variables by the patient itself,
(ii) the readability and ease of follow-up of the given
recommendations, and (iii) the guideline ability to
deal with the guidance of the three pregnancy stages
(before, during, and after pregnancy).
After analysing the different guidelines for GDM
management, the Queensland Clinical Guideline
(Queensland Clinical Guidelines, 2015) was selected
to be used as the backbone guideline, extended with
the knowledge of several other guidelines in the
domain.
But, as it is known, following multiple CPGs in
parallel could result in statements that may interact
with each other, such as giving conflicting
recommendations. Furthermore, the impetus to
deliver customised care based on patient-specific
information, results in the need to be able to offer
guidelines in an integrated manner. In order to deal
with these problems, a harmonized patient-centered
CPG was created and formatted as CIG, enabling the
development of a CIG-driven CDSS.
2.3 Computer Interpretable
Guidelines (CIGs)
Computer Interpretable Guidelines (CIGs) are formal
representations of CPGs that can be executed to
provide guideline-based decision support. One of the
several well-known approaches for formally
representing CIGs are the “Task-Network Models”
(TNMs). These models structure the dependencies
among actions as hierarchical networks that when
fulfilled in a satisfactory way provide a
recommendation.
There exist several proposals to cope with
different clinical modelling challenges (Peleg et al.,
2003), such as GLIF (Patel et al., 1998), PROforma
(Sutton and Fox, 2003), Asbru (Seyfang et al., 2002)
or EON (Tu and Musen, 2001).
These types of CIG formalisms have been used in
many projects over the past years. For instance,
PROforma representation was used by Isern et al.
(Isern et al., 2012) for their proposal of ontology-
driven execution of CPGs. Eccher et al. (Eccher et al.,
2014) implemented and evaluated an Asbru-based
DSS for adjuvant treatment in breast cancer. In Peleg
et al. (Peleg et al., 2014) web-based interactive
clinical algorithms were developed based on GLIF
formalism for the sequencing of tasks to analyse
patients with particular clinical conditions.
In this work, a simplified version of TNM was
implemented based on inference rules (i.e. IF-THEN
type rules), which is explained in Section 3.2.
2.4 Clinical Decision Support
Systems (CDSS)
Realizing the potential of using CIGs and ontologies,
most of the approaches in this field over the past years
focus on guideline development and implementation
for decision support. For instance, the work described
in (Galopin et al., 2015) proposes an ontological
reasoning method based on semantic web techniques
to bring more flexibility to CDSS and offer the ability
to deal with patients suffering from multiple
pathologies by including several modelled CPGs.
Another approach was done in (Riaño et al., 2012),
where the contents of an ontology for the care of
chronically ill patients were adapted to create
individual intervention plans describing health-care
general treatments and also to use it as the knowledge
base of a decision support tool.
CDSS aim aiding clinicians in their decision-
making process by providing the needed tools to
analyse clinical data with latest evidence in the
shortest time (Garg et al., 2005). However, they can
also support patients in the management of different
diseases. Advances in mobile communication for
health care (m-Health) allow the design and
development of patient-centric models to improve
patient’s self-management capabilities.
Recently, many studies have reported that
computer-assisted expert systems, such as CDSS,
might help diabetes practitioners and patients to make
reliable diagnoses and management decisions (Balas
et al., 2004; García-Sáez et al., 2014; Sun and
Costello, 2018; Wilkinson et al., 2013). On the other
hand, lifestyle plays an essential role in controlling
diabetes, in both the prevention and management of
the disease. Many reports in clinical research
(Mottola, 2007; Padayachee and Coombes, 2015;
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
106
Silva-Zolezzi et al., 2017) support the theory that
healthy eating and regular exercise are beneficial in
both preventing GDM and improving pregnancy
outcomes in women with GDM.
Taking this into account, among the different
CDSS designed in recent years, the ones related to the
management of GDM were analysed. For instance, in
the work of Caballero-Ruiz et al. (Caballero-Ruiz et
al., 2017) a web-based telemedicine platform for its
use as CDSS for the management of GDM was
designed to remotely evaluate patients, allowing them
to upload their data at home. In addition, mobile
applications allow using automatic processing tools
to provide real-time advice based on monitoring data.
Peleg et al. (Peleg et al., 2017) presented a
personalized and patient-centric CDSS for the
monitoring and evaluation of atrial fibrillation and
gestational diabetes. A newer study (Miremberg et
al., 2018) demonstrated the positive effect of a
smartphone-based daily feedback system among
women with GDM for improving patient compliance
to treatment and a better control of glycaemic levels.
With the aim of going beyond the current state of
the art of CPG-based CDSS, the objective of this
project was to design a patient-centered mobile CDSS
for the management of GDM with the novelty of
integrating SWTs to the system.
3 METHODOLOGY
In this chapter, the methodology followed for the
development of a semantically validated mobile
CDSS for giving guidance to women in the
management of Gestational Diabetes Mellitus
(GDM) is described.
3.1 GDM Ontology
In this section, an ontology formalizing all concepts
and knowledge coming from the different CPGs
related to GDM prevention, management and
treatment is presented. For this work, the Protégé
editor was used to define the different gestational
diabetes related clinical concepts and the
relationships among them. This ontology was built
using NCI Thesaurus terminology to assure a
semantic interoperability of the knowledge. The big
amount of biomedical concepts that NCI Thesaurus
contains and the fact that it is open access makes it an
appropriate terminology to be used for the GDM
ontology. Besides, the Unified Medical Language
7
https://www.nlm.nih.gov/research/umls/
System (UMLS)
7
code for each of the terminologies
was also specified in the ontology. UMLS integrates
the most notable vocabularies (e.g. SNOMED CT,
ICD-10, etc.) in its repository. The result was then
validated with a reasoner (Section 3.1.2) for ensuring
a consistent ontology.
3.1.1 Ontology Formalization
All the conditions and rules expressed in the CIG
contain variables and properties that are organized in
an ontology with their specific names. This ontology
is composed by a total number of 166 classes, which
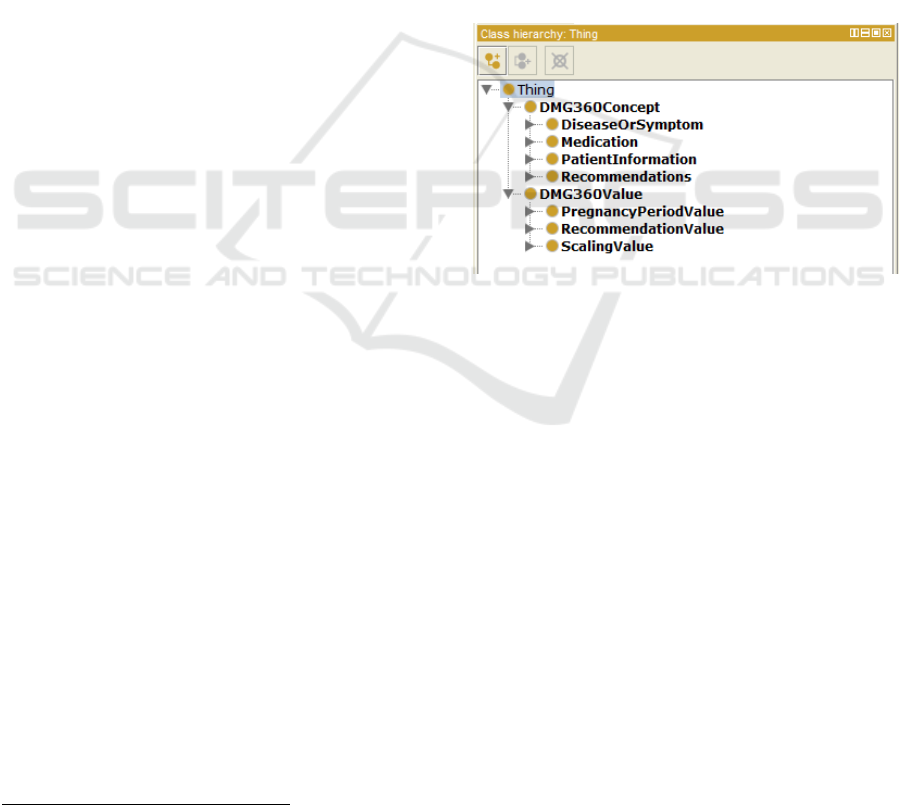
are separated in two main groups (Figure 1). The first
group is under the class named DMG360Concept that
comprises all the necessary variables’ names for
creating the CIG. The second group, named
DMG360Value, compiles the classes that define the
possible values of the classes from the first group.
Figure 1: The two main groups of classes in the ontology:
DMG360Concept and DMG360Value.
As can be seen in Figure 1, the group of concepts
has four subclasses, and each of them comprises other
subclasses. The first concept in the list (i.e.
DiseaseOrSymptom) is composed by two subclasses:
the Disease class, which contains two diseases
definition (DiabetesMellitus and
PolycysticOvarianSyndrome); and the Symptom
class, with a list of 15 different symptoms as
subclasses. The second concept (i.e. Medication)
includes three subclasses corresponding to three
different medications: Antipsychotics,
Corticosteroids, and Insulin. The third one (i.e.
PatientInformation) is the concept with the biggest
number of subclasses, containing different
information about the patient (e.g. information about
maternal age, diabetes family history, ethnicity,
physical activity, body mass index (BMI), etc.). The
last concept (i.e. Recommendations) is related to the
recommendations that are given to the user, which
Development of a Gestational Diabetes Computer Interpretable Guideline using Semantic Web Technologies
107
can vary depending on the stage of pregnancy, as
explained in Chapter 3.2. In total, seven different
types of recommendations are specified.
Furthermore, two different properties are defined
within the ontology to relate the different classes and
their values: a data property and an object property.
Both are used to specify the range of values that can
be taken by these classes, thus they are both defined
using the noun hasRange. These ranges can handle
either common data types (i.e. integer, Boolean…)
when linked by a data property, or other possible
values expressed as classes in the DMG360Value
group when linked by an object property. For
example, the CurrentPhysicalActivityLevel class is
restricted to have a value corresponding to any
subclass of the ScalingValue class (i.e. High, Low, or
Moderate).
In addition, the clinical concepts’ names,
definitions and codes were extracted from NCI
Thesaurus repository and defined within the model
using annotation properties for its semantic
standardization. Five different annotation properties
were defined: (i) NCI_label, for giving the label of the
class as stated in the NCI Thesaurus, (ii)
NCI_definition, which contains the definition of the
concept by the NCI, (iii) NCI_code, with the unique
code of the term, (iv) UMLS_CUI, containing the
corresponding UMLS code, and (v) NCI_version,
specifying the version of the NCI Thesaurus
repository used.
For the ontology design, OWL language was
used. Having all the knowledge mapped, the model
was exported in RDF language for the integration
with the CDSS using the Jena API.
3.1.2 Ontology Validation
To validate the defined relationships between the
variables and their possible values in the ontology, a
tool called Reasoner was used. The reasoners offered
by Protégé (i.e. FaCT ++, HermiT or Pellet) are
programs that evaluate the consistency of an ontology
by identifying relationships between classes. In this
project, the FaCT++ reasoner was used.
After the triggering of this reasoner without
obtaining any unsatisfactory relationship between the
classes, the hierarchy of the designed GDM ontology
was validated.
Moreover, the SPARQL Query tool in Protégé
was used for the validation of the requests that could
be done to the ontology in the process of its
integration with the CDSS. For that, the SPARQL
query language
8
was applied, a language that can be
used to express queries across diverse data sources,
whether the data is stored natively as RDF or viewed
as RDF via middleware. Simple queries such as
requesting data/object properties of a class, finding
subclasses of a class, or showing the list of all classes
in the ontology were tested.
3.2 GDM CIG
As stated in the state of the art, CPGs must be
formalized in a computer interpretable way to provide
guideline-based decision support and allow the
evaluation of a patient in a computerized way. In this
work, several CPGs with the clinical statements that
describe the procedures to be followed in each
clinical setup for the GDM management were studied
and a more complete guideline that contained all the
needed information for monitoring patients before,
during and after pregnancy was created. This
extended CPG was then formalized as IF-THEN rules
in a computerized way (i.e. as a CIG) containing the
concepts defined in the ontology.
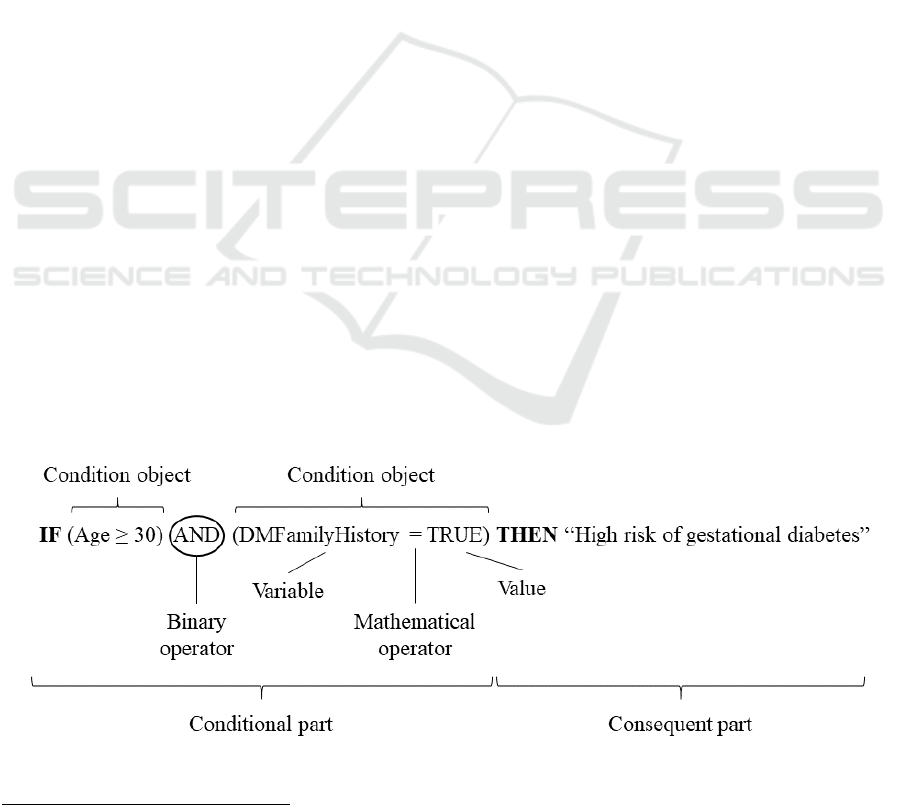
Figure 2: Components of a Rule object used in this work.
8
https://www.w3.org/TR/sparql11-query/
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
108
To create the rules for this project, conditions
(i.e. clinical statements to be accomplished) and
their respective consequences (i.e. the
recommendations to be given to the patient) were
identified and extracted from the selected CPGs in
the GDM domain. Once this knowledge was
formalized in our extended CPG, a translation into
computer interpretable language was done. For that,
statements were translated into IF-THEN kind rules,
where the conditional part is preceded by the IF
expression and the consequent part by the THEN
expression.
Each of the statements describing the criteria to
be followed for guiding patients in this setup were
defined as Rule objects, which encompasses (i) a
conditional part composed by one or more
conditions linked by (ii) a binary operator (i.e. AND,
or OR) and (iii) a consequent part containing the
recommendation for the patient, as can be seen in
Figure 2. Each of the conditions is based on a
Condition object, which stores (i) the name of the
clinical variable to be evaluated (i.e. concept defined
in the ontology), (ii) the mathematical operator (i.e.
>, ≥, =, <, ≤) and (iii) the value or the threshold of
the clinical variable to be evaluated. When the
conditions of a rule are matching a patient’s clinical
information, a recommendation is provided to the
patient.
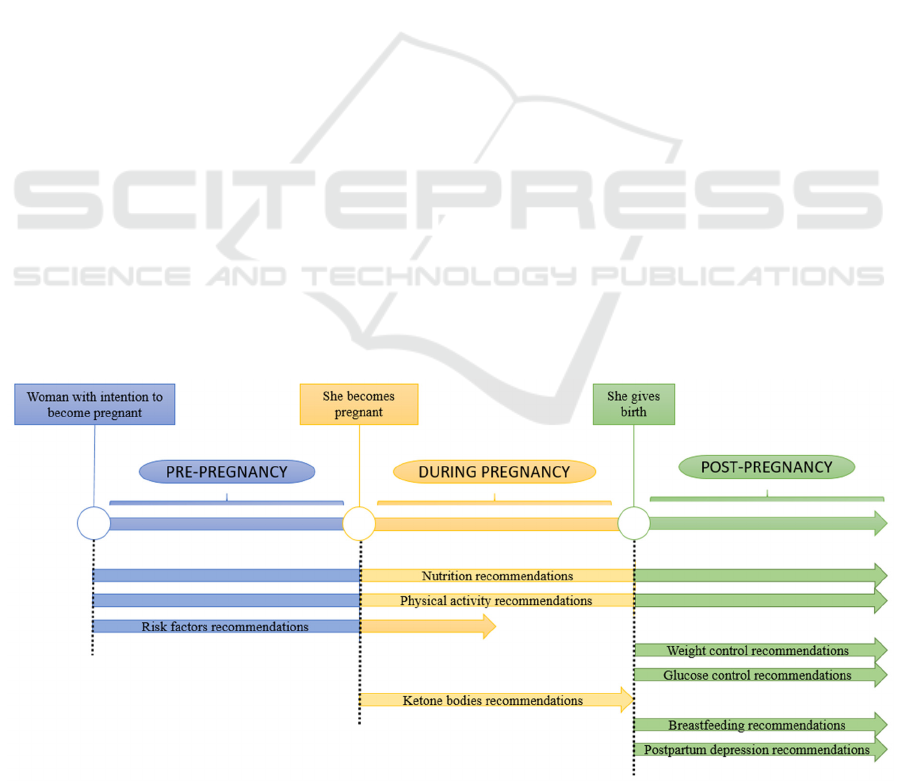
Depending on the patient clinical stage, different
types of interventions for the management of GDM
were identified in the rule formalization process. For
the stage before pregnancy, information about
nutrition, physical activity and GDM risk factors
were formalized. For the pregnancy period, guides
related to nutrition, weight, physical activity, risk
factors, and glucose control were covered. And
finally, in the case of the post-pregnancy stage,
nutrition, weight control, physical activity,
breastfeeding, glucose control, and postpartum
depression recommendations were included. The
schema describing all this information is represented
in Figure 3.
When formalizing this knowledge in the
extended CPG and in its computerized version as
CIG, the pipeline presented in Figure 3 was
followed. The extended CIG contains 96 different
rules distributed in three stages: 13 for the
pre-pregnancy stage, 45 for the GDM management
during pregnancy, and 16 for post-pregnancy
stage. In addition, 22 characterization rules were
also formalized, whose action is the change of
variable values instead of providing a
recommendation. These rules were not introduced
into the CDSS as part of the rule base to be executed
by the engine, but they were implemented in the
mobile app itself.
3.3 Integration of the Ontology within
the CDSS
Once the CIG was formalized with all computer
interpretable rules and the GDM ontology was
validated, the next step was to integrate both into the
CDSS. For this objective, an authoring tool
previously designed (Muro et al., 2019) was used.
This tool enables an intuitive Graphical User
Interface (GUI) for including new clinical knowledge
Figure 3: The three stages of pregnancy and the different recommendations giving in each of them.
Development of a Gestational Diabetes Computer Interpretable Guideline using Semantic Web Technologies
109
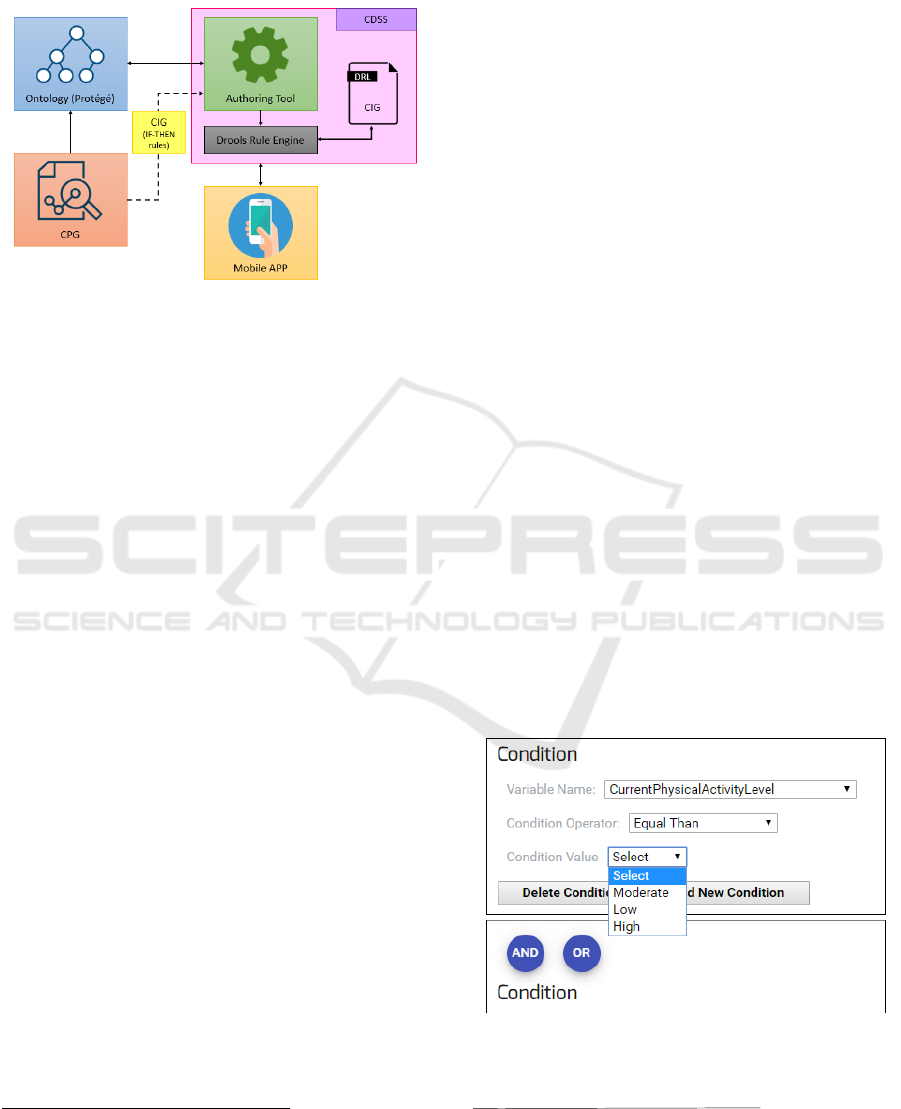
in a computerized way into the CDSS rule base. A
high-level project’s architecture representation is
shown in Figure 4.
Figure 4: High level representation of the architecture of the
project.
The development made within this project
extended the authoring tool by introducing a new
functionality: the syntactical and semantical
validation of the formalized knowledge through an
ontology. A research on different APIs (i.e. Protégé-
OWL API
9
, OWL API
10
, Apache Jena
11
, or RDF4J
12
)
for the integration of ontologies in this kind of
systems was done. After comparing them, Jena API
was selected for carrying out the integration of the
ontology with the Java based CDSS, because of its
ease of use and compatibility advantages with our
system.
In order to have an interaction between the GUI
and the CDSS, different web services were developed
as linkers to transmit data between them. These web
services were used by the authoring tool (i) to get the
list of variables (classes) for defining the evaluated
variable name within a Condition object, (ii) to get the
possible values of the specific variable selected in the
Condition object, (iii) to get the list of
recommendations for completing the consequent part
of the Rule object, and (iv) to post each generated rule
to the backend of the system. This backend is
composed by different modules based on Drools
13
, (i)
a rule engine and (ii) a rule file generator in Drools
Rule Language (.drl) extension.
To create the GDM CIG, each of the rules or
conditions were introduced manually in the authoring
tool using its GUI. In this process, four main blocks
are fulfilled. First, the name of the rule is defined.
Then, the conditions of the rule are introduced. Next,
9
https://protegewiki.stanford.edu/wiki/ProtegeOWL_API
10
https://github.com/owlcs/owlapi/wiki
11
https://jena.apache.org/documentation/ontology/
the recommendation for the introduced rule is
specified. And finally, the rule is sent to the backend.
In the second step for introducing the conditions
of each rule, the Condition objects are constructed
using the information obtained from querying the
integrated ontology in the authoring tool. For
requesting the filling out options, the previously
described web services are used. In this context, the
different types of ontology classes were used for
different purposes, as explained below.
The classes of the ontology that correspond to the
variables used for defining the conditional part of the
CIG are under the DMG360Concepts class. These
variables are requested and given as options for the
first parameter of a Condition object in the authoring
tool GUI.
Once the variable name of the Condition object is
specified, the respective condition operator is
selected. Then, in the third box for specifying the
value or threshold of the selected variable, the
authoring tool uses the web service for requesting its
possible values, and here is where the second type of
ontology classes interact. In the case of variables with
data properties, the backend of the authoring tool
defines the type of data that needs to be used by
changing the format of the box in the GUI. For the
case of the variables with object properties, a list
containing subclasses of the group DMG360Value is
given for being selected by the clinician.
Once the first condition is introduced, the binary
operator can be selected from the given options (i.e.
AND, or OR) and as many as required conditions can
be defined within the conditional part of a single rule
following the same procedure (see an example in
Figure 5).
Figure 5: Example of the definition of rule conditional
statements in the AT.
12
http://rdf4j.org/
13
https://www.drools.org/
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
110

The third step of the introduction of the rule
corresponds to the tab of the GUI for introducing
recommendations, which permits the user to select
the recommendation from a drop-down list. This list
is the result of the request made by the authoring tool
for the subclasses of the class RecommendationValue
from the DMG360Value group in the ontology. When
a recommendation is selected by the user using its
name or abbreviation, the text corresponding to that
recommendation is displayed above the selection box.
Each of the texts for the respective recommendation
class are specified in the ontology using annotation
properties. The authoring tool uses a web service for
obtaining the information from those annotations
when a recommendation name is selected from the
dropdown list.
Finally, the last step of the introduction of rules
uses the fourth web service for sending the rules to
the backend of the system and generating the .drl file,
as explained above. This last web service takes all the
values introduced by the clinician in the authoring
tool and sends them to the backend, where they are
processed and uploaded to the clinical database. As
the structure of the objects generated in the authoring
tool are the same as in the Rule object, a direct
mapping can be done. The generated Rule object is
then sent to the database, where it is stored, and then
retrieved for sending it to the backend and generating
the .drl file.
3.4 Patient-centered Mobile CDSS
Once the integration of the semantically validated
CIG was implemented within the CDSS as a .drl file,
a patient-centered mobile application was developed
to interact with it. The user profiles defined for this
application are (i) women that want to be pregnant
and are monitoring their health status for it, (ii)
women already pregnant, or (iii) women that have just
given birth, and they all want to receive
recommendations for managing the gestational
diabetes. In the next lines, some examples of the
different screens of the designed app and their
functionalities are described.
First, a logging system was developed with
registering or signing in options. This logging will
save the basic personal data (i.e. sex, ethnicity…) that
is supposed to be static during the monitoring period
to avoid introducing it each time the user logs into the
application.
Once the user is logged in, a main menu is
displayed providing the possible user profiles to start
the GDM management depending on their needs.
Each of the pregnancy stages will provide specific
recommendations, differing ones from the others as
each period has a different focus on what to evaluate
and how to treat the patient to reach her objective (see
Chapter 3.2): in pre-pregnancy period,
recommendations for nutrition, physical activity and
GDM risk factors are given. During pregnancy,
nutrition, weight, physical activity, risk factors, and
glucose control related guidance is given. And for
post-pregnancy period, nutrition, weight control,
physical activity, breastfeeding, glucose control, and
postpartum depression recommendations are given.
This information is managed by the app through a
menu, where for each stage different
recommendations on the above stated topics can be
provided.
Each time the user selects an option in the
different menus, the variable corresponding to that
specific topic is changed. For example, in the case of
the menu for the post-pregnancy stage, when the user
touches the image for obtaining recommendation
concerning the glucose control, the variable
GlucoseControlRecommendations is set to true.
If required, the application will retrieve more
clinical information by questionnaires (see Figure 6).
These questionnaires will ask for some questions that
have possible answers to be selected. These answers
are related to some clinical variables defined in the
GDM ontology (mostly Boolean or Object property
type variables), but sometimes it can require to the
user introducing numerical variables’ values (e.g. the
blood glucose level). Every time she answers a
question required in the app, this clinical information
will be stored as part of her profile to be evaluated by
the CDSS later.
Once all the questions for completing the needed
patient profile are answered, they are sent to the
CDSS in the backend, getting back the corresponding
recommendation(s) to be displayed in the
recommendation screen of the mobile application
(see Figure 7).
4 PRACTICAL GDM SCENARIO
In this chapter a practical scenario is presented to
show up how the whole architecture works. The
selected patient profile represents a pregnant woman
that would like to receive recommendations related to
her glucose control. For that, she will be using the
mobile application designed in this project. The
presented clinical case is a non-diabetic pregnant
woman that has suffered a decompensation in her
Blood Glucose Level (BGL) with a glucose value of
136 mg/dL before eating (pre-prandial BGL). In
Development of a Gestational Diabetes Computer Interpretable Guideline using Semantic Web Technologies
111
addition, her glucose levels showed higher values
than the maximum threshold more than twice during
the same week and in different intervals. Hence, her
clinical data would be as follows:
Table 2: Clinical data of the patient profile for the use case.
Variable name Value
DuringPregnancyPeriod TRUE
GlucoseControlRecommendations TRUE
GDM FALSE
Type1Diabetes FALSE
Type2Diabetes FALSE
NormalBGLResults FALSE
Insulin FALSE
TwoOrMoreDecomp1Week TRUE
InDifferentIntervals TRUE
ExceedMaximumValues TRUE
PrePrandialBGL 136
In the process for introducing these values in the
mobile application, first the diabetes related questions
are done to the user (i.e. the type of diabetes, and the
glucose level value are requested). Then, some other
questions are done to the user like (i) if she is taking
insulin, (ii) if she had more than one decompensation
of blood glucose levels in the same week, (iii) if they
were in different intervals, and (iv) if they exceeded
the maximum values. The screen of the app showing
these questions is visualized in Figure 6.
Figure 6: Questions of the questionnaire for glucose control
in the mobile app.
Once this data is gathered by the system, it is sent
to the backend to be evaluated by the CDSS. The
rule(s) fitting the values of the variables sent by the
user are triggered getting as result the
recommendation corresponding to the matching rule.
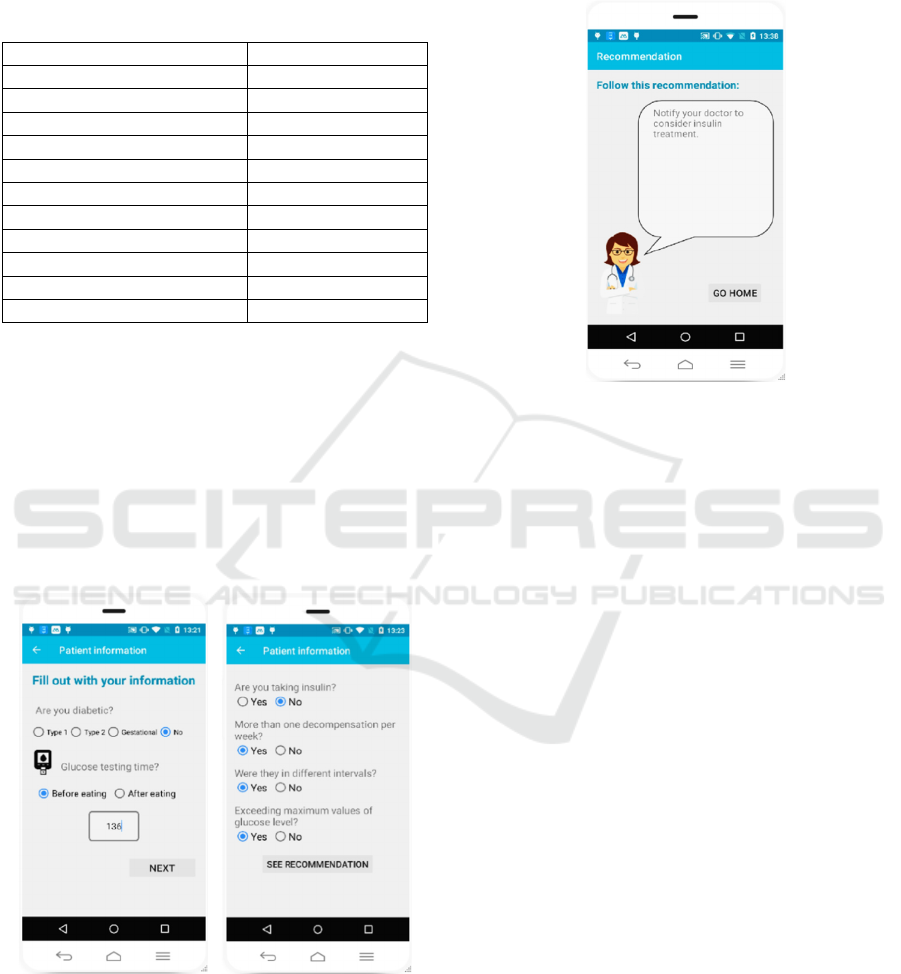
In this particular case, the recommendation obtained
was: “Notify your doctor to consider insulin
treatment” (see Figure 7).
Figure 7: Recommendation screen in the mobile app.
Once the user receives the recommendation, she
can follow it to solve her glucose level problem in this
case. Besides, she can also go back to the menu and
select other options for receiving recommendations
about more interventions for the management of
GDM.
5 CONCLUSIONS AND FUTURE
WORK
CPGs have been promoted as a powerful method for
standardization and improvement of medical care
quality and personalization of healthcare. However,
applying these guidelines to promote evidence-based
and up-to-date clinical practice is a known challenge
due to lack of digitalization of clinical guidelines. To
overcome these issues, the use of semantic web
technologies along with CDSS is proposed, in order
to avoid the lack of semantic interoperability and
promote the interoperability among systems in a
standardized way.
In this paper, an architecture that allows the
formalization of CPGs into Computer Interpretable
Guidelines (CIGs) supported by an ontology in the
gestational diabetes domain is presented. This CIG
has been implemented within a CDSS to give support
to the patients before, during, and after pregnancy. In
addition, a mobile application has been developed for
guiding patients based on up-to-date evidence based
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
112

clinical guidelines. This application is able to provide
users different recommendations based on their
clinical information.
As future work, applying tools for obtaining
information from patients’ electronic health records
could optimize the efficiency of the designed mobile
CDSS. In the same way, a more flexible way of
gathering the user’s clinical data will be implemented
(e.g. using wearables for obtaining patient data
without needing to introduce them manually).
Likewise, a way to facilitate the ontology generation
for clinicians will also be researched. Finally, it has
been also envisioned the future inclusion of feedback
tools within the mobile application in order to gather
the user appreciation of the system, as well as the
possibility to submit the system to the evaluation of
clinical specialists.
ACKNOWLEDGEMENTS
This work has been developed under the research
project DMG360 (2017-2018 / Exp. number ZE-
2017/00011, in collaboration with INIT Health, SL),
which has been funded by the Department of
Economic Development and Infrastructure of the
Basque Government under the Hazitek program.
REFERENCES
American Diabetes Association, 2016. Classification and
Diagnosis of Diabetes. Diabetes Care 39, S13–S22.
https://doi.org/10.2337/dc16-S005
Balas, E. A., Krishna, S., Kretschmer, R. A., Cheek, T. R.,
Lobach, D. F., Boren, S. A., 2004. Computerized
knowledge management in diabetes care. Med. Care
42, 610–621.
Bellamy, L., Casas, J.-P., Hingorani, A.D., Williams, D.,
2009. Type 2 diabetes mellitus after gestational
diabetes: a systematic review and meta-analysis. Lancet
Lond. Engl. 373, 1773–1779. https://doi.org/10.1016/
S0140-6736(09)60731-5
Caballero-Ruiz, E., García-Sáez, G., Rigla, M., Villaplana,
M., Pons, B., Hernando, M. E., 2017. A web-based
clinical decision support system for gestational
diabetes: Automatic diet prescription and detection of
insulin needs. Int. J. Med. Inf. 102, 35–49.
https://doi.org/10.1016/j.ijmedinf.2017.02.014
Chiefari, E., Arcidiacono, B., Foti, D., Brunetti, A., 2017.
Gestational diabetes mellitus: an updated overview. J.
Endocrinol. Invest. 40, 899–909. https://doi.org/
10.1007/s40618-016-0607-5
Eccher, C., Seyfang, A., Ferro, A., 2014. Implementation
and evaluation of an Asbru-based decision support
system for adjuvant treatment in breast cancer. Comput.
Methods Programs Biomed. 117, 308–321.
https://doi.org/10.1016/j.cmpb.2014.06.021
El-Sappagh, S., Kwak, D., Ali, F., Kwak, K.-S., 2018.
DMTO: a realistic ontology for standard diabetes
mellitus treatment. J. Biomed. Semant. 9.
https://doi.org/10.1186/s13326-018-0176-y
Galopin, A., Bouaud, J., Pereira, S., Seroussi, B., 2015. An
Ontology-Based Clinical Decision Support System for
the Management of Patients with Multiple Chronic
Disorders. Stud. Health Technol. Inform. 216, 275–279.
García-Sáez, G., Rigla, M., Martínez-Sarriegui, I., Shalom,
E., Peleg, M., Broens, T., Pons, B., Caballero-Ruíz, E.,
Gómez, E.J., Hernando, M.E., 2014. Patient-oriented
Computerized Clinical Guidelines for Mobile Decision
Support in Gestational Diabetes. J. Diabetes Sci.
Technol. 8, 238–246. https://doi.org/10.1177/1932296
814526492
Garg, A. X., Adhikari, N. K. J., McDonald, H., Rosas-
Arellano, M. P., Devereaux, P. J., Beyene, J., Sam, J.,
Haynes, R. B., 2005. Effects of computerized clinical
decision support systems on practitioner performance
and patient outcomes: a systematic review. JAMA 293,
1223–1238. https://doi.org/10.1001/jama.293.10.1223
Grimshaw, J. M., Russell, I. T., 1993. Effect of clinical
guidelines on medical practice: a systematic review of
rigorous evaluations. Lancet Lond. Engl. 342, 1317–
1322.
Hu, Q., Huang, Z., Gu, J., 2015. Semantic representation of
evidence-based medical guidelines and its use cases.
Wuhan Univ. J. Nat. Sci. 20, 397–404.
https://doi.org/10.1007/s11859-015-1112-y
Huang, Z., ten Teije, A., van Harmelen, F., Aït-Mokhtar, S.,
2014. Semantic Representation of Evidence-Based
Clinical Guidelines, in: Miksch, S., Riaño, D., ten Teije,
A. (Eds.), Knowledge Representation for Health Care.
Springer International Publishing, Cham, pp. 78–94.
https://doi.org/10.1007/978-3-319-13281-5_6
Institute of Medicine, 1990. Clinical Practice Guidelines:
Directions for a New Program. National Academies
Press (US), Washington (DC).
Isern, D., Sánchez, D., Moreno, A., 2012. Ontology-driven
execution of clinical guidelines. Comput. Methods
Programs Biomed. 107, 122–139. https://doi.org/
10.1016/j.cmpb.2011.06.006
Kolias, V.D., Stoitsis, J., Golemati, S., Nikita, K.S., 2014.
Utilizing Semantic Web Technologies in Healthcare.
Springer Int. Publ. Switz. 16. https://doi.org/10.1007/
978-3-319-06844-2_2
Kredo, T., Bernhardsson, S., Machingaidze, S., Young, T.,
Louw, Q., Ochodo, E., Grimmer, K., 2016. Guide to
clinical practice guidelines: the current state of play. Int.
J. Qual. Health Care 28, 122–128. https://doi.org/
10.1093/intqhc/mzv115
Latoszek-Berendsen, A., Tange, H., Herik, H. J. van den,
Hasman, A., 2010. From Clinical Practice Guidelines to
Computer-interpretable Guidelines. Methods Inf. Med.
49, 550–570. https://doi.org/10.3414/ME10-01-0056
Miremberg, H., Ben-Ari, T., Betzer, T., Raphaeli, H.,
Gasnier, R., Barda, G., Bar, J., Weiner, E., 2018. The
impact of a daily smartphone-based feedback system
Development of a Gestational Diabetes Computer Interpretable Guideline using Semantic Web Technologies
113

among women with gestational diabetes on compliance,
glycemic control, satisfaction, and pregnancy outcome:
a randomized controlled trial. Am. J. Obstet. Gynecol.
218, 453.e1-453.e7. https://doi.org/10.1016/j.ajog.
2018.01.044
Mottola, M.F., 2007. The role of exercise in the prevention
and treatment of gestational diabetes mellitus. Curr.
Sports Med. Rep. 6, 381–386. https://doi.org/10.1007/
s11932-007-0056-1
Muro, N., Larburu, N., Torres, J., Kerexeta, J., Artola, G.,
Arrúe, M., Macía, I., Seroussi, B., 2019. Architecture
for a Multimodal and Domain-Independent Clinical
Decision Support System Software Development Kit.
Presented at the 41st Annual International Conference
of the IEEE Engineering in Medicine and Biology
Society (EMBC), Berlin, Germany.
Musen, M.A., 2015. The Protégé Project: A Look Back and
a Look Forward. AI Matters 1, 4–12. https://doi.org/
10.1145/2757001.2757003
Noy, N. F., McGuinness, D. L., 2001. Ontology
Development: A Guide to Creating Your First Ontology
25.
Padayachee, C., Coombes, J. S., 2015. Exercise guidelines
for gestational diabetes mellitus. World J. Diabetes 6,
1033–1044. https://doi.org/10.4239/wjd.v6.i8.1033
Patel, V. L., Allen, V. G., Arocha, J. F., Shortliffe, E. H.,
1998. Representing Clinical Guidelines in GLIF. J. Am.
Med. Inform. Assoc. JAMIA 5, 467–483.
Peleg, M., Fox, J., Patkar, V., Glasspool, D., Chronakis, I.,
South, M., Nassar, S., Gaglia, J. L., Gharib, H., Papini,
E., Paschke, R., Duick, D. S., Valcavi, R., Hegedüs, L.,
Garber, J. R., 2014. A Computer-Interpretable Version
of the AACE, AME, ETA Medical Guidelines for
Clinical Practice for the Diagnosis and Management of
Thyroid Nodules. Endocr. Pract. Off. J. Am. Coll.
Endocrinol. Am. Assoc. Clin. Endocrinol. 20, 352–359.
https://doi.org/10.4158/EP13271.OR
Peleg, M., Shahar, Y., Quaglini, S., Fux, A., García-Sáez,
G., Goldstein, A., Hernando, M.E., Klimov, D.,
Martínez-Sarriegui, I., Napolitano, C., Parimbelli, E.,
Rigla, M., Sacchi, L., Shalom, E., Soffer, P., 2017.
MobiGuide: a personalized and patient-centric
decision-support system and its evaluation in the atrial
fibrillation and gestational diabetes domains. User
Model. User-Adapt. Interact. 27, 159–213.
https://doi.org/10.1007/s11257-017-9190-5
Peleg, M., Tu, S., Bury, J., Ciccarese, P., Fox, J., Greenes,
R.A., Hall, R., Johnson, P.D., Jones, N., Kumar, A.,
Miksch, S., Quaglini, S., Seyfang, A., Shortliffe, E.H.,
Stefanelli, M., 2003. Comparing Computer-
interpretable Guideline Models: A Case-study
Approach. J. Am. Med. Inform. Assoc. 10, 52–68.
https://doi.org/10.1197/jamia.M1135
Queensland Clinical Guidelines, 2015. Guideline:
Gestational diabetes mellitus 38.
Riaño, D., Real, F., López-Vallverdú, J.A., Campana, F.,
Ercolani, S., Mecocci, P., Annicchiarico, R.,
Caltagirone, C., 2012. An ontology-based personaliza-
tion of health-care knowledge to support clinical
decisions for chronically ill patients. J. Biomed. Inform.
45, 429–446. https://doi.org/10.1016/j.jbi.2011.12.008
Seyfang, A., Miksch, S., Marcos, M., 2002. Combining
diagnosis and treatment using ASBRU. Int. J. Med. Inf.
68, 49–57.
Silva-Zolezzi, I., Samuel, T. M., Spieldenner, J., 2017.
Maternal nutrition: opportunities in the prevention of
gestational diabetes. Nutr. Rev. 75, 32–50.
https://doi.org/10.1093/nutrit/nuw033
Sullivan, S. D., Umans, J. G., Ratner, R., 2012. Gestational
Diabetes: Implications for Cardiovascular Health. Curr.
Diab. Rep. 12, 43–52. https://doi.org/10.1007/s11892-
011-0238-3
Sun, S., Costello, K. L., 2018. Designing Decision-Support
Technologies for Patient-Generated Data in Type 1
Diabetes. AMIA. Annu. Symp. Proc. 2017, 1645–1654.
Sutton, D. R., Fox, J., 2003. The Syntax and Semantics of
the PROforma Guideline Modeling Language. J. Am.
Med. Inform. Assoc. JAMIA 10, 433–443.
https://doi.org/10.1197/jamia.M1264
The HAPO Study Cooperative Research Group, 2008.
Hyperglycemia and Adverse Pregnancy Outcomes. N.
Engl. J. Med. 358, 1991–2002. https://doi.org/10.1056/
NEJMoa0707943
Tu, S.W., Musen, M.A., 2001. Modeling data and
knowledge in the EON guideline architecture. Stud.
Health Technol. Inform. 84, 280–284.
Wilkinson, M.J., Nathan, A.G., Huang, E.S., 2013.
Personalized Decision Support in Type 2 Diabetes
Mellitus: Current Evidence and Future Directions.
Curr. Diab. Rep. 13, 205–212. https://doi.org/10.1007/
s11892-012-0348-6
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
114
