
A Regression-like Classification System for Geometric Semantic Genetic
Programming
Illya Bakurov
1
, Mauro Castelli
1
, Francesco Fontanella
2
and Leonardo Vanneschi
1
1
NOVA Information Management School (NOVA IMS), Universidade Nova de Lisboa,
Campus de Campolide, 1070-312, Lisbon, Portugal
2
Department of Electrical Engineering and Information, Universit
´
a degli Studi di Cassino e del Lazio Meridionale,
03043, Cassino, Italy
Keywords:
Classification, Regression, Geometric Semantic Genetic Programming.
Abstract:
Geometric Semantic Genetic Programming (GSGP) is a recent variant of Genetic Programming, that is gain-
ing popularity thanks to its ability to induce a unimodal error surface for any supervised learning problem.
Nevertheless, so far GSGP has been applied to the real world basically only on regression problems. This
paper represents an attempt to apply GSGP to real world classification problems. Taking inspiration from Per-
ceptron neural networks, we represent class labels as numbers and we use an activation function to constraint
the output of the solutions in a given range of possible values. In this way, the classification problem is turned
into a regression one, and traditional GSGP can be used. In this work, we focus on binary classification;
logistic constraining outputs in [0,1] is used as an activation function and the class labels are transformed into
0 and 1. The use of the logistic activation function helps to improve the generalization ability of the system.
The presented results are encouraging: our regression-based classification system was able to obtain results
that are better than, or comparable to, the ones of a set of competitor machine learning methods, on a rather
rich set of real-life test problems.
1 INTRODUCTION
Geometric Semantic Genetic Program-
ming (GSGP) (Moraglio et al., 2012) is a recent
development of Genetic Programming (GP) (Koza,
1992). GSGP uses particular genetic operators,
called Geometric Semantic Operators (GSOs) and it
has gained noteworthy popularity among researchers
in the last few years. This popularity is probably
because GSOs can induce a unimodal error surface
(i.e., with no locally optimal solutions) for any
supervised learning problem. This property bestows
a remarkable optimization power on GSGP. Further-
more, in several references (see for instance (Castelli
et al., 2015a)), it was shown that, in some particular
circumstances, GSOs are also able to limit over-
fitting. Thanks to these important properties, and
also to a very efficient implementation presented
in (Castelli et al., 2015a), GSGP was successful in
several different real-life applications in recent years
(a rather complete summary of the applications is
reported in (Vanneschi et al., 2014)).
However, looking at the different applications
that have been developed with GSGP, it is possi-
ble to notice a clear trend: those applications are
mainly regression and forecasting problems. In other
words, almost no relevant results showing success-
ful applications of GSGP on classification problems
were presented. This does not happen by chance:
even though in the contribution that originally in-
troduced GSGP (Moraglio et al., 2012) a version
of GSOs for classification was presented, that version
of the operators is not very easy to use. More specif-
ically, those operators assume that GSGP is evolving
individuals that are classification rules. Even though
systems evolving rules, like decision trees, using GP
have been presented so far, this is not the most typical
use of GP. GP individuals are, typically, mathematical
functions of the inputs, returning as output numbers.
As such, GP is naturally more prone to be used for
regression than for classification.
In this paper, we intend to fill this gap, proposing
a GSGP-based approach for classification for the first
time, where individuals are numeric expressions, as
is usual in GP. The approach is straightforward and
inspired by the output layer of Artificial Neural Net-
works (ANNs), where an activation function is ap-
plied to the output. Similarly to ANNs, a logistic ac-
40
Bakurov, I., Castelli, M., Fontanella, F. and Vanneschi, L.
A Regression-like Classification System for Geometric Semantic Genetic Programming.
DOI: 10.5220/0008052900400048
In Proceedings of the 11th International Joint Conference on Computational Intelligence (IJCCI 2019), pages 40-48
ISBN: 978-989-758-384-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

tivation is applied to the output of a GSGP individual,
and compared to the target of a binary classification,
appropriately transformed into binary values (with 0
representing one class label, and 1 representing the
other). It is worth noticing that, since the problem is
“de facto” transformed into a regression and the lo-
gistic function is increasing monotonically, the prop-
erty of GSOs of inducing a unimodal error surface is
maintained. Despite its simplicity, we think that, if
successful, this idea may have a significant impact,
paving the way for new GSGP contributions for real-
life classification problems.
The paper is organized as follows: in Section 2,
we describe GSGP; Section 3 presents our idea of a
regression-like classification system based on GSGP;
Section 4.3 contains our experimental study. More
specifically, in the first part of this section, we present
experiments aimed at tuning two important system
parameters, while in the second part we present an ex-
perimental comparison of the proposed approach with
Support Vector Machines, one of the most popular
and powerful classification algorithms. The compari-
son is made on 10 complex real-life datasets. Finally,
Section 5 concludes the work, and presents ideas for
future investigation.
2 GEOMETRIC SEMANTIC
GENETIC PROGRAMMING
Let X = {
−→
x
1
,
−→
x
2
,...,
−→
x
n
} be the set of input data (train-
ing instances, observations or fitness cases) of a sym-
bolic regression problem, and
−→
t = [t
1
,t
2
,...,t
n
] the
vector of the respective expected output or target val-
ues (in other words, for each i = 1,2,...,n, t
i
is the
expected output corresponding to input
−→
x
i
). A GP
individual (or program) P can be seen as a function
that, for each input vector
−→
x
i
returns the scalar value
P(
−→
x
i
). Following (Moraglio et al., 2012), we call se-
mantics of P the vector
−→
s
P
= [P(
−→
x
1
),P(
−→
x
2
),...,P(
−→
x
n
)].
This vector can be represented as a point in an n-
dimensional space, that we call semantic space. Re-
mark that the target vector
−→
t itself is a point in the
semantic space.
As explained above, GSGP is a variant of GP
where the traditional crossover and mutation are re-
placed by new operators called Geometric Semantic
Operators (GSOs). The objective of GSOs is to de-
fine modifications on the syntax of GP individuals that
have a precise effect on their semantics. More in par-
ticular, as schematically shown in Figure 1, GSOs are:
• Geometric Semantic Crossover. This operator
generates only one offspring, whose semantics
stands in the line joining the semantics of the two
parents in the semantic space.
• Geometric Semantic Mutation. With this opera-
tor, by mutating an individual i, we obtain another
individual j such that the semantics of j stands
inside a ball of a given predetermined radius, cen-
tered in the semantics of i.
One of the reasons why GSOs became so popular in
the GP community (Castelli et al., 2015d; Castelli
et al., 2013; Castelli et al., 2015c; Ruberto et al.,
2014; Castelli et al., 2015e; Castelli et al., 2016b;
Castelli et al., 2015b; Vanneschi et al., 2017; Bakurov
et al., 2018; Bartashevich et al., 2018) is related to
the fact that GSOs induce an unimodal error surface
(on training data) for any supervised learning prob-
lem, where fitness is calculated using an error mea-
sure between outputs and targets. In other words, us-
ing GSOs the error surface on training data is guaran-
teed to not have any locally optimal solution. This
property holds, for instance, for any regression or
classification problem, independently of how big and
how complex data are (Vanneschi et al., 2014; Castelli
et al., 2016a). The definitions of the GSOs are, as
given in (Moraglio et al., 2012), respectively:
Geometric Semantic Crossover (GSC). Given
two parent functions T
1
,T
2
: R
n
→ R, the geomet-
ric semantic crossover returns the real function
T
XO
= (T
1
· T
R
) + ((1 − T
R
) · T
2
), where T
R
is a ran-
dom real function whose output values range in the
interval [0,1].
Geometric Semantic Mutation (GSM). Given a par-
ent function T : R
n
→ R, the geometric semantic mu-
tation with mutation step ms returns the real function
T
M
= T + ms · (T
R1
− T
R2
), where T
R1
and T
R2
are ran-
dom real functions.
Even though this is not in the original definition
of GSM, later contributions (Castelli et al., 2015a;
Castelli et al., 2014) have clearly shown that limiting
the codomain of T
R1
and T
R2
in a predefined interval
(for instace [0,1], as is done for T
R
in GSC) helps
improving the generalization ability of GSGP. As
in several previous works (Vanneschi et al., 2014;
Castelli et al., 2015a), here we constrain the outputs
of T
R
, T
R1
and T
R2
by wrapping them in a logistic
function. Only the definitions of the GSOs for
symbolic regression problems are given here, since
they are the only ones used in this work. For the
definition of GSOs for other domains, the reader is
referred to (Moraglio et al., 2012). Figure 1 shows a
graphical representation of the mapping between the
syntactic and semantic space given by the GSOs.
Using these operators, the semantics of the off-
spring is completely defined by the semantics of the
A Regression-like Classification System for Geometric Semantic Genetic Programming
41
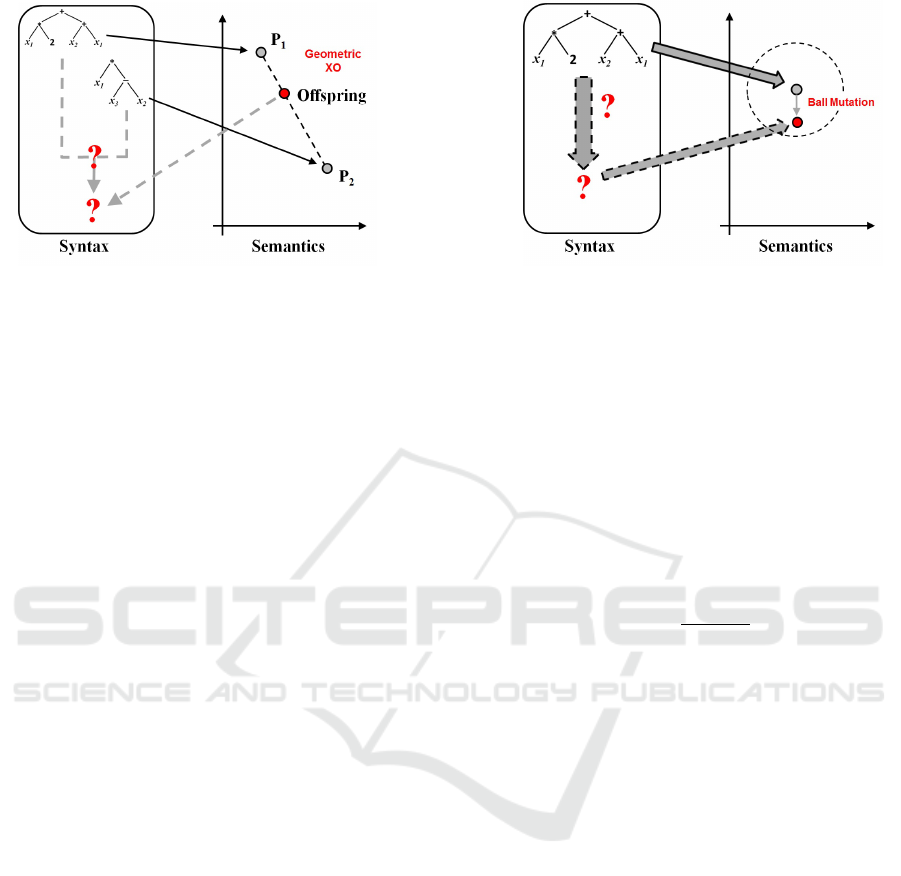
(a) (b)
Figure 1: Geometric semantic crossover (plot (a)) (respectively geometric semantic mutation (plot (b))) performs a trans-
formation on the syntax of the individual that corresponds to geometric crossover (respectively geometric mutation) on the
semantic space. In this figure, the unrealistic case of a bidimensional semantic space is considered, for simplicity.
parents: the semantics of an offspring produced by
GSC will lie on the segment between the semantics
of both parents (geometric crossover), while GSM de-
fines a mutation such that the semantics of the off-
spring lies within the ball of radius ms that surrounds
the semantics of the parent (geometric mutation).
As reported in (Moraglio et al., 2012), the prop-
erty of GSOs of inducing a unimodal error surface
has a price. The price, in this case, is that GSOs al-
ways generate larger offspring than the parents, and
this entails a rapid growth of the size of the individ-
uals in the population. To counteract this problem,
in (Castelli et al., 2015a) an implementation of GSOs
was proposed that makes GSGP not only usable in
practice, but also significantly faster than standard GP.
This is possible by means of a smart representation
of GP individuals, that allows us not to store their
genotypes during the evolution. This is the imple-
mentation used here. Even though this implementa-
tion is efficient, it does not solve the problem of the
size of the final model: the genotype of the final so-
lution returned by GSGP can be reconstructed, but it
is so large that it is practically impossible to under-
stand. This turns GSGP into a “black-box” system,
as many other popular machine learning systems are,
including deep neural networks.
3 REGRESSION-LIKE
CLASSIFICATION WITH GSGP
In this work, we use symbolic regression GSOs
to solve binary classification problems. The used
method is inspired by the functioning of the Percep-
tron neural network (Vanneschi and Castelli, 2019).
The two class labels that characterize the problem are
transformed into two numeric values. The values 0
and 1 have been used in this work. In this way, the
classification problem can be imagined as a regres-
sion problem, containing only 0 and 1 as possible tar-
get values. The output of a GP individual P is given
as input to an activation function, and the fitness of P
is the Root Mean Square Error (RMSE) between the
output of the activation function and the binary tar-
get. As customary in Perceptrons, we have used the
logistic activation function:
L(x) =
1
1 + e
−αx
(1)
This function has two asymptotes, respectively in 0
and 1, that are the target values we have used. Us-
ing this activation function should help us to improve
the generalization ability of the system, since faith-
fully approximating the target values 0 and 1 entails
a separation of data belonging to different classes. In
other words, the evolutionary process automatically
pushes towards the generation of GP individuals that
return large positive values for observations labeled
with 1, and large negative values for observations la-
beled with 0. At the same time, a good individual
should not return small values for any of the obser-
vations, since those small outputs would immediately
give a deteriorating contribution to the error.
This effect of data separation can be tuned by
modifying constant α in Equation (1). In order to have
a visual intuition of this, Figure 2 shows two different
logistic functions, one with α = 1 (plot (a)), and the
other with α = 0.1 (plot (b)).
From this figure, it is possible to see that, in order
to obtain the same approximation of an output value
equal to 1 (respectively 0), when α = 0.1 the output
has to be larger (respectively smaller), compared to
the case α = 1. So, for the same value of the er-
ror, data belonging to different classes are better sep-
arated for smaller values of α. On the other hand, of
ECTA 2019 - 11th International Conference on Evolutionary Computation Theory and Applications
42

(a) α = 1 (b) α = 0.1
Figure 2: Graphical representation of logistic functions (Equation (1)) with two different values of the α parameter.
Plot (a): α = 1; Plot (b): α = 0.1.
course, using small values of α also has a drawback:
given a prefixed value of the training error ε, finding
individuals with an error equal to ε will be computa-
tionally harder for smaller values of α. All this con-
sidered, finding the appropriate value of α, represent-
ing a good compromise between generalization abil-
ity and computational cost, is a challenge and various
preliminary tuning experiments have been needed, in
order to obtain the value used here. Once a model op-
timizing the RMSE on training data has been found,
it can be used for generalization on unseen data, sim-
ply using a threshold equal to zero on the output: ob-
servations with positive outputs will be predicted as
belonging to one of the two classes, while the other
observations will be categorized in the other class.
4 EXPERIMENTAL STUDY
4.1 Test Problems
In order to assess the effectiveness of the proposed ap-
proach, we considered ten binary classification prob-
lems, which present different characteristics in terms
of the number of attributes and instances. The related
datasets are publicly available on the UCI (Dua and
Karra Taniskidou, 2017) and openML (Vanschoren
et al., 2013) repositories. Table 1 reports, for each one
of these problems, the number of input features (vari-
ables) and data instances (observations) in the respec-
tive datasets and the repository from which the data
have been downloaded.
Since for each feature the range of values may
vary widely, data was normalized by using the Z-
normalization method: for each feature f
i
, we first
computed the mean µ
i
and the standard deviation σ
i
on the training set, then we applied the following
transformation:
z
i j
=
x
i j
− µ
i
σ
i
where x
i j
is the value of the i-th feature from the j −th
Table 1: Description of the benchmark problems. For each
dataset, the number of features (independent variables), the
number of instances (observations) that were reported.
Dataset #Features #Instances Repository
Fertility 9 100 openML
Hill 100 1212 openML
Ilpd 10 579 openML
Liver 6 345 UCI
Mammo 5 830 UCI
Musk 166 476 UCI
Qsar 41 1055 UCI
Retinopathy 19 1151 UCI
Spam 57 3037 UCI
Spect 22 266 UCI
Table 2: Parametrization used in the experiments.
Parameters Values
Population size 500
# generations 2000
Tree initialization Ramped Half-and-Half,
maximum depth 6
Crossover rate 0.0
Mutation rate 1.0
Function set +,−,∗, and protected /
Terminal set Input variables, random con-
stants
tournament size 4
Elitism Best individual always sur-
vives
# random trees 500
sample (both from the training or the test set) and z
i j
is the corresponding normalized value.
4.2 Experimental Settings
During the experimental study, we performed two sets
of experiments. In the first set we investigated the
classification performances of the proposed approach
as function of α and R, which represent the steepness
of the curve and width of the range for the random
constants in the terminal set, respectively. In the sec-
A Regression-like Classification System for Geometric Semantic Genetic Programming
43

ond set of experiments, instead, we compared the re-
sults achieved by our GSGP-based binary classifica-
tion system with those of an SVM classifier.
For all the considered test problems, 30 indepen-
dent runs of each studied system were executed. For
each problem, the related dataset was randomly split
into an equally sized and statistically independent
training and test set. The RMSE (see Section 3) on
the training set was used to compute the fitness of
each individual, whereas the error rate on the test set
1
was used to assess the classification performance on
unseen data of the best individual found at the end
of each run. All the results reported in the following
were computed averaging these classification results.
As for the parameters, we performed some prelimi-
nary trials to set them. These parameters were used
for all the experiments described below and are re-
ported in Table 2.
4.3 Experimental Results
As mentioned in previously, we performed two sets
of experiments. The first to find the best values for
the parameters α and R, and the second to compare
our results with those achieved by an SVM classifier.
Both sets of experiments are described as follows.
In the first set of experiments, we investigated how
the values ofα and R affect the classification perfor-
mance of our system since both values affect the final
output of the GP individuals. The former, which rep-
resents the limits of the range of the random constants
[−R,+R], affects the values the random constants of
the terminal set can assume, thus affecting the input
values given as input to the GP trees. As for α, as
mentioned in Section 3, it affects the separation of
samples belonging to different classes, with small α
values ensuring a better separation than larger ones
(see Figure 2). For this reason, we performed a facto-
rial experiment to investigate the relationship between
these two parameters. More specifically, we consid-
ered the following sets of parameters:
S
α
= {0.0001,0.001,0.01,0.05,0.1,0.2, 0.5}
S
R
= {1,2,5,10,20,50}
As for the remaining parameters, we used those
shown in Table 2. The plots in Figures 3 and 4 show
the test error as a function of the R for the different
α values tested. From the Figures it can be noted
1
Given an individual GP individual I representing the
function f
I
: R
n
→ R, where R
n
is the feature space, the test
sample x is labeled as belonging to the class 0 if f (x) > 0,
to the class 1 otherwise. In practice, the logistic function
is only used for training the system. This because in any
binary classification problem, the classifier must provide as
output a class label (0 or 1), instead of a numerical value.
that for all the considered datasets, too low α values
achieve the worse performance. In particular, the val-
ues 0.0001 and 0.001 achieve the worst performance,
while for most of the datasets, the value 0.01 per-
forms worse than the higher values. Moreover, it is
worth noting that for these three values the ”perfor-
mance ranking”, the performance differences among
these values, is kept for all the datasets and for all the
R values. These results prove that too low α values do
not improve the ”data separation” effect mentioned in
Section 3. This is most probably because these val-
ues produce too flat curves (similar to straight lines
with a low slope). In practice, these α values produce
similar output values for a large range of input values
returned by the GP individuals, thus reducing the se-
lective pressure of the evolutionary process. It is also
worth noting that the curves for these values exhibit a
high variability with respect to R, even if there is no
monotonic trend, that would prove that higher (lower)
R values perform better. As for the remaining α val-
ues (0.05,0.1,0.2,0.5) from the plots, it can be noted
that:
(i) in most of the cases they achieve similar results;
(ii) even when their performances are significantly
different, the related curves tend to be over-
lapped;
(iii) on several datasets (see for example Musk in
Figure 3(c) or Spam in Figure 4(c)) the value 0.5
tends to perform better than the smaller values.
Maybe a better explanation for these results is that
these α values allow better differentiation between
samples belonging to different classes, with the posi-
tive (negative) ones labeled as 1 (0) and correctly clas-
sified by GP individuals yielding positive (negative)
outputs. In fact, for an individual I the error for a
given sample x is computed as
(L( f
I
(x)) −l
x
)
2
(2)
where f
I
(x) is the function encoded by I, L the
(logistic) activation function (see Equation 1) and l
x
the label of x (0 or 1). Then, according to the above
equation, the effect of higher α values is twofold:
they allow reducing the RMSE for correct classifica-
tion even when f
I
returns small values and increasing
the RMSE even for misclassifications characterized
by small values. In practice, those higher α values
better reward/penalize correctly/incorrectly classified
instances.
To better explain this concept, we drew the logistic
function for these values (Figure 5) and we also com-
puted the output values of the logistic functions for
these remaining α for some samples. From the table it
is worth noting that for small f (x) output values (see
ECTA 2019 - 11th International Conference on Evolutionary Computation Theory and Applications
44
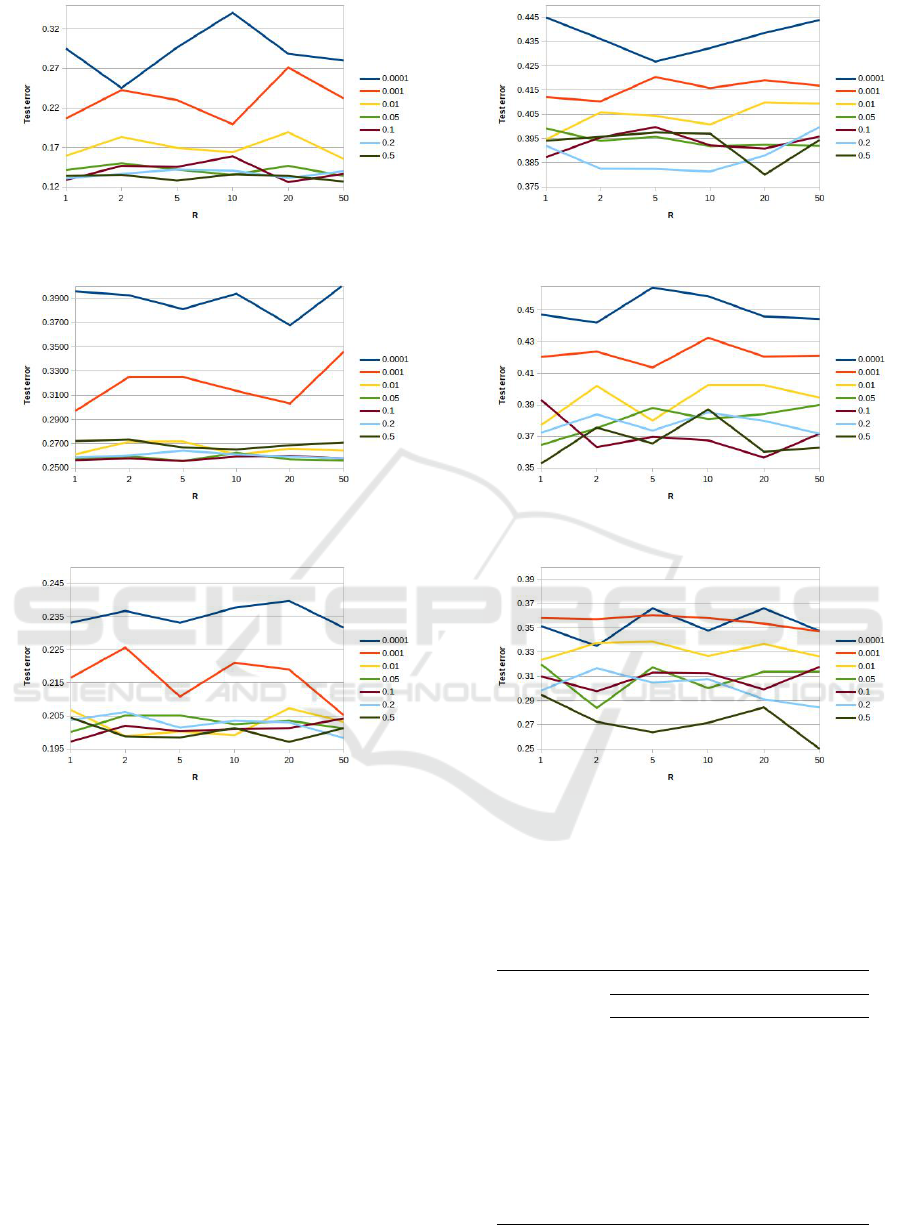
(a) Fertility (b) Hill
(c) Ilpd (d) Liver
(e) Mammo (f) Musk
Figure 3: Test error as a function of the limits of the range of the random constants [−R,+R] for the first six datasets of
Table 1.
for examples the samples 1 and 3), L with α = 0.05 re-
turns similar values both for correctly (sample #1) and
incorrectly classified (sample #3) samples, whereas
with α = 0.5 L returns quite different values for these
samples. Similar considerations can be done for the
other samples. For example, for the misclassified
sample #8, with f
I
(x) = −11.2, L returns 0.41 with
α = 0.05, whereas it returns 0.00 with α = 0.5.
4.4 Comparison with SVMs
In order to test the effectiveness of the proposed ap-
proach, we compared its results with those achieved
by a Support Vector Machine (SVM). Support Vec-
tor Machines (SVMs) are supervised learning meth-
Table 3: Output values of the logistic functions for some
samples, where f
I
(x) is the output value of a generic GP
individual I. Rows in bold represent misclassified samples.
# l
x
f
I
(x) (L( f
I
(x)) −l
x
)
2
(Eq. (2))
0.05 0.1 0.2 0.5
1 0 -1.6 0.23 0.21 0.18 0.10
2 0 -16.5 0.093 0.026 0.001 6.8e-8
3 0 1.2 0.27 0.28 0.31 0.42
4 0 7.2 0.35 0.45 0.65 0.95
5 1 1.4 0.23 0.22 0.19 0.11
6 1 3.5 0.21 0.17 0.11 0.02
7 1 -2 0.28 0.30 0.36 0.53
8 1 -11.2 0.41 0.57 0.82 0.99
9 1 6.1 0.18 0.12 0.05 2.04e-3
A Regression-like Classification System for Geometric Semantic Genetic Programming
45
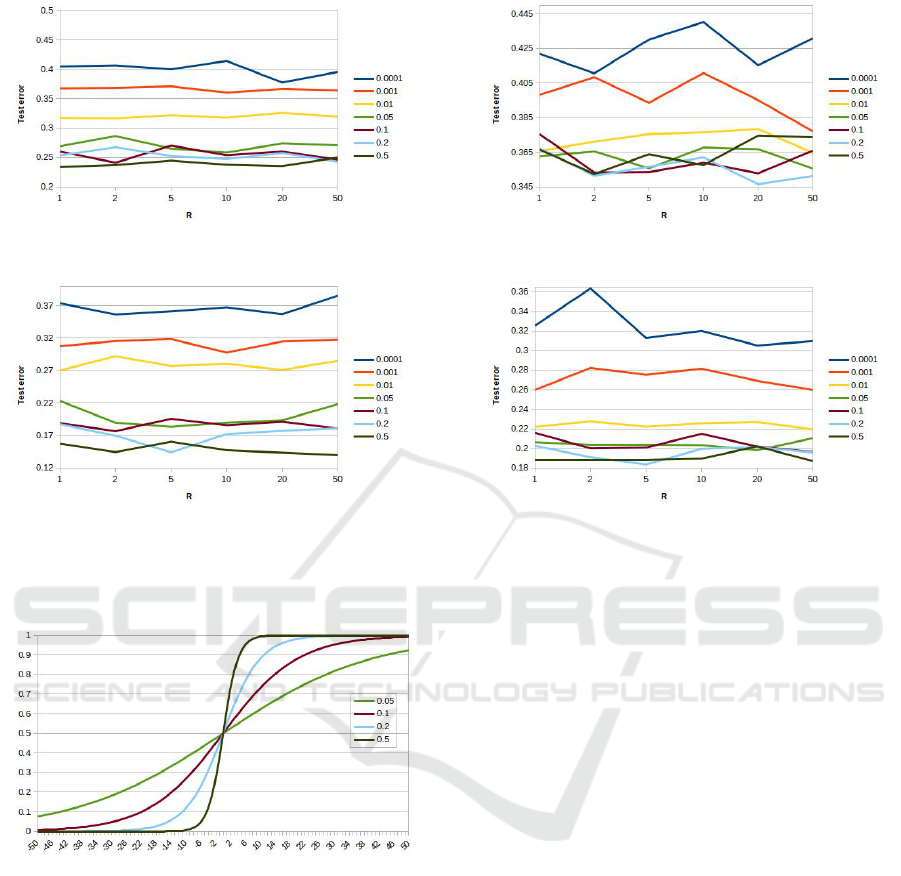
(a) Qsar (b) Retinopathy
(c) Spam (d) Spect
Figure 4: Test error as a function of the limits of the range of the random constants [−R,+R] for the last four datasets of Table
1.
Figure 5: The logistic functions for the α values: 0.05, 0.1,
0.2 and 0.5.
ods that are based on the concept of decision planes,
which linearly separates, in the feature space, objects
belonging to the different classes (Cortes and Vapnik,
1995). Intuitively, given two classes to be discrimi-
nated in a given feature space, a good separation is
achieved by the hyperplanes that have the largest dis-
tance to the nearest training points belonging to dif-
ferent classes; in general, the larger the margin, the
lower the generalization error of the classifier. While
the basic idea of the SVM applies to linear classifiers,
they can be easily adapted to non-linear classifica-
tion tasks by using the so called ”kernel trick”, which
implies the mapping of the original features into a
higher dimensional feature space. Differently from
other classifier schemes, whose learning usually im-
plies stochastic and suboptimal gradient descent pro-
cedures, e.g., NN and DT, given a training set, the
SVM learning optimization problem can be solved by
using quadratic programming algorithms which pro-
vide exact solutions. SVMs have shown to be ef-
fective in solving many real world applications like
handwriting recognition, text categorization and im-
age classification (Byun and Lee, 2002).
As suggested by (Lin and Lin, 2003; Keerthi and Lin,
2003), we chose the radial basis function (RBF) as
kernel in our experiments:
K(x, y) = e
−γkx−yk
2
where x and y are two generic samples in the feature
space.
As for the implementation, we used that provided
by the LIBSVM public domain software (Chang and
Lin, 2011). Moreover, to perform a fair comparison
with the GSGP, for each considered problem, we per-
formed 30 runs. As mentioned before, SVMs are de-
terministic, i.e. for a given training set the algorithm
always returns the same set of support vectors. For
this reason, to get averaged results on unseen data, for
each run the data was randomly shuffled and split into
a training and a test set, equally sized. The results
reported in the following have been averaged on the
ECTA 2019 - 11th International Conference on Evolutionary Computation Theory and Applications
46

accuracy obtained on the 30 generated test sets.
To statistically validate the comparison results,
we performed the non-parametric Wilcoxon rank-sum
test. The comparison results are shown in Table 4,
which also shows the p-value for the Wilcoxon test.
From the table it can be observed that on eight of the
ten considered problems, our GSGP-based classifier
outperforms the SVM, while as for the remaining two
(Fertility and Mammo), the differences are not statis-
tically significant (p-value > 0.05). Moreover, from
the table it can be also noted that in most of the cases
performance differences increase with the number of
features. This seems to suggest that the evolutionary
search process implemented by our approach is able
to: (i) effectively identify the features that better dis-
criminate samples belonging to different classes; (ii)
effectively find the functions (expressions) of these
features that characterize the classes of the problem
at hand.
5 CONCLUSIONS AND FUTURE
WORK
A new classification approach based on Geometric
Semantic Genetic Programming (GSGP) was pre-
sented in this paper. Despite the simplicity of the idea,
the presented results showed that this approach was
able to outperform Support Vector Machines (SVMs)
on 8 different complex real-life problems, returning
comparable results on the remaining 2 test problems.
These results encourages us to pursue the idea, and we
believe that this work could pave the way to a set of
applicative successes, comparable with the ones that
GSGP has obtained on regression and forecasting in
recent years (Vanneschi et al., 2014).
An important part of our current work is an at-
tempt of extending the proposed approach to multi-
Table 4: Comparison results (accuracy, expressed in per-
centage) between the proposed approach and the SVM clas-
sifier. Rows in bold highlight problems with no statistical
significant differences.
Dataset SVM GSGP p-value
Fertility 87.07 87.40 0.7024
Hill 51.00 61.28 1.73e-6
Ilpd 71.16 74.44 1.90e-6
Liver 58.84 64.73 2.34e-6
Mammo 80.23 80.29 0.7584
Musk1 61.08 75.00 2.34e-6
Qsar 67.01 76.63 7.68e-6
Retinopathy 59.40 65.35 3.52e-6
Spam 69.86 86.01 1.73e-6
Spect 79.27 81.65 2.20e-3
class classification. Our objective is, if possible, to be
able to solve, with success, complex multi-class clas-
sification tasks, maintaining the simplicity of the idea
presented here. Furthermore, in the future, we plan
to integrate the proposed approach with local search
strategies and with regularization algorithms, in order
to further stress, possibly at the same time, its opti-
mization power on training data, and its generaliza-
tion ability. Inheriting ideas from SVMs, like maxi-
mum separation hyperplane and kernel trick, is also
a possibility that we are considering to extend the
work. Last but not least, we are planning to imple-
ment a parallel and distributed version of the system,
with highly heterogeneous populations. In this way,
different variants of the algorithm could be executed
in parallel, like different parameter settings, different
representations for the solutions, different activation
functions and different fitness functions, just to men-
tion a few.
ACKNOWLEDGMENTS
This work was supported by national funds through
FCT (Fundac¸
˜
ao para a Ci
ˆ
encia e a Tecnologia)
under project DSAIPA/DS/0022/2018 (GADgET)
and project PTDC/CCI-INF/29168/2017 (BINDER).
Mauro Castelli acknowledges the financial support
from the Slovenian Research Agency (research core
funding No. P5-0410).
REFERENCES
Bakurov, I., Vanneschi, L., Castelli, M., and Fontanella, F.
(2018). Edda-v2–an improvement of the evolution-
ary demes despeciation algorithm. In International
Conference on Parallel Problem Solving from Nature,
pages 185–196. Springer.
Bartashevich, P., Bakurov, I., Mostaghim, S., and Van-
neschi, L. (2018). Pso-based search rules for aerial
swarms against unexplored vector fields via genetic
programming. In International Conference on Par-
allel Problem Solving from Nature, pages 41–53.
Springer.
Byun, H. and Lee, S.-W. (2002). Applications of support
vector machines for pattern recognition: A survey. In
Proceedings of the First International Workshop on
Pattern Recognition with Support Vector Machines,
pages 213–236, London, UK. Springer-Verlag.
Castelli, M., Castaldi, D., Giordani, I., Silva, S., Vanneschi,
L., Archetti, F., and Maccagnola, D. (2013). An ef-
ficient implementation of geometric semantic genetic
programming for anticoagulation level prediction in
pharmacogenetics. In Portuguese Conference on Arti-
ficial Intelligence, pages 78–89. Springer.
A Regression-like Classification System for Geometric Semantic Genetic Programming
47

Castelli, M., Silva, S., and Vanneschi, L. (2015a). A
c++ framework for geometric semantic genetic pro-
gramming. Genetic Programming and Evolvable Ma-
chines, 16(1):73–81.
Castelli, M., Trujillo, L., and Vanneschi, L. (2015b). Energy
consumption forecasting using semantic-based ge-
netic programming with local search optimizer. Com-
putational intelligence and neuroscience, 2015:57.
Castelli, M., Trujillo, L., Vanneschi, L., Silva, S., et al.
(2015c). Geometric semantic genetic programming
with local search. In Proceedings of the 2015 An-
nual Conference on Genetic and Evolutionary Com-
putation, pages 999–1006. ACM.
Castelli, M., Vanneschi, L., and De Felice, M. (2015d).
Forecasting short-term electricity consumption using
a semantics-based genetic programming framework:
The south italy case. Energy Economics, 47:37–41.
Castelli, M., Vanneschi, L., Manzoni, L., and Popovi
ˇ
c, A.
(2016a). Semantic genetic programming for fast and
accurate data knowledge discovery. Swarm and Evo-
lutionary Computation, 26:1–7.
Castelli, M., Vanneschi, L., and Popovi
ˇ
c, A. (2015e). Pre-
dicting burned areas of forest fires: an artificial intel-
ligence approach. Fire ecology, 11(1):106–118.
Castelli, M., Vanneschi, L., and Popovi
ˇ
c, A. (2016b). Pa-
rameter evaluation of geometric semantic genetic pro-
gramming in pharmacokinetics. International Journal
of Bio-Inspired Computation, 8(1):42–50.
Castelli, M., Vanneschi, L., and Silva, S. (2014). Prediction
of the unified parkinson’s disease rating scale assess-
ment using a genetic programming system with ge-
ometric semantic genetic operators. Expert Systems
with Applications, 41(10):4608–4616.
Chang, C.-C. and Lin, C.-J. (2011). LIBSVM: A library for
support vector machines. ACM Transactions on Intel-
ligent Systems and Technology, 2:27:1–27:27. Soft-
ware available at http://www.csie.ntu.edu.tw/
˜
cjlin/libsvm.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
Machine Learning, 20(3):273–297.
Dua, D. and Karra Taniskidou, E. (2017). UCI machine
learning repository.
Keerthi, S. S. and Lin, C.-J. (2003). Asymptotic behaviors
of support vector machines with gaussian kernel. Neu-
ral Computation, 15(7):1667–1689.
Koza, J. R. (1992). Genetic Programming: On the Pro-
gramming of Computers by Means of Natural Selec-
tion. MIT Press, Cambridge, MA, USA.
Lin, H.-T. and Lin, C.-J. (2003). A study on sigmoid ker-
nels for svm and the training of non-psd kernels by
smo-type methods. Technical report, Department of
Computer Science, National Taiwan University.
Moraglio, A., Krawiec, K., and Johnson, C. (2012). Ge-
ometric semantic genetic programming. In Coello,
C., Cutello, V., Deb, K., Forrest, S., Nicosia, G., and
Pavone, M., editors, Parallel Problem Solving from
Nature - PPSN XII, volume 7491 of Lecture Notes in
Computer Science, pages 21–31. Springer Berlin Hei-
delberg.
Ruberto, S., Vanneschi, L., Castelli, M., and Silva, S.
(2014). Esagp–a semantic gp framework based on
alignment in the error space. In European Conference
on Genetic Programming, pages 150–161. Springer.
Vanneschi, L., Bakurov, I., and Castelli, M. (2017). An ini-
tialization technique for geometric semantic gp based
on demes evolution and despeciation. In Evolutionary
Computation (CEC), 2017 IEEE Congress on, pages
113–120. IEEE.
Vanneschi, L. and Castelli, M. (2019). Multilayer percep-
trons. In Ranganathan, S., Gribskov, M., Nakai, K.,
and Sch
¨
onbach, C., editors, Encyclopedia of Bioinfor-
matics and Computational Biology, pages 612 – 620.
Academic Press, Oxford.
Vanneschi, L., Silva, S., Castelli, M., and Manzoni, L.
(2014). Geometric semantic genetic programming for
real life applications. In Genetic programming theory
and practice xi, pages 191–209. Springer.
Vanschoren, J., van Rijn, J. N., Bischl, B., and Torgo,
L. (2013). Openml: Networked science in machine
learning. SIGKDD Explorations, 15(2):49–60.
ECTA 2019 - 11th International Conference on Evolutionary Computation Theory and Applications
48
