
Associative Classification in Big Data through a G3P Approach
J. M. Luna, F. Padillo and S. Ventura
Department of Computer Science and Numerical Analysis, University of Cordoba, Spain
Keywords:
Big Data, Associative Classification, Evolutionary Computation.
Abstract:
The associative classification field includes really interesting approaches for building reliable classifiers and
any of these approaches generally work on four different phases (data discretization, pattern mining, rule
mining, and classifier building). This number of phases is a handicap when big datasets are analysed. The aim
of this work is to propose a novel evolutionary algorithm for efficiently building associative classifiers in Big
Data. The proposed model works in only two phases (a grammar-guided genetic programming framework is
performed in each phase): 1) mining reliable association rules; 2) building an accurate classifier by ranking
and combining the previously mined rules. The proposal has been implemented on Apache Spark to take
advantage of the distributed computing. The experimental analysis was performend on 40 well-known datasets
and considering 13 algorithms taken from literature. A series of non-parametric tests has also been carried
out to determine statistical differences. Results are quite promising in terms of reliability and efficiency on
high-dimensional data.
1 INTRODUCTION
Nowadays, data storage is getting cheaper and
cheaper what implies an increment in the efforts for
analyzing and extracting valuable information from
large datasets. This issue has motivated recent re-
search studies on Big Data (Chen et al., 2012), which
encompasses a set of techniques to face up problems
derived from the management and analysis of huge
quantities of data. In data analysis, two different
tasks are considered: descriptive tasks, which depict
intrinsic and important properties of data (Agrawal
et al., 1993); and predictive tasks, which predict out-
put variables for unseen data (Han and Kamber, 2011)
by learning a mapping between a set of input vari-
ables and the output variable. Focusing on predic-
tive tasks, different methodologies can be considered
to build accurate models that predict the output vari-
able: rule-based systems (Han and Kamber, 2011),
decision trees (Quinlan, 1993) and support vector ma-
chines (Cortes and Vapnik, 1995), just to list a few.
From all these methodologies, rule-based classifiers
provide a high-level of interpretability and, therefore,
classification results can be explained since rules tend
to be easily understood and interpreted by the end-
user. An example of such systems is classification
based on association rule mining, generally known
as associative classification (AC) (Ventura and Luna,
2018), which integrates a descriptive task (association
rule mining (Agrawal et al., 1993)) in the process of
inferring a new classifier (Liu et al., 1998).
AC algorithms generally operate in four phases.
First, data transformation in which continuous at-
tributes are preprocessed and defined in a discrete do-
main. Second, the attribute values are combined and
characterized by an occurrence value beyond a given
threshold. Third, association rules (the consequent
is fixed to the class variable) are produced from the
set of frequent attributes previously obtained. Finally,
rules are ranked and post-processed to build an accu-
rate classifier. However, these numerous phases, spe-
cially when working on Big Data, become unfeasible
to be addressed by existing methodologies even when
advanced techniques in distributed computing (Dean
and Ghemawat, 2008) are considered. At this point,
a reduction in the number of required phases has
been recently addressed by considering evolutionary
algorithms (EAs) (Alcal
´
a-Fdez et al., 2011). Here,
rules were directly mined without a previous step of
extracting patterns (combination of attribute values).
Nevertheless, even for a lower number of phases, the
computational complexity in Big Data is still a hand-
icap since it exponentially increases with the number
of variables (2
k
−1 solutions can be found from k vari-
ables). Recent approaches (MRAC (Bechini et al.,
2016), MRAC+ (Bechini et al., 2016), DAC (Ven-
turini et al., 2017), etc.) have dealt with the prob-
lem through current advances in distributed comput-
94
Luna, J., Padillo, F. and Ventura, S.
Associative Classification in Big Data through a G3P Approach.
DOI: 10.5220/0007688400940102
In Proceedings of the 4th International Conference on Internet of Things, Big Data and Security (IoTBDS 2019), pages 94-102
ISBN: 978-989-758-369-8
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

ing even though they were based on classical algo-
rithms and only provided an improvement in runtime.
The aim of this paper is therefore to propose a
new grammar-guided genetic programming (G3P) al-
gorithm for AC on Big Data by considering Spark.
G3P has been already studied in mining association
rules (Ventura and Luna, 2016), and it has proved to
obtain excellent results in both introducing subjective
knowledge into the mining process and constraining
the search space by including syntax constraints. An
important feature of the proposed algorithm, which
really improves the state-of-the-art, is the running on
just two phases: 1) mining reliable association rules;
and 2) building an accurate classifier. In the first
stage, the best rules for each class are obtained by
means of multiple and independent evolutionary pro-
cesses (no discretization step is required since the use
of a grammar enables continuous features to be en-
coded). In the second stage, the set of the previ-
ously mined rules are ranked and combined to form
an accurate classifier. Since rules for each class is ob-
tained, it is guaranteed that minority/majority classes
are equally considered. This is an additional major
feature of the proposal since many AC approaches are
focused on improvements of classification accuracy,
not paying attention to the imbalance problem. In
an experimental analysis, the proposal has been com-
pared to multiple AC approaches as well as traditional
classification algorithms. In this study, both sequen-
tial and trending MapReduce AC algorithms are con-
sidered and compared. Experiments were performed
on a total of 40 datasets and results were validated by
non-parametric statistical tests.
The rest of the paper is organized as follows. Sec-
tion 2 presents the most relevant definitions and re-
lated work; Section 3 describes the proposed algo-
rithm; Section 4 presents the experimental results, and
some concluding remarks are outlined in Section 5.
2 PRELIMINARIES
In this section, the associative classification task is
formally defined as well as some Big Data architec-
tures.
2.1 Associative Classification
In 1998, Liu et al. (Liu et al., 1998) connected associ-
ation rule mining (ARM) and classification rule min-
ing to give rise to the task known as associative clas-
sification (AC). The aim of this task was to build an
accurate and high interpretable classifier by means of
rules obtained from ARM techniques. In order to ob-
tain this kind of classifiers many methods have been
proposed along the years (Thabtah, 2007), almost all
of them being based on exhaustive search ARM algo-
rithms (Agrawal et al., 1993). In general, existing AC
approaches work on four different steps, which is a
real handicap when computationally expensive prob-
lems are addressed. For instance, the extraction of
patterns (itemsets) and association rules from them
implies two different and computationally hard prob-
lems (the number of solutions exponentially increases
with the number of items in data) (Padillo et al.,
2017). To overcome these and other problems (work-
ing on continuous domains), some researchers have
focused on the application of evolutionary algorithms
(EAs) for performing the AC task. At this point, the
combination of EAs with emerging paradigms like
MapReduce to process high volumes of data in an ac-
curate and efficient fashion is a trending topic (Padillo
et al., 2017).
2.2 Big Data Architectures
MapReduce (Dean and Ghemawat, 2008) is a re-
cent paradigm, firstly implemented by Hadoop (Lam,
2010), of distributed computing for Big Data in which
programs are composed of two main stages, that is,
map and reduce. In the map phase each mapper pro-
cesses a subset of input data and produces a set of
hk, vi pairs. Finally, the reducer takes this new list
to produce the final values. A major drawback of
Hadoop, however, is it imposes an acyclic data flow
graph, and there are applications (iterative or inter-
active analysis (Zaharia et al., 2010)) that cannot be
efficiently modeled through this kind of graph. Be-
sides, Hadoop cannot keep intermediate data in mem-
ory for faster performance. To solve these downsides,
Apache Spark has risen up for solving all the defi-
ciencies of Hadoop, introducing an abstraction called
RDD (Resilient Distributed Datasets) to store data in
main memory as well as a new approach that consid-
ers micro-batch technology.
3 G3P PROPOSAL
The proposed grammar-guided genetic programming
(G3P) algorithm has been designed to be parallel
without affecting the final accuracy. A major fea-
ture of the proposed model, named G3P-AC, is it
works in two stages: 1) mining reliable association
rules; 2) building an accurate classifier by ranking
and combining the previously mined rules. The first
step is responsible for extracting the best set of rules
Associative Classification in Big Data through a G3P Approach
95

G = (Σ
N
, Σ
T
, P, S) with:
S = <Rule>
Σ
N
= {<Rule>, <Antecedent>, <Consequent>, <Condition>, <Nominal>,
<Numerical>}
Σ
T
= {attribute, value, AND, =, IN, Min value, Max value, class}
P = {<Rule> ⇐ <Antecedent>, <Consequent> ;
<Antecedent> ⇐ <Condition> (AND <Condition>)* ;
<Consequent> ⇐ class = value ;
<Condition> ⇐ <Numerical> | <Nominal> ;
<Numerical> ⇐ name IN Min value, Max value ;
<Nominal> ⇐ name = value ;
}
Figure 1: Context-free grammar defined for the rule extraction stage of the proposed algorithm. The grammar is expressed in
extended BNF notation.
for each class by means of several evolutionary pro-
cesses (one per class). Such evolutionary processes
follow a normal generational genetic algorithm with
elitism in which the best solutions found until that
moment are kept unaltered. The quality of the so-
lutions is measured by a fitness function (taking val-
ues in the range [0, 1] where the higher the better) that
was designed to obtain a good trade-off between re-
liability (confidence of the rule) and frequency with
regard to the class, that is, F(R) = con f idence(R) ×
support(R)/support(Y ) for a rule R ≡ X → Y . This
first stage starts by encoding a set of individuals
through a number of production rules from a context-
free grammar (see Figure 1). Thanks to this grammar,
an expert in the domain may determine the maximum
or minimum length of the rules (in the form of class
association rules), and the kind of conditions each rule
should include (Ventura and Luna, 2016). This evolu-
tionary process works in an iterative fashion through
a number of generations previously fixed by the end-
user. However, this iterative process may finish with-
out reaching the maximum number of generations in
situations where the average fitness value of individ-
uals within the elite does not change for a number of
generations. Additionally, in order to guarantee the
quality of the solutions and to avoid redundant solu-
tions a pattern weighting scheme is used in the elite
population (Herrera et al., 2011). Finally, in order to
produce new individuals, a tournament selector is ap-
plied in each iteration of the evolutionary process and
new individuals are then obtained by the traditional
G3P genetic operators, producing a new population
that will update both the previous one and the elite.
The genetic operators used to produce new solutions
are two well-known operators that have proved to ob-
tain promising results in G3P (McKay et al., 2010).
The second stage of the proposed algorithm fol-
lows an evolutionary process that is similar to the
one of the first stage. In this second stage, how-
ever, the aim is to filter, sort and arrange previously
mined rules to form a final classifier. Unlike the pre-
vious stage, individuals are represented in a differ-
ent fashion since now each individual represents a
set of rules that will form the final classifier (not a
single rule as in the first stage). It is important to
remark that no rule can be produced in this phase
and it only works with those previously obtained in
the first phase. This second phase considers a gram-
mar (see Figure 2) to customize the shape of the final
classifer. Here, the quality of the solutions (sets of
rules that represents a classifier) is calculated as the
average accuracy in training for each class. Given
an individual ind, the fitness function F is defined
as F(ind) = (1/l) ∗ (
∑
m
n=1
accuracy
c
/m). Here, m is
the number of classes, l the number of rules included
in ind and accuracy
c
is the accuracy in the training
set for the class c. It was designed to avoid classi-
fiers including a large number of rules and to penal-
ize classifiers that ignore minority class. Similarly to
the previous stage, the genetic operators used to pro-
duce new solutions are two well-known operators that
have proved to obtain promising results (McKay et al.,
2010).
These two stages have been designed to be run on
a cluster of computers that includes a central com-
puter (driver program) acting as point of coordination,
and several additional nodes that collaboratively work
G = (Σ
N
, Σ
T
, P, S) with:
S = Classifier
Σ
N
= {<Classifier>, <Rules>, <DefaultClass>}
Σ
T
= {rule, class, =, value}
P = {<Classifier> ⇐ <Rules> <DefaultClass> ;
<Rules> ⇐ rule (rule)* ;
<DefaultClass> ⇐ class = value ;
}
Figure 2: Context-free grammar defined for the rule selec-
tion stage of the proposed algorithm. The grammar is ex-
pressed in extended BNF notation. The terminal symbol
rule represents an individual from the previous phase (see
grammar shown in Figure 1).
IoTBDS 2019 - 4th International Conference on Internet of Things, Big Data and Security
96

Algorithm 1: Proposed algorithm on Spark - Rule extraction stage.
procedure Driver
1: pool rules ←
/
0
2: for all c in classes do
3: new Thread() do
4: P
0,c
← Generate a random population of n rules following the grammar with class c
5: elite
c
←
/
0
6: for i = 0 to #Generations do
7: MapReduce to evaluate rules from (P
i,c
)
8: elite
c
← Maintain elitism using P
i,c
∪ elite
c
9: Stop if mean(elite
c
) has not changed in a number of generations specified by the user
10: selected invididuals ← Tournament selector to P
i,c
11: for all pair in selected individuals do
12: o f f spring ← pair
13: if Rand number(0,1) > Prob
cro
then
14: o f f spring ← Cross o f f spring
15: end if
16: for all individual of the o f f spring do
17: if Rand number(0,1) > Prob
mut
then
18: o f f spring ← Mutate individual
19: end if
20: end for
21: end for
22: P
i+1,c
← o f f spring ∪ best n individuals from elite
c
23: end for
24: pool rules ← pool rules ∪ elite
c
25: end
26: end for
end procedure
procedure Map(instance, P
i,c
)
1: for all r in P
i,c
do
2: measures ← r.evaluate(instance)
3: emit(r, measures)
4: end for
end procedure
procedure Reduce(r, measures)
1: finalMeasures ← (0, 0, 0) // Support antecedent, consequent and rule
2: for all measure in measures do
3: for i = 0 to 2 do
4: f inalMeasures[i] ← f inalMeasures[i] + measure[i]
5: end for
6: end for
7: fitness ← calculateFitness( f inalMeasures)
8: emit(r, f itness)
end procedure
with the driver. The two phases (rule extraction and
rule selection) are described as follows:
• Rule Extraction. The driver program starts by
creating as many threads as classes exist in data
(see Driver procedure of Algorithm 1). After
that, each thread enqueues several MapReduce
jobs (which will run on several compute nodes)
to evaluate its population. In the Map proce-
dure (see Algorithm 1), the input for each map-
per is a chunk of data and the population. A
group of pairs hkey,valuei are generated by each
mapper, key being the rule, value representing a
tuple of support values (antecedent, consequent
and whole rule). Reducers (see Reduce pro-
cedure, Algorithm 1), on the contrary, receive
the previously created hkey, valuei pairs as in-
put. Here, the global support values for an-
tecedent (support(X)), consequent (support(Y ))
and rule (support(R)) are obtained for each in-
dividual. Once, these three measures have being
calculated, the fitness function is obtained as F(R)
= con f idence(R) × support(R)/support(Y ) and
the rules (individuals) are returned to their respec-
tive threads. Each thread continues its evolution-
ary process until a new population is required to
be evaluated and the previous process repeated.
Associative Classification in Big Data through a G3P Approach
97
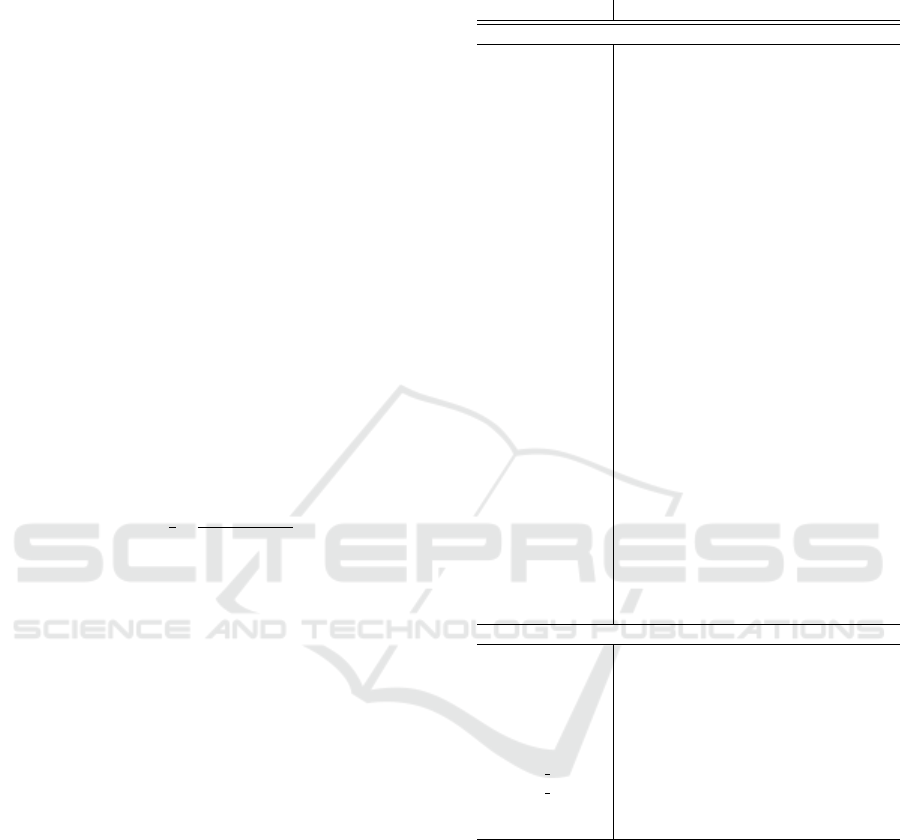
∑
m
i=0
numberGenerations(c
i
) represents the num-
ber of MapReduce jobs, where m is the number of
classes, and numberGenerations(c
i
) is the num-
ber of generations for the i-th class. Once all the
threads end, a pool of rules is obtained by gath-
ering rules for each class (obtained by different
threads). This pool of rules is saved on distributed
structures of storage as RDD for Spark, enabling
a distributed fast access as well as a large quantity
of results to be saved.
• Rule Selection. In this second phase, the eval-
uation process is the only procedure to be paral-
lelized. In this regard, Algorithm 2 shows pseudo-
code for the evaluation process through a MapRe-
duce Job. The Map procedure (see Algorithm 2)
receives two elements as input: a subset of the
dataset, and the population. A group of pairs
hkey,valuei is generated by each mapper, where
the key is the rule-set, and the value is a tuple
with the accuracy values per class. The reducer
procedure (see Algorithm 2), on the contrary, re-
ceives the previously created hkey,valuei pairs as
input. Its goal is to calculate the total accuracy
values per class (considering the whole dataset).
After that, the fitness function is calculated as
F(rule − set) =
1
l
∗
∑
m
n=1
accuracy
c
m
and the rule-set
is returned. The output of the reducer is the eval-
uated population considering the whole dataset.
4 EXPERIMENTAL ANALYSIS
The aim of this section is to study the behaviour
of our proposal on more than 40 well-known
datasets (see Table 1)— all of them are available at
KEEL (Triguero et al., 2017) repository. Results are
compared to multiple AC algorithms by considering
a series of non-parametric tests. All the experiments
have been run on a HPC cluster comprising 12 com-
pute nodes, with two Intel E5-2620 microprocessors
at 2 GHz and 24 GB DDR memory. Cluster operating
system was Linux CentOS 6.3. As for the specific de-
tails of the used software, the experiments have been
run on Spark 2.0.0. To quantify the usefulness of
the solutions in this experimental analysis both accu-
racy rate (Han and Kamber, 2011) and Cohen’s kappa
rate (Ben-David, 2008) are considered. A 10-fold
stratified cross-validation has been used, and each al-
gorithm has been executed 5 times. Thus, the results
for each dataset are the average result of 50 different
runs. The algorithms used in this experimental anal-
ysis are divided into two main groups, that is, clas-
sical and Big Data algorithms. As for classical algo-
rithms, it includes CBA (Liu et al., 1998), CBA2 (Liu
Table 1: List of datasets (in alphabetical order) used for the
experimental study. They have been categorized into two
categories: classical datasets and Big Data datasets.
Datasets #Attributes #Instances #Classes
Classical datasets
Appendicitis 7 106 2
Australian 14 690 2
Banana 2 5,300 2
Breast 9 277 2
Cleveland 13 297 5
Contraceptive 9 1,473 3
Flare 11 1,066 6
German 20 1,000 2
Hayes-roth 4 160 3
Heart 13 270 2
Iris 4 150 3
Lymphography 18 148 4
Magic 10 19,020 2
Mammographic 5 830 2
Monk-2 6 432 2
Mushroom 22 5,644 2
Page-blocks 10 5,472 5
Phoneme 5 5,404 2
Pima 8 768 2
Post-operative 8 87 3
Saheart 9 462 2
Spectfheart 44 267 2
Splice 60 3,190 3
Tae 5 151 3
Tic-tac-toe
9 958 2
Titanic 3 2,201 2
Vehicle 18 846 4
Wine 13 178 3
Winequality 11 4,898 7
Wisconsin 9 683 2
Big Data datasets
Census 40 299,285 2
CoverType 54 581,012 2
Hepmass 28 10,500,000 2
Higgs 28 11,000,000 2
Poker 10 1,025,010 11
Kddcup1999 41 4,898,431 23
KDD99 2 41 4,856,151 2
KDD99 5 41 4,856,151 5
Record-Linkage 12 5,749,132 2
Sussy 18 5,000,000 2
et al., 2001), CMAR (Li et al., 2001), CPAR (Yin
and Han, 2003) and FARCHD (Alcal
´
a-Fdez et al.,
2011). Additionally, four classical rule-based classifi-
cation algorithms have been considered: C4.5 (Quin-
lan, 1993), RIPPER (Cohen, 1995), CORE (Tan et al.,
2006) and OneR (Holte, 1993). As for AC algo-
rithms designed for Big Data, the following are con-
sidered: MRAC (Bechini et al., 2016), MRAC+ (Be-
chini et al., 2016), DAC (Venturini et al., 2017) and
DFAC-FFP (Segatori et al., 2018).
IoTBDS 2019 - 4th International Conference on Internet of Things, Big Data and Security
98

Algorithm 2: Proposed algorithm on Spark - Rule selection stage.
procedure Algorithm
1: P
0
← Initialize a random population of n individuals (classifiers) including rules from pool rules
2: for i = 0 to #Generations do
3: for all ruleset in population do
4: MapReduce to evaluate ruleset
5: end for
6: best individual ← Best individual from P
i
7: Stop if best individual(P
i
) has not improved in a number of generations specified by the user
8: selected invididuals ← Apply tournament selector to P
i
9: for all pair in selected individuals do
10: o f f spring ← pair
11: if Rand number(0,1) > Prob
cro
then
12: o f f spring ← Cross o f f spring
13: end if
14: for all individual of the o f f spring do
15: if Rand number(0,1) > Prob
mut
then
16: o f f spring ← Mutate individual
17: end if
18: end for
19: end for
20: P
i+1
← o f f spring ∪ best invididual
21: end for
22: de f ault class ← Class whose frequency of occurrence is the highest
23: Classify using best individual and de f ault class
end procedure
procedure Map(chunk, ruleset)
1: accuracy class ← (0, ..., 0) // One per class in data
2: for all instance in chunk do
3: accuracy class[instance.class] ← accuracy class[instance.class] + Accuracy of ruleset in instance
4: end for
5: emit(ruleset, accuracy class)
end procedure
procedure Reduce(ruleset, accuracy class)
1: f inal accuracy class ← (0, ..., 0) // As many as classes exist in dataset
2: for all accuracy class in accuracy class do
3: for i = 0 until number classes do
4: f inal accuracy class[i] ← accuracy class[i]
5: end for
6: end for
7: f itness ← ruleset.calculateFitness( f inal accuracy class)
8: emit(ruleset, f itness)
end procedure
4.1 Quality of the Solutions
All the classical algorithms were run on the classical
datasets and the rankings for both accuracy and kappa
measures are shown in Table 2. Focusing on accu-
racy (see Table 2a), it is obtained that CMAR obtained
the worst result and it may be caused by the fact that
CMAR is based on exhaustive search algorithms that
cannot be directly run on continuous domains, requir-
ing therefore a discretization step that implies data-
loss. Besides, CMAR optimizes the confidence mea-
sure in isolation, generating very specific classifiers
that are not able to correctly predict unseen examples.
On the other hand, AC algorithms such as FARCHD,
CPAR and our proposal (G3P-AC) obtained the best
results with really small differences among them. Fi-
nally, focusing on the Kappa metric (see Table 2b),
very similar results were obtained and the three best
algorithms in ranking were FARCHD, CPAR and our
proposal.
The next step in this experimental analysis is the
study of whether there exist statistical differences
among all the algorithms and, therefore, several non-
parametric tests were carried out. First, a Friedman
test has been performed on the accuracy measure, ob-
taining a X
2
F
= 77.55 with a critical value of 21.66 and
a p-value = 4.93
−13
. In the same way, a X
2
F
= 70.66
with a critical value of 21.66 and a p-value = 1.12
−11
has been obtained for the Kappa metric. In both cases,
and considering a value α = 0.01, it is not possible
Associative Classification in Big Data through a G3P Approach
99
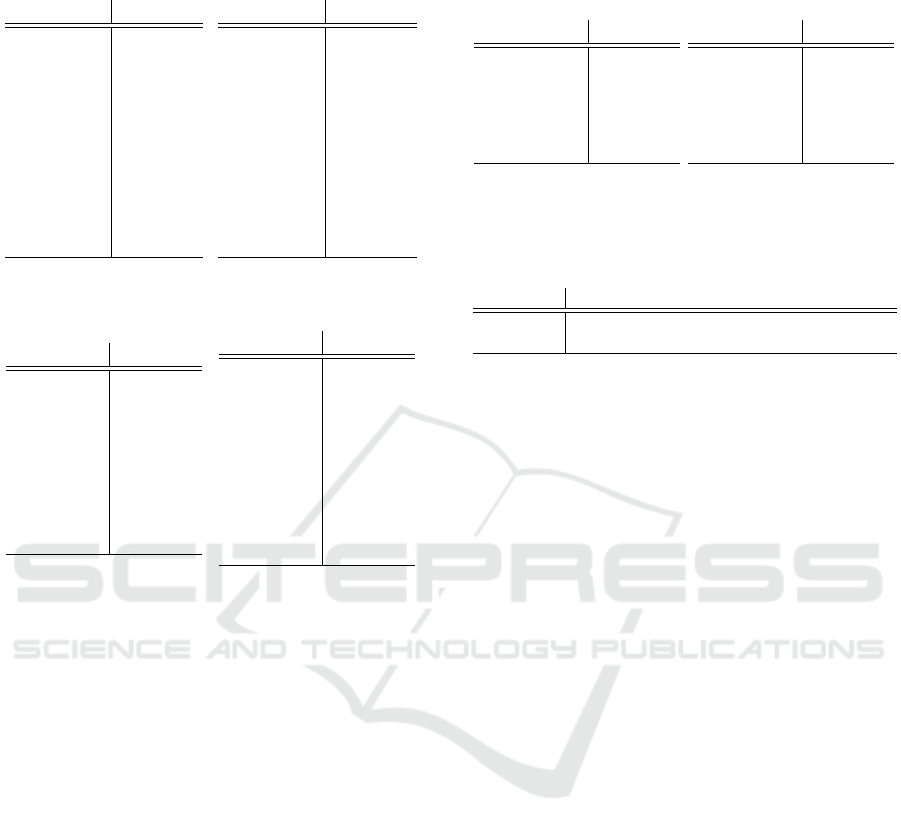
Table 2: Average ranking for each algorithm (sorted in de-
scending order) according to the Friedman test.
Algorithm Ranking
CMAR 7.916
OneR 7.350
CORE
7.150
CBA 6.233
Ripper 5.950
CBA2 4.816
C4.5 4.233
FARCHD 3.983
CPAR 3.683
G3P-AC 3.683
(a) Accuracy measure.
Algorithm Ranking
OneR 7.850
CMAR 7.400
CORE
7.383
CBA 6.100
Ripper 4.966
CBA2 4.766
C4.5 4.383
CPAR 4.283
FARCHD 4.150
G3P-AC 3.716
(b) Kappa measure.
Table 3: p-values for the Holland test with α = 0.01.
Vs G3P-AC
CBA 0.035
CBA2 0.933
CMAR 0.000
FARCHD 0.999
C4.5 0.998
Ripper 0.100
CORE 0.000
OneR 0.000
(a) Accuracy measure.
Vs G3P-AC
CBA 0.060
CBA2 0.965
CMAR 0.000
CPAR 0.999
FARCHD 1.000
C4.5 0.999
Ripper 0.890
CORE
0.000
OneR 0.000
(b) Kappa measure.
to assert that all the algorithm equally behave. In
this regard, a post-hoc test is performed (see Table 3)
by considering α = 0.01. Focusing on the accuracy
measure and taking our proposal as control, the post-
hoct test (see Table 3a) denotes some statistical dif-
ferences with regard to CMAR, CORE and OneR. It
should be noted that our proposal and CPAR did not
obtain any statistical differences so th post-hoc test
was not performed on them. In this sense, a Wilcoxon
signed rank test has been carried out on CPAR and
our proposal, obtaining a Z-value = −0.6582 with p-
value = 0.50926. It is therefore not possible to assert
that, at a significance level of α = 0.01, there is a sig-
nificant difference between them. Finally, the same
process is carried out for the kappa measure, taking
the algorithm that achieved the best ranking as con-
trol (see Table 3b). Results revealed that there are sta-
tistical differences with regard to CMAR, CORE and
OneR.
Finally, the same analysis related to accuracy
and kappa measures was carried out on Big Data
approaches (see Table 4). In order to analyze
whether exists any statistical difference, several non-
parametric tests are carried out. Focusing on accu-
racy, the Friedman test revealed a X
2
F
= 23.12 with
a critical value of 13.277 and a p-value = 0.0001.
Table 4: Average ranking for each algorithm (sorted in de-
scending order) according to the Friedman test when Big
Data datasets are considered.
Algorithm Ranking
DAC 4.350
MRAC 4.150
MRAC+ 2.700
DFAC-FFP 2.150
G3P-AC 1.650
(a) Accuracy measure.
Algorithm Ranking
DAC 4.600
MRAC 4.100
MRAC+ 2.600
DFAC-FFP 1.900
G3P-AC 1.800
(b) Kappa measure.
Table 5: Comparison of our G3P-AC proposal and the other
algorithms for both accuracy and kappa measures. p-values
for Holland test with α = 0.01.
MRAC MRAC+ DAC DFAC-FFP
Accuracy 0.004 0.447 0.001 0.821
Kappa 0.009 0.697 0.001 0.888
Considering the Kappa measure, results for Friedman
was X
2
F
= 26.32 with a critical value of 13.277 and
a p-value = 2.72 · 10
−5
. For both measures, with
α = 0.01, it is possible to assert not all the algo-
rithms equally behave. A post-hoc test is therefore
performed to determine the algorithms that present
some differences. According to the results (see Ta-
ble 5 including Holland test with α = 0.01), DAC and
MRAC behave statistically worse than the rest of al-
gorithms. On the contrary, our G3P-AC algorithm,
DFAC-FFP and MRAC+ did not present any statisti-
cal difference in terms of quality.
4.2 Efficiency
In this second analysis, it is interesting to analyse the
efficiency of the proposal when it is compared to other
AC algorithms. First, we compare our proposal based
on Spark (see Table 6) to classical methods similarly
we did in the previous section for the reliability of
the model. The Friedman test revealed a X
2
F
= 175.85
with a critical value of 9 and a p-value = 2.2 · 10
−16
.
Considering a α = 0.01 it is possible to assert not all
the algorithms equally behave. Focusing on the effi-
ciency and taking our proposal as control, the post-
hoct test (see Table 7) denotes some statistical dif-
ferences with regard to CMAR, CPAR, Ripper and
C4.5, our proposal performing worse. On the con-
trary, the Spark proposal does not obtain any statis-
tical difference with regard to other proposals. At
this point, it is important to remark that the proposal
(based on Spark) was designed to be run on truly big
datasets so the behaviour on small or classical datasets
was expected. It is obvious that those algorithms like
C4.5, Ripper, CPAR, CMAR, etc that were designed
to work on small data should be better in performance
IoTBDS 2019 - 4th International Conference on Internet of Things, Big Data and Security
100
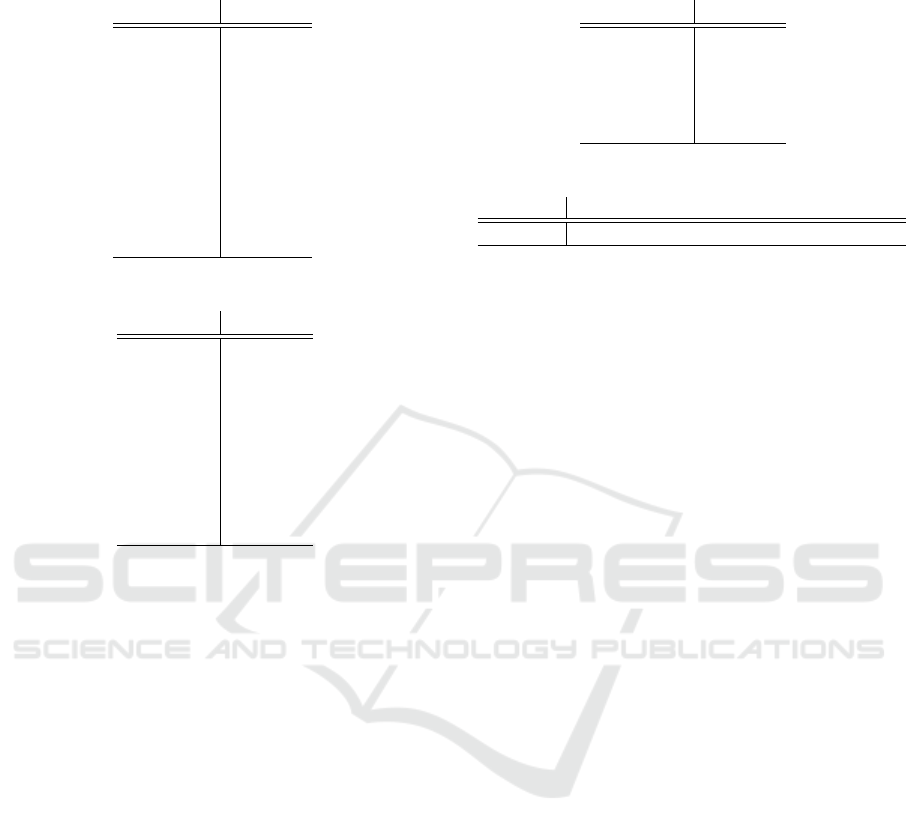
Table 6: Average ranking for each algorithm (sorted in de-
scending order) according to the Friedman test.
Algorithm Ranking
FARCHD 8.600
CORE 8.050
OneR
8.317
G3P-AC 6.933
CBA 6.100
CBA2 4.967
CMAR 3.800
CPAR 3.467
Ripper 2.900
C4.5 1.867
Table 7: p-values for the Holland test with α = 0.01.
Vs G3P-AC
FARCHD 0.396
CORE 0.799
OneR 0.617
CBA 0.868
CBA2 0.194
CMAR 0.002
CPAR 0.000
Ripper 0.000
C4.5 0.000
than those based on MapReduce that require to dis-
tribute and gather information. As a result, this exper-
imental analysis demonstrate that the use of MapRe-
duce approaches are desiderable only for Big Data
and should be avoid for classical datasets.
Finally, it is essential to demonstrate the perfor-
mance of our proposal when Big Data are analyzed.
In this regard, the proposal is compared to AC al-
gorithms for Big Data (MRAC, MRAC+, DAC and
DFAC-FFP) on a series of datasets of high dimen-
sionality (see Table 1). The experimental results
(see Table 8) considering the Friedman test revealed
a X
2
F
= 28.26 with a critical value of 4 and a p-
value = 1.1 ·10
−5
. Considering a α = 0.01 it is possi-
ble to assert not all the algorithms equally behave and
a post-hoct test (see Table 9) is performed taking our
proposal as control. Results denote that our proposal
performs statistically better than DAC and DFAC-FFP
in efficiency. With regard to MRAC+ and MRAC, no
statistical difference was found, but our proposal ob-
tained the best ranking. Thus, it is demonstrated the
promising behaviour of our proposal when it is ap-
plied to truly Big Data.
Table 8: Average ranking for each algorithm (sorted in de-
scending order) according to the Friedman test.
Algorithm Ranking
DFAC-FFP 4.500
DAC 4.200
MRAC
2.850
MRAC+ 1.950
G3P-AC 1.500
Table 9: p-values for the Holland test with α = 0.01.
MRAC+ MRAC DAC DFAC-FFP
G3P-AC 0.774 0.251 0.001 0.000
5 CONCLUSIONS
In this work a grammar-guided genetic programming
algorithm for associative classification in Big Data,
named G3P-AC, has been proposed. The novelty of
this approach is that it is eminently designed to be as
parallel as possible without affecting the accuracy and
interpretability of the classifier. This proposal, imple-
mented in Apache Spark, obtained really promising
results in terms of the reliability of the resulting pre-
dictive model. In fact, it performs better than many
of existing AC algorithms (classical and Big Data ap-
proaches) as well as classical classification algorithms
(not based on associative classification). According
to the efficiency, it has been demonstrated that the
proposal should be avoid when small datasets are re-
quired to be analysed but it performs really well on
Big Data.
ACKNOWLEDGEMENTS
This research was supported by the Spanish Min-
istry of Economy and Competitiveness and the Euro-
pean Regional Development Fund, projects TIN2017-
83445-P
REFERENCES
Agrawal, R., Imieli
´
nski, T., and Swami, A. (1993). Min-
ing association rules between sets of items in large
databases. SIGMOD Rec., 22(2):207–216.
Alcal
´
a-Fdez, J., Alcal
´
a, R., and Herrera, F. (2011). A fuzzy
association rule-based classification model for high-
dimensional problems with genetic rule selection and
lateral tuning. IEEE Transactions on Fuzzy Systems,
19(5):857–872.
Associative Classification in Big Data through a G3P Approach
101

Bechini, A., Marcelloni, F., and Segatori, A. (2016). A
mapreduce solution for associative classification of
big data. Information Sciences, 332:33–55.
Ben-David, A. (2008). Comparison of classification accu-
racy using cohen’s weighted kappa. Expert Systems
with Applications, 34(2):825 – 832.
Chen, H., Chiang, R., and Storey, V. (2012). Business
intelligence and analytics: From big data to big im-
pact. MIS Quarterly: Management Information Sys-
tems, 36(4):1165–1188.
Cohen, W. (1995). Fast effective rule induction. In Ma-
chine Learning: Proceedings of the Twelfth Interna-
tional Conference, pages 1–10.
Cortes, C. and Vapnik, V. (1995). Support vector networks.
Machine Learning, 20:273–297.
Dean, J. and Ghemawat, S. (2008). MapReduce: Simplified
Data Processing on Large Clusters. Communications
of the ACM - 50th anniversary issue: 1958 - 2008,
51(1):107–113.
Han, J. and Kamber, M. (2011). Data Mining: Concepts
and Techniques. Morgan Kaufmann.
Herrera, F., Carmona, C. J., Gonz
´
alez, P., and del Jesus,
M. J. (2011). An overview on subgroup discovery:
foundations and applications. Knowledge and Infor-
mation Systems, 29(3):495–525.
Holte, R. (1993). Very simple classification rules per-
form well on most commonly used datasets. Machine
Learning, 11:63–91.
Lam, C. (2010). Hadoop in Action. Manning Publications
Co., Greenwich, CT, USA, 1st edition.
Li, W., Han, J., and Pei, J. (2001). Cmar: Accurate and effi-
cient classification based on multiple class-association
rules. In 2001 IEEE International Conference on Data
Mining(ICDM01), pages 369–376.
Liu, B., Hsu, W., and Ma, Y. (1998). Integrating classifica-
tion and association rule mining. In 4th International
Conference on Knowledge Discovery and Data Min-
ing(KDD98), pages 80–86.
Liu, B., Ma, Y., and Wong, C. (2001). Classification Using
Association Rules: Weaknesses and Enhancements,
pages 591–601. Kluwer Academic Publishers.
McKay, R. I., Hoai, N. X., Whigham, P. A., Shan, Y., and
O’Neill, M. (2010). Grammar-based genetic program-
ming: a survey. Genetic Programming and Evolvable
Machines, 11:365–396.
Padillo, F., Luna, J. M., and Ventura, S. (2017). Exhaustive
search algorithms to mine subgroups on big data us-
ing apache spark. Progress in Artificial Intelligence,
6(2):145–158.
Quinlan, R. (1993). C4.5: Programs for Machine Learning.
Morgan Kaufmann Publishers, San Mateo, CA.
Segatori, A., Bechini, A., Ducange, P., and Marcelloni,
F. (2018). A distributed fuzzy associative classi-
fier for big data. IEEE Transactions on Cybernetics,
48(9):2656–2669.
Tan, K., Yu, Q., and Ang, J. (2006). A coevolutionary algo-
rithm for rules discovery in data mining. International
Journal of Systems Science, 37(12):835–864.
Thabtah, F. A. (2007). A review of associative classification
mining. Knowledge Engineering Review, 22(1):37–
65.
Triguero, I., Gonz
´
alez, S., Moyano, J. M., Garc
ˆ
ıa, S., Al-
cal
´
a-Fdez, J., Luengo, J., Fern
´
andez, A., del Jes
´
us,
M. J., S
´
anchez, L., and Herrera, F. (2017). Keel 3.0:
an open source software for multi-stage analysis in
data mining. International Journal of Computational
Intelligence Systems, 10(1):1238–1249.
Ventura, S. and Luna, J. M. (2016). Pattern Mining with
Evolutionary Algorithms. Springer International Pub-
lishing.
Ventura, S. and Luna, J. M. (2018). Supervised Descriptive
Pattern Mining. Springer International Publishing.
Venturini, L., Baralis, E., and Garza, P. (2017). Scaling as-
sociative classification for very large datasets. Journal
of Big Data, 4(1).
Yin, X. and Han, J. (2003). Cpar: Classification based
on predictive association rules. In 3rd SIAM Inter-
national Conference on Data Mining(SDM03), pages
331–335.
Zaharia, M., Chowdhury, M., Franklin, M. J., Shenker, S.,
and Stoica, I. (2010). Spark: Cluster computing with
working sets. In Proceedings of the 2nd USENIX
Conference on Hot Topics in Cloud Computing, Hot-
Cloud’10, Berkeley, CA, USA.
IoTBDS 2019 - 4th International Conference on Internet of Things, Big Data and Security
102
