
Coordinated Image- and Feature-space Visualization for Interactive
Magnetic Resonance Spectroscopy Imaging Data Analysis
Muhammad Jawad, Vladimir Molchanov and Lars Linsen
Westf
¨
alische Wilhelms-Universit
¨
at M
¨
unster, Germany
Keywords:
Multidimensional Data Visualization, Medical Visualization, Coordinated Views, Spectral Imaging Analysis.
Abstract:
Magnetic Resonance Spectroscopy Imaging (MRSI) is a medical imaging method that measures per voxel a
spectrum of signal intensities. It allows for the analysis of chemical compositions within the scanned tissue,
which is particularly useful for tumor classification and measuring its infiltration of healthy tissue. Common
analysis approaches consider one metabolite concentration at a time to produce intensity maps in the image
space, which does not consider all relevant information at hand. We propose a system that uses coordinated
views between image-space visualizations and visual representations of the spectral (or feature) space. Co-
ordinated interaction allows for analyzing both aspects and relating the analysis results back to the other for
further investigations. We demonstrate how our system can be used to analyze brain tumors.
1 INTRODUCTION
Magnetic Resonance Spectroscopy Imaging (MRSI)
is an in-vivo medical imaging method for measuring
chemical compositions of scanned tissues. The com-
positions in the form of metabolite concentrations can
be computed from spectral information of chemical
resonance (Gujar et al., 2005). While T1- or T2-
weighted Magnetic Resonance Imaging (MRI) data
allows for the detection of tumors and to determine
their shape, MRSI provides additional information on
their metabolic activity. Such information, on the one
hand, allows for a classification of the tumor with re-
spect to its malignancy (from benign to malignant),
and, on the other hand, for an investigation, whether
the tumor has already started infiltrating surrounding
“healthy” tissue (Burnet et al., 2004).
The metabolic information is extracted from the
spectrum for each voxel of the MRSI data in a pre-
processing step, see Section 3.2. Afterwards, the in-
formation at hand is the metabolic concentration of all
extracted metabolites per voxel. The set of metabo-
lites form a multidimensional space, which we refer
to as the spectral or feature space. Each voxel is re-
flected by a point in this multidimensional space. On
the other hand, the voxels have a spatial arrangement
in the image space.
Common analysis tools for MRSI data in clini-
cal settings visualize the spatial distribution of indi-
vidual metabolite concentrations using color mapping
of an image slice. Thus, only a single metabolite is
investigated at a time. Therefore, a lot of informa-
tion is being neglected and the interplay of metabo-
lites concentrations cannot be analyzed. We propose
a novel tool that integrates all information at hand
for a comprehensive analysis of MRSI data. Our ap-
proach is based on coordinated views of visualiza-
tion in image and feature space. For image space,
we also use slice-based visualizations, as they avoid
occlusion and it is common to only scan a few slices
in MRSI. MRSI visualizations are overlaid with MR
images and combined with automatic MRI segmen-
tation results. For feature space, multidimensional
data visualization methods are used. Given the image
segmentation result, the multidimensional data are la-
beled accordingly and we apply interaction methods
to separate the labeled classes. Using star-coordinates
encoding, the separations can be related back to the
individual dimension, basically allowing for conclu-
sions, which metabolite concentration allow for class
separation. The methodology is detailed in Section 4.
In Section 5, we apply our methods to a scenario
for MRSI data analysis to investigate brain tumors.
In 2012, WHO reported 256,213 brain cancer cases,
of which 189,382 deaths have been recorded (Board,
PDQ Adult Treatment Editorial, 2018). We show how
our coordinated views allow for analyzing the brain
tumor’s chemical composition as well as the infiltra-
tion of surrounding tissue that when based on MRI
data may be classified as non-tumor region.
118
Jawad, M., Molchanov, V. and Linsen, L.
Coordinated Image- and Feature-space Visualization for Interactive Magnetic Resonance Spectroscopy Imaging Data Analysis.
DOI: 10.5220/0007571801180128
In Proceedings of the 14th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2019), pages 118-128
ISBN: 978-989-758-354-4
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 RELATED WORK
Nunes et al. (Nunes et al., 2014a) provided a sur-
vey of existing methods for analyzing MRSI data.
They conclude that no approach exists that analyzes
all metabolic information. They report that MRSI
data visualization packages provided with commer-
cial scanners such as SyngoMR and SpectroView
only provide color mapping of individual metabolic
concentrations (or a ratio of metabolic concentra-
tions) in image space. Other tools such as Java-based
Magnetic Resonance User Interface (jMRUI) (Stefan
et al., 2009) and Spectroscopic Imaging Visualization
and Computing (SIVIC) (Crane et al., 2013) that are
also widely used in clinical practice provide a simi-
larly restricted functionality.
Raschke et al. (Raschke et al., 2014) proposed
an approach to differentiate between tumor and non-
tumor regions by showing the relation of concentra-
tions of two selected metabolites in a scatterplot. By
plotting the regression line, they identified abnormal
regions by data points that are far from the line. Sim-
ilarly, Rowland et al. (Rowland et al., 2013) use scat-
terplots to inspect three different pairs of metabolites
tumor analysis. These procedures only target selected
metabolites, which are chosen a priori.
An approach to exploit the metabolic informa-
tion better was presented by Maudsley et al. (Maud-
sley et al., 2006). They developed the Metabolite
Imaging and Data Analysis System (MIDAS) for
MRSI pre-processing and visualization. Users can
view histograms of metabolites, but relations between
metabolites cannot be studied. Feng et al. (Feng
et al., 2010) presented the Scaled Data Driven Sphere
(SDDS) technique, where information of multiple
metabolite concentrations is combined in a glyph-
based visualization using mappings to color and size.
Obviously, the amount of dimensions that can be
mapped is limited. They overlay the images with
the glyphs and link them to parallel coordinate plots.
Users can make selections in the parallel coordinate
plot and observe respective spatial regions in the im-
age space. As a follow-up of their survey, Nunes et
al. (Nunes et al., 2014b) presented a system that cou-
ples the existing systems ComVis (Matkovic et al.,
2008) and MITK (Wolf et al., 2004). They use scatter-
plot, histogram, and parallel coordinate plot visualiza-
tions for analyzing the metabolic features space. The
visualizations provide linked interaction to image-
space representations such that brushing on metabolic
data triggers highlighting in image space.
We build upon the idea of using coordinated inter-
actions in feature and image space, but enhance the
functionality significantly: We incorporate segmen-
tation results, provide means to separate the labeled
data in feature space, allow for a comparative visual-
ization of classes, and provide a single tool that inte-
grates all information and allows for fully coordinated
interaction in both directions.
3 BACKGROUND
In this section, we provide background on the imaging
method, describe the executed pre-processing steps,
describe the data at hand after imaging and pre-
processing, and the driving questions.
3.1 Imaging
MRSI is an in-vivo medical imaging method, where
per voxel a whole spectrum of intensities is recorded.
While MRI only measures water intensities per voxel,
MRSI measures intensities at different radio frequen-
cies. Most commonly
1
H MRSI is used, where the
signal of hydrogen protons (
1
H) in different chemi-
cals is measured in the form of intensity peaks. As
this measurement is performed for different frequen-
cies, different chemicals can be detected within each
voxel. The intensity peaks within the frequency spec-
trum are quantified as parts per million (ppm).
Measuring entire spectra in MRSI comes at the
expense of much lower spatial resolution when com-
pared to MRI. Using 1.5T or 3T scanners yields voxel
sizes of about 10mm × 10mm × 8mm (Nunes et al.,
2014a; McKnight et al., 2001). 7T scanners yield-
ing higher resolutions are barely used in clinical prac-
tice due to high costs (Scheenen et al., 2008). Typ-
ically, MR images are taken (using T1 or T2 relax-
ation) in addition to MRS images within the same ses-
sion. The MR images provide anatomical information
at a higher resolution.
3.2 Pre-processing
The main goal of the pre-processing step is to quan-
tify the various chemicals, referred to as metabo-
lites, within each voxel from the respective intensity
spectrum. MRSI is not yet standardized (e.g., us-
ing a DICOM format), but there is open source and
proprietary software available for pre-processing and
metabolite quantification like LCModel (Provencher,
1993), jMRUI (Stefan et al., 2009), and Totally Au-
tomatic Robust Quantitation in NMR (TARQUIN)
(Reynolds et al., 2006).
Common pre-processing steps provided by the
tools are eddy current compensation, offset correc-
tion, noise filtering, zero filling, residual water sup-
Coordinated Image- and Feature-space Visualization for Interactive Magnetic Resonance Spectroscopy Imaging Data Analysis
119

pression, and phase and base line correction. These
steps are executed in the measured time domain or
in the frequency domain after performing a Fourier
transform. Signal strength in time domain indicates
the metabolites’ concentration, while the area under
the metabolite curve is used to compute the concen-
tration in frequency domain. The computed values are
similar when measurements with high signal-to-noise
ratio are provided (Vanhamme et al., 2001). For the
actual metabolite concentration, mathematical mod-
els are used and fitted to the measured data. Different
tools use different fitting approaches.
For the pre-processing of our data, we used TAR-
QUIN, which is an open-source GUI-based soft-
ware for in-vivo spectroscopy data pre-processing and
quantification. It operates in time domain and uses a
non-negative least-squares method for model fitting to
compute the metabolite concentrations. We incorpo-
rate TARQUIN in our data preparation step due to free
availability, friendly user interface, automatic quan-
tification, support for multi-voxel spectroscopy, and
being able to process various file formats and to ex-
port the results in various formats (Reynolds et al.,
2006; Wilson et al., 2011).
3.3 Data
The data we use in this paper for our applica-
tion scenario was acquired using an
1
H MRSI tech-
nology on a 3T Siemens scanner (TR/TE/flip =
1700ms/135ms/90
◦
). Two MRSI series are taken,
each having a 160mm × 160mm × 1mm field of view.
In addition, an MRI volume is captured and regis-
tered with the MRSI volume. The MRI volume is
224mm×256mm×144mm with 1 mm slice thickness.
We clip the MRI volume to the MRSI volume. The
resolution of the MRI volume is much higher such
that each MRSI voxel stretches over 10mm × 10mm ×
12mm MRI voxels. The data are courtesy of Miriam
Bopp and Christopher Nimsky from the University
Hospital Marburg, Germany. We used TARQUIN for
MRSI pre-processing and metabolite quantification.
The quantification process resulted in 33 metabolites
that are listed in Table 1.
After metabolite quantification, we can summa-
rize the available data as two slabs of MRSI voxels
with registered MRI voxels. For each MRSI voxel,
we have computed concentrations of 33 metabolites,
leading to a 33-dimensional feature space, where each
point in the feature space represents the chemical
composition of one voxel. In addition, we know for
each MRSI voxel, which are the matching MRI vox-
els (single intensity values). In the following, we will
propose methods for visualization in image space and
Table 1: Metabolites delivered by a quantification from
brain MRSI data using TARQUIN.
No. Name No. Name No. Name
1 Ala 12 Lip09 23 PC
2 Asp 13 Lip13a 24 PCr
3 Cr 14 Lip13b 25 Scyllo
4 GABA 15 Lip20 26 Tau
5 GPC 16 MM09 27 TNAA
6 Glc 17 MM12 28 TCho
7 Gln 18 MM14 29 TCr
8 Glth 19 MM17 30 Glx
9 Glu 20 MM20 31 TLM09
10 Ins 21 NAA 32 TLM13
11 Lac 22 NAAG 33 TLM20
feature space, where coordinated interaction within
the linked views is used to analyze the MRSI data.
3.4 Driving Questions
MRSI data are mainly acquired to analyze tumors. In
our application scenario, we look into brain tumors.
The first driving question is the tumor classification,
i.e., one would want to finds out what type of tumor
it is and how its malignancy is rated. To do so, one
should analyze the MRSI voxels belonging to the tu-
mor. The question one would like to answer with
MRSI measurements is then: What are the chemi-
cal compositions of the tumor? The second important
question is whether the tissue surrounding the tumor
is already infiltrated by the tumor. MRSI allows for a
more detailed analysis of the surrounding tissue. The
question to be answered is then: How do the chemical
composition in areas surrounding the tumor compare
to those of the tumor and to those of healthy tissue?
Finally, one may want to see whether tumors in other
regions occur, i.e., one would ask: Are there other ar-
eas that have a similar chemical composition as the
tumor?
4 METHODOLOGY
4.1 Image Space Visualization
In order to analyze image regions such as tumors or
surrounding tissue, one would need to be able to vi-
sually inspect those regions and interactively select
them. We support this using a slice-based viewer,
which we implemented using the VTK (Schroeder
et al., 2006) and ITK (Johnson et al., 2013) libraries.
Within the slice, we have two visualization layers.
The first layer represents the MRI volume. It pro-
IVAPP 2019 - 10th International Conference on Information Visualization Theory and Applications
120

vides the anatomical context for the MRSI data analy-
sis, see Figure 1. A greyscale luminance color map is
used, as this is a common standard in MRI visualiza-
tion. The second layer represents the MRSI volume.
Here, individual voxels can be selected interactively
by brushing on the image regions. Selected voxels
are highlighted by color, see Figure 7(left).
Figure 1: Anatomical context in slice-based image-space
MRI visualization.
In addition to manual selection of MRSI voxels
based on visual inspection, we also support an au-
tomatic image segmentation method. The automatic
image segmentation predefines regions for quick se-
lection and region analysis. The automatic segmenta-
tion method partitions the MR image. A large range
of algorithms exist each having certain advantages
and drawbacks. For the data at hand, the best re-
sults in our tests were obtained by the Multiplicative
Intrinsic Component Optimization (MICO) (Li et al.,
2014) segmentation method for auto characterization
of various tissues in brain MRI. MICO could handle
MRI with low signal-to-noise ratio that is due to mag-
netic field disturbance and patient movement during
the scanning process. MICO performs well in bias
field estimation and in discarding intensity inhomo-
geneity.
If we impose the MR image segmentation result
on the MRSI voxels, we have to deal with partial vol-
ume effects, as one MRSI voxel corresponds to many
MRI voxels, which may be classified differently. We
propose to use a visual encoding that conveys the par-
tial volume effect. Instead of simply color-coding
the MRSI voxel by the color for the dominant class
among the MRI voxels, we render square glyphs that
are filled with different colors, where the color por-
tions reflect the percentages how often the MRI vox-
els are assigned to the respective class. Figure 2
shows a respective slice-based visualization, where
the segmentation result is shown in the MRSI layer
overlaying the MRI layer. The mixture of colors indi-
cate the uncertainty of the anatomical region segmen-
tation at the MRSI resolution. In the image, we show
the glyphs for each of the dominant class, i.e., the five
columns (from left to right) show MRSI voxels, where
the respective MRI voxels have primarily been classi-
fied as background, cerebrospinal fluid (CSF), tumor,
gray matter, and white matter, respectively. The col-
ors used for the respective classes are shown above
the images. Results are shown for both MRSI slabs.
4.2 Feature Space Visualization
The feature-space visualization problem is that of a
multidimensional data visualization problem, where
dimensionality is in the range of tens, while the num-
ber of data points is in the range of hundreds. Hence,
we need an approach that scales sufficiently well in
both aspects. Moreover, the tasks require us to ob-
serve patterns such as clusters of data points, i.e., sets
of data points that are close to each other and different
from other data points. Many multidimensional data
visualization approaches exist and we refer to a recent
survey for their descriptions (Liu et al., 2017). They
can be distinguished by the performed data transfor-
mations, by the visual encodings, and by their map-
pings to visual interfaces. Point-based visual encod-
ings scale well in the number of data points and al-
low for an intuitive detection of clusters. Among
them, projection-based dimensionality reduction ap-
proaches use data transformations to a visual inter-
face supporting the handling of high dimensionality.
Linear projections have the advantage over non-linear
projections that the resulting plots can be easily re-
lated back to the original dimensions by the means
of star coordinates. Thus, we propose to use star-
coordinates plots (Kandogan, 2000) to visualize the
feature space.
A projection from an n-dimensional feature space
to a 2D visual space is obtained by a projection ma-
trix of dimensions 2 × n, where each of the n columns
represent the tip of the n star-coordinates axes. The
default set-up is to locate the tips equidistantly on
the unit circle, see Figure 3. The tips can be moved
to change the projection matrix, which allows for an
interactive multidimensional data space analysis sup-
porting the detection of trends, clusters, outliers, and
correlations (Teoh and Ma, 2003).
When assuming an image-space segmentation of
the voxels as described in the preceding section, each
voxel is assigned to a class. Hence, we are dealing
with labeled multidimensional data. A common task
in the visual analysis of labeled multidimensional data
is to find a projected view with a good separation of
the classes. This is also of our concern, as we want
Coordinated Image- and Feature-space Visualization for Interactive Magnetic Resonance Spectroscopy Imaging Data Analysis
121
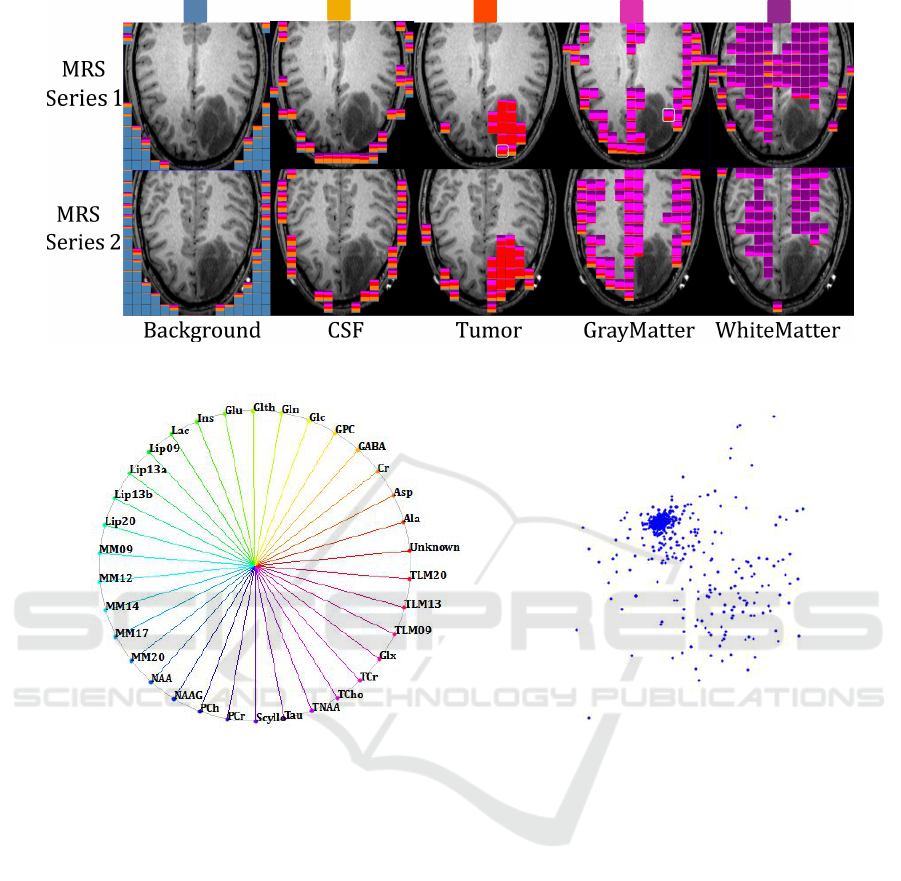
Figure 2: MRI segmentation visualized at MRSI resolution using glyphs with color ratios that reflect class distribution of each
voxel.
Figure 3: Default star-coordinates configuration (left) and respective projection of 33-dimensional feature space to a 2D visual
space. Each dimension represents a metabolite concentration, each point corresponds to a MRSI voxel.
to separate tumor from healthy tissues. Molchanov et
al. (Molchanov and Linsen, 2014) proposed an ap-
proach for intuitive interactive class separation in lin-
early projected views. We adopt their idea for our pur-
poses. The idea is to use a control point for each class,
e.g., being the classes’ medians. Classes can then be
separated by dragging control points apart. Since the
visual representation is restricted to linear projections,
the position to which the control points are dragged
can, in general, not be exactly obtained in a linear
projection. Molchanov et al. proposed to use a least-
squares approach to find the best match to the desired
interaction. Using this idea, the user just needs to
move the control points of the classes in an intuitive
manner, where the number of classes is usually low
(five in the case of brain imaging data), instead of
interacting with all star-coordinates axes, which be-
comes tedious for larger number of dimensions (33 in
our application scenario). Figure 4 illustrates this by
showing the star-coordinates configuration to the left
and the linear projection of the labeled multidimen-
sional data to the right, where the control points that
can be interactively moved are the ones with a black
frame. Since the visualization would be too cluttered
when overlaying the two views, we decided to show
them in a juxtaposed manner. Further argumentation
for juxtaposed views of star coordinates is provided
by Molchanov and Linsen (Molchanov and Linsen,
2018).
Since the points in our projected view represent
MRSI voxels, while the image segmentation is per-
formed on the higher-resolution MR image, we have
partial volume effects as discussed in the previous
section. The uncertainty of the labeling result shall
also be conveyed in our projected view. For example,
if some points are identified as outliers of a cluster,
one should be able to reason if the labeling of the re-
spective voxel is uncertain or not. If it was uncertain,
the outlyingness may be due to a wrong labeling deci-
sion. To visually convey the labeling uncertainty, the
IVAPP 2019 - 10th International Conference on Information Visualization Theory and Applications
122

Figure 4: Interactive visual analysis of labeled multidimensional data: star-coordinates configuration (left) and respective
projected view (right) obtained by interacting with control points (black frames) of the classes induced by image segmentation.
probabilities of each point belonging to each class la-
bel shall be represented. Since the probabilities sum
up to one, they are well represented by a pie chart, i.e.,
each point in the projected view is displayed by a pie
chart. Figure 5 shows a projected view with labeling
uncertainty conveyed by pie charts.
While interacting with the control points in the
projected view, the star-coordinates axes are up-
dated accordingly. Hence, when separating classes
in the projected view, one can observe in the star-
coordinates view, which axes are mainly responsible
for the separation, i.e., which axes are the ones that
allow for such a separation. The dimensions that are
associated with these axes are subject to further in-
vestigations, as one of our tasks was to detect the
metabolic compositions of tumors and surrounding
tissues. For selected metabolites, we use different sta-
tistical plots supporting different analysis steps.
If we are interested in investigating the metabolic
compositions of two classes for selected metabolites,
we use juxtaposed box plots to show the statistical in-
formation of the metabolic concentrations of all vox-
els that were assigned to each class. The box plots
convey the median, the interquartile range, as well as
minimum and maximum, see Figure 12.
If we are interested in investigating the interplay
of two metabolites, we can analyze their correlation
using a 2D scatterplot. Scatterplots are most effec-
tive in showing correlation and detecting outliers, see
Figure 6(right). Correlation analysis is supported
by computing and displaying a regression line. In
case more than two metabolites shall be analyzed si-
multaneously, parallel coordinate plots allow for a
good correlation analysis and scale better than scat-
terplot matrices in the number of dimensions, see Fig-
ure 7(right).
4.3 Coordinated Interaction in Image
and Feature Space
The interactive visual analysis becomes effective
when using coordinated interaction of the image- and
feature-space views. In the image space, one can
brush the MRSI voxels in the slice-based visualiza-
tion and group them accordingly. Hence, interactive
labeling is supported. Of course, one can also use the
labeling that is implied by the MRI segmentation (in-
cluding the uncertainty visualization). The resulting
labeling is transferred to the projected feature-space
visualization by using the same colors in both visu-
alizations, see image-space visualization in Figure 2
and respective projected feature-space visualization
in Figures 4 and 5. This coordinated interaction al-
lows for investigating whether image segments form
coherent clusters in feature space, which dimensions
of the feature space allow for a separation of the clus-
ters, and whether there are some outliers. Of course,
the same holds true when using the statistical plots
as a coordinated feature-space view. Figure 6 shows
a brushing in image space and investigation of the
selection in a scatterplot visualization of two (previ-
ously selected) dimensions, while Figure 7 shows a
brushing in image space and investigation of the se-
lection in a parallel coordinate plot of seven (previ-
ously selected) dimensions.
The coordinated interaction of image and feature
space is bidirectional. Thus, the user may also brush
in the feature space, e.g., by selecting a group of
points in the feature space that form a cluster, and
observe the spatial distribution of the selection in the
image space. Again, one can use any of the feature-
space visualizations or a combination thereof. For ex-
ample, in Figure 8, the four voxels to the left are cho-
sen (voxels having low concentration of TNAA rela-
tive to TCho and TCr) in the bar chart and the respec-
tive highlighting in the image space convey that these
Coordinated Image- and Feature-space Visualization for Interactive Magnetic Resonance Spectroscopy Imaging Data Analysis
123
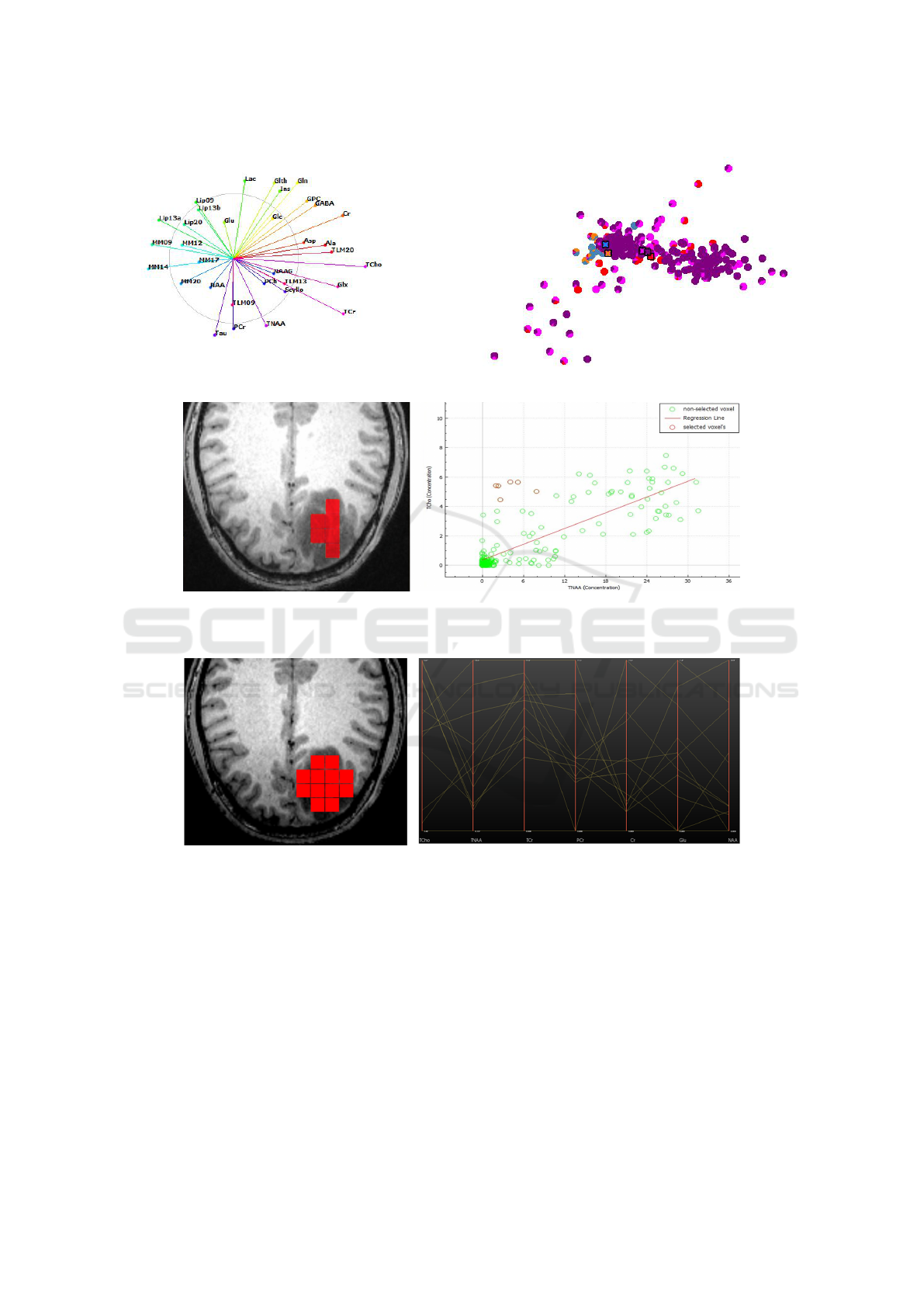
Figure 5: Pie-chart representation of uncertainty for projected points.
Figure 6: Correlation analysis of the metabolites TCho and TNAA in a scatterplot (right) and detection of outliers. Voxels
corresponding to the interactively selected outliers are shown in the image-space visualization (left), which conveys their
relation to the tumor.
Figure 7: (left) Interactive selection of MRSI voxels in image space and (right) linked view for correlation analysis of seven
selected metabolites (TCho, TNAA, TCr, PCr, Cr, Glu, and NAA) using parallel coordinate plot for the selected voxels.
voxels belong to the tumor region.
We also support a heatmap visualization as com-
monly used when observing the spatial distribution of
individual metabolite concentrations. The user may
select a single metabolite or a combination of metabo-
lites. Figure 9 shows heatmaps of TCho and TNAA
concentrations and In Figure 10, a heatmap of the log-
arithm of the ratio of TCho/TNAA is shown.
The full potential of our system is reached by us-
ing coordinated interaction in both ways simultane-
ously. For example, one can select the tumor voxels
in the image space, can investigate the respective set
of points in feature space possibly forming a cluster,
detect further points that fall into the cluster, select
those further points, and observe their spatial distri-
bution. These newly selected voxels may be voxels
surrounding the tumor, in case the tumor has already
infiltrated surrounding regions, or may be voxels that
form a region elsewhere, in case there is a second tu-
mor.
IVAPP 2019 - 10th International Conference on Information Visualization Theory and Applications
124
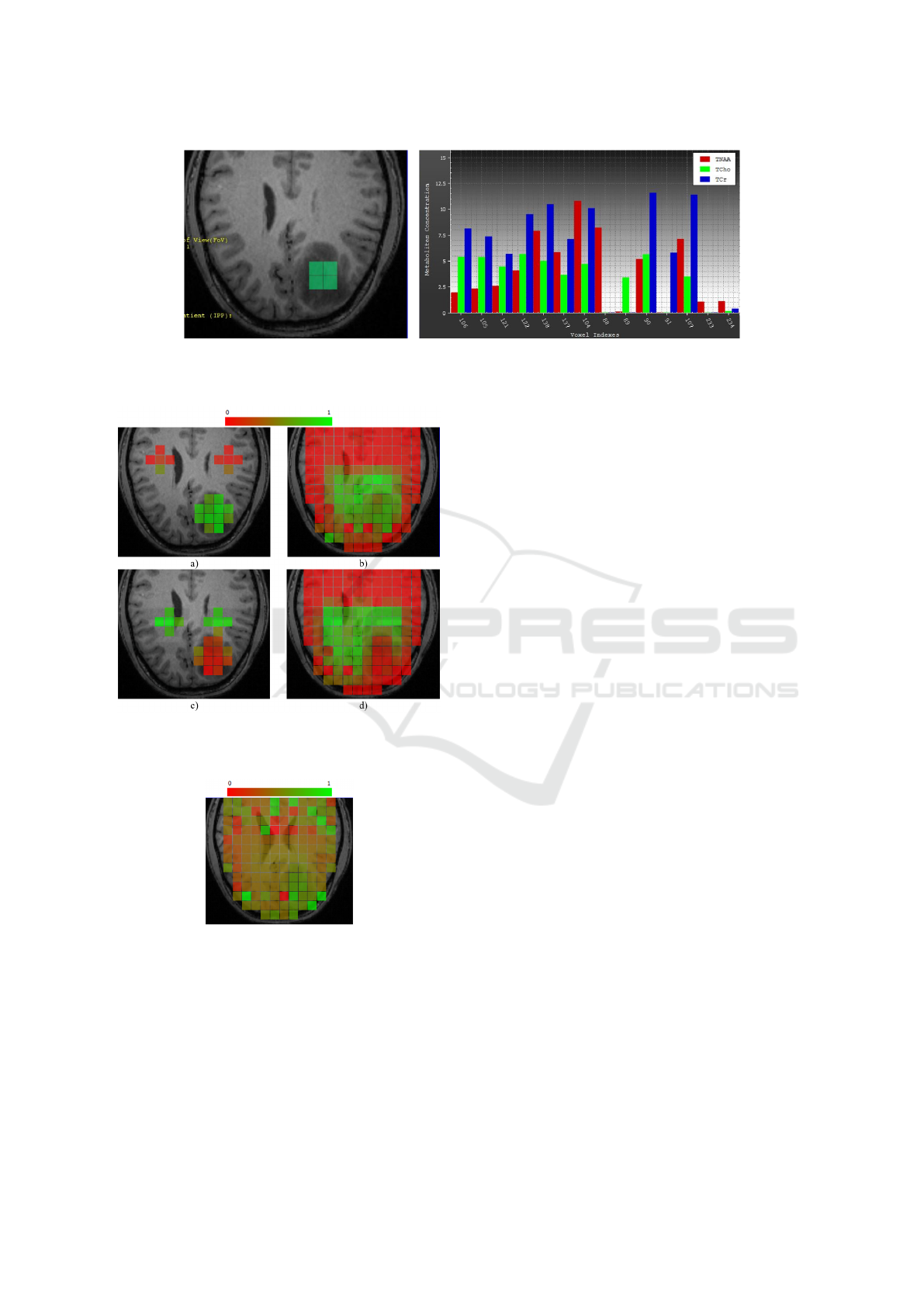
Figure 8: Concentration of three selected metabolites shown side by side in the bar chart (right). Four voxels with low
concentration of TNAA relative to TCho and TCr are selected by brushing. The coordinated image-space view (left) highlights
the selected voxels, which form the core of the tumor.
Figure 9: Heatmaps of TCho (top) and TNAA concentration
(bottom) for all brain voxels (right) and selected regions of
interest (left).
Figure 10: Heatmap of ratio of TCho/TNAA concentrations
(in logarithmic scaling).
5 RESULT AND DISCUSSION
In our application scenario, we applied the developed
methods of Section 4 to the data acquired from a 26-
year old male patient with a brain tumor. MRSI and
MRI head scans were obtained as described in Sec-
tion 3.3. We preprocessed the data as described in
Section 3.2. Then, we first investigated the projected
feature space using the default star-coordinates lay-
out as in Figure 3. We observe no obvious clus-
ters in the projected space. Thus, we next applied
an automatic segmentation of the MR image and im-
posed the segmentation onto the MRSI voxels using
our uncertainty-aware visualization in Figure 2. The
extracted segments represent grey matter, white mat-
ter, CSF, the tumor, and background. This segmen-
tation implies a labeling as shown in Figure 4, where
the colors match with the ones in Figure 2. We ap-
plied the interactive technique to separate the classes
in feature space using the interaction with the classes’
control points. In particular, we applied it to separate
the tumor class (red) from the other classes. We ob-
serve that certain dimensions of our 33-dimensional
feature space are affected strongly by this interactive
optimization. Hence, the respective metabolites may
be the ones that distinguish tumor from the other seg-
ments. In Figure 4, we see that the axes labeled NAA,
Glu, TNAA, PCr, TCr, TCho, and GPC are longest.
These metabolites are candidates for further investi-
gations. In Figure 6, we select TNAA and TCho con-
centrations, which had the longest axes in Figure 4
and plot all voxels in a scatterplot. We select a group
of outliers (red) with high TCho and low TNAA con-
centrations and observe that these form the core of
the tumor region. Please note that by just looking at
one of the two metabolites the voxels would have not
been outliers. In Figure 9, we use heatmaps to in-
vestigate the spatial distributions of TCho and TNAA
concentrations, respectively. When looking at the en-
tire brain region (right image column), it is hard to
detect structures. However, when selecting regions of
interest such as a white matter region and the tumor
region, we can spot the differences (left image col-
umn). The two columns apply the red-to-green color
map to the minimum-to-maximum range of selected
voxels only. In Figure 10, a heatmap of the logarithm
of the ratio of the two metabolites is shown.
Coordinated Image- and Feature-space Visualization for Interactive Magnetic Resonance Spectroscopy Imaging Data Analysis
125
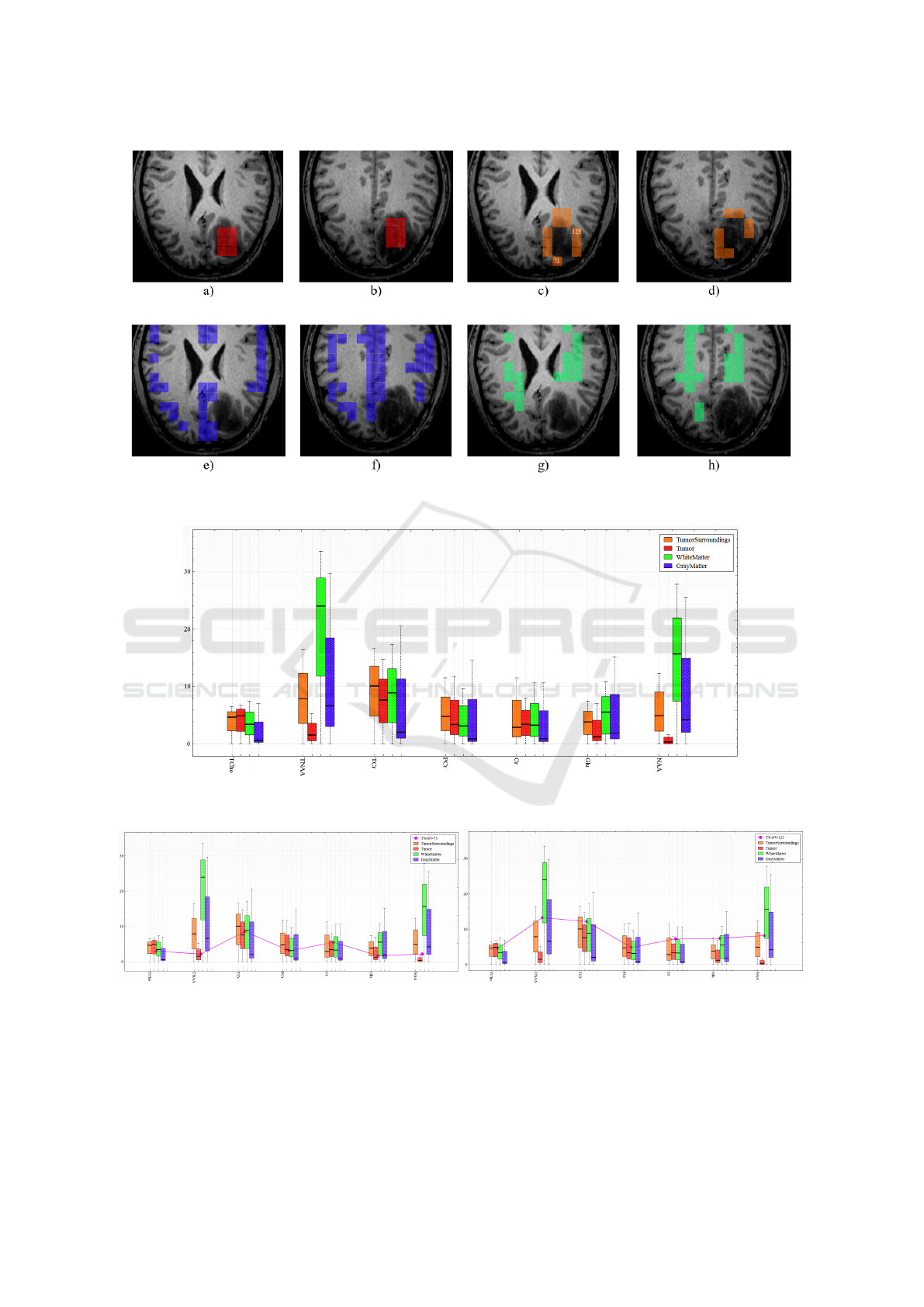
Figure 11: Interactive labeling of voxels within both MRSI slabs representing tumor (red), voxels in the vicinity of the tumor
(orange), grey matter (blue), and white matter (green).
Figure 12: Juxtaposed box plots to compare class statistics for selected classes (voxels labeled as white matter, grey matter,
tumor, and surrounding tumor) and selected metabolites.
Figure 13: Metabolite concentration of voxels 73 (left) and 123 (right) in vicinity of the tumor selected in Figure 11 in
comparison to the concentrations of the tumor, white matter, and grey matter classes.
When investigating the projected feature space in
Figure 4, we observe that the classes are actually not
well separated. We further investigate the pie chart
visualization of the labeling result, see Figure 5 to
observe quite a few voxels that are rather uncertain
with respect to the automatic labeling, which may
be due to the partial volume effect. Also, when se-
lecting all voxels that contain parts of the tumor as
IVAPP 2019 - 10th International Conference on Information Visualization Theory and Applications
126

in Figure 7, the distribution of values in the parallel
coordinate plot appear rather diverse. Thus, we de-
cided to manually select regions of low-uncertainty
voxels and create new labels, see Figure 11. Fig-
ure 12 shows the juxtaposed box plots of the four se-
lected voxel groups. We observe that the tumor re-
gion (red) is quite different from the other regions
in NAA and TNAA concentrations, where TNAA is
the sum of NAA and NAAG. We also observe that
the voxels surrounding the tumor (orange) behave like
gray/white matter voxels rather than tumor voxels for
these metabolites, which makes us believe that these
voxels are not yet infiltrated by the tumor. How-
ever, this may not be true for individual voxels of that
group. Since the tumor size and shape of its bound-
ary is important for diagnosis and treatment, we se-
lected individual voxels at the border of the tumor
and investigated the metabolite concentrations and in-
dividually compared their metabolite concentrations
to those of tumor as well as white and grey matter.
Figure 13 shows such a comparison for the voxels
labeled 73 and 123 in Figure 11. These two voxels
showed high uncertainties in the automatic segmenta-
tion result. We can observe in Figure 13 that voxel 73
matches well the tumor class, while voxel 123 does
not. Hence, we conclude that the tumor may already
have infiltrated the area at voxel 73, while it may have
not yet done so for voxel 123.
We invited two MRSI experts with many years
of experience of acquiring and analyzing MRSI data
to our lab to show them our tool on a large multi-
touch display. The visual encodings were mostly in-
tuitive to them and they were quickly able to suggest
interactive analysis steps themselves. Only the pro-
jected view needed some explanations, but the intu-
itive interaction with the control points made them
adopt the concept quickly. They also quickly brought
in their expertise into the analytical workflow by ex-
cluding some metabolites that they knew would not
be important for the given tasks such as lipids and
macromolecules and by interpreting correctly combi-
nations such as TNAA being a combination of NAA
and NAAG. In the session, we jointly looked into
the metabolic composition of voxels in the vicinity
of the tumors as documented above. To test the re-
producibility of the analysis, it would be desirable to
test our tool with a large number of experts on a large
number of data sets. We hope that we can conduct
such a study in future work, but acknowledge that it
will be challenging to recruit a large number of ex-
perts.
6 CONCLUSIONS AND FUTURE
WORK
We presented a comprehensive tool for the analysis
of MRSI data and applied it to brain tumor investi-
gations. Using coordinated views of image and fea-
ture space visualizations, effective analysis steps can
be performed that allow for a comprehensive inves-
tigation of all data facets. We showed how relevant
metabolites can be identified and how image regions
can be detected and compared. In this paper, we fo-
cused on the analysis of a tumor region. Future direc-
tions include the analysis of different tumor types, for
which MRSI information from many patient shall be
combined, eventually leading to a cohort analyses.
ACKNOWLEDGEMENTS
This work was supported in part by DFG grant MO
3050/2-1.
REFERENCES
Board, PDQ Adult Treatment Editorial (2018). Adult cen-
tral nervous system tumors treatment PDQ
R
. In PDQ
Cancer Information Summaries. National Cancer In-
stitute (US).
Burnet, N. G., Thomas, S. J., Burton, K. E., and Jefferies,
S. J. (2004). Defining the tumour and target volumes
for radiotherapy. Cancer Imaging, 4(2):153–161.
Crane, J. C., Olson, M. P., and Nelson, S. J. (2013). SIVIC:
open-source, standards-based software for DICOM
MR spectroscopy workflows. Journal of Biomedical
Imaging, 2013:12.
Feng, D., Kwock, L., Lee, Y., and Taylor II, R. M. (2010).
Linked exploratory visualizations for uncertain MR
spectroscopy data. In Park, J., Hao, M. C., Wong,
P. C., and Chen, C., editors, Visualization and Data
Analysis 2010. SPIE.
Gujar, S. K., Maheshwari, S., Bj
¨
orkman-Burtscher, I.,
and Sundgren, P. C. (2005). Magnetic reso-
nance spectroscopy. Journal of neuro-ophthalmology,
25(3):217–226.
Johnson, H. J., McCormick, M., Ib
´
a
˜
nez, L., and Consor-
tium, T. I. S. (2013). The ITK Software Guide. Kit-
ware, Inc., third edition.
Kandogan, E. (2000). Star coordinates: A multi-
dimensional visualization technique with uniform
treatment of dimensions. In In Proceedings of
the IEEE Information Visualization Symposium, Late
Breaking Hot Topics, pages 9–12.
Li, C., Gore, J. C., and Davatzikos, C. (2014). Multi-
plicative Intrinsic Component Optimization (MICO)
for MRI bias field estimation and tissue segmentation.
Magnetic resonance imaging, 32(7):913–923.
Coordinated Image- and Feature-space Visualization for Interactive Magnetic Resonance Spectroscopy Imaging Data Analysis
127

Liu, S., Maljovec, D., Wang, B., Bremer, P., and Pascucci,
V. (2017). Visualizing high-dimensional data: Ad-
vances in the past decade. IEEE Transactions on Vi-
sualization & Computer Graphics, 23(3):1249–1268.
Matkovic, K., Freiler, W., Gracanin, D., and Hauser, H.
(2008). ComVis: A coordinated multiple views sys-
tem for prototyping new visualization technology. In
2008 12th International Conference Information Visu-
alisation, pages 215–220. IEEE.
Maudsley, A., Darkazanli, A., Alger, J., Hall, L., Schuff, N.,
Studholme, C., Yu, Y., Ebel, A., Frew, A., Goldgof,
D., et al. (2006). Comprehensive processing, display
and analysis for in vivo MR spectroscopic imaging.
NMR in Biomedicine, 19(4):492–503.
McKnight, T. R., Noworolski, S. M., Vigneron, D. B., and
Nelson, S. J. (2001). An automated technique for
the quantitative assessment of 3D-MRSI data from
patients with glioma. Journal of Magnetic Reso-
nance Imaging: An Official Journal of the Interna-
tional Society for Magnetic Resonance in Medicine,
13(2):167–177.
Molchanov, V. and Linsen, L. (2014). Interactive design
of multidimensional data projection layout. In N.
Elmqvist, M. Hlawitschka, and J. Kennedy, editors,
EuroVis - Short Papers. The Eurographics Associa-
tion.
Molchanov, V. and Linsen, L. (2018). Shape-preserving star
coordinates. IEEE Transactions on Visualization &
Computer Graphics, pre-print.
Nunes, M., Laruelo, A., Ken, S., Laprie, A., and B
¨
uhler,
K. (2014a). A survey on visualizing magnetic reso-
nance spectroscopy data. In Proceedings of the 4th
Eurographics Workshop on Visual Computing for Bi-
ology and Medicine, pages 21–30. Eurographics As-
sociation.
Nunes, M., Rowland, B., Schlachter, M., Ken, S., Matkovic,
K., Laprie, A., and B
¨
uhler, K. (2014b). An integrated
visual analysis system for fusing MR spectroscopy
and multi-modal radiology imaging. In Visual Analyt-
ics Science and Technology (VAST), 2014 IEEE Con-
ference on, pages 53–62. IEEE.
Provencher, S. W. (1993). Estimation of metabolite concen-
trations from localized in vivo proton NMR spectra.
Magnetic resonance in medicine, 30(6):672–679.
Raschke, F., Jones, T., Barrick, T., and Howe, F. (2014). De-
lineation of gliomas using radial metabolite indexing.
NMR in Biomedicine, 27(9):1053–1062.
Reynolds, G., Wilson, M., Peet, A., and Arvanitis, T.
(2006). An algorithm for the automated quantitation
of metabolites in in vitro NMR signals. Magnetic res-
onance in medicine, 56(6):1211–1219.
Rowland, B., Deviers, A., Ken, S., Laruelo, A., Ferrand,
R., Simon, L., and Laprie, A. (2013). Beyond the
metabolic map: an alternative perspective on MRSI
data. ESMRMB 2013, 30th Annual Scientific Meeting,
page 270.
Scheenen, T. W., Heerschap, A., and Klomp, D. W. (2008).
Towards
1
H-MRSI of the human brain at 7T with
slice-selective adiabatic refocusing pulses. Mag-
netic Resonance Materials in Physics, Biology and
Medicine, 21(1-2):95–101.
Schroeder, W., Martin, K., Lorensen, B., Avila, Sobierajski,
L., Avila, R., and Law, C. (2006). The Visualization
Toolkit. Kitware, Inc., fourth edition.
Stefan, D., Di Cesare, F., Andrasescu, A., Popa, E.,
Lazariev, A., Vescovo, E., Strbak, O., Williams, S.,
Starcuk, Z., Cabanas, M., et al. (2009). Quantita-
tion of magnetic resonance spectroscopy signals: the
jMRUI software package. Measurement Science and
Technology, 20(10):104035.
Teoh, S. T. and Ma, K.-L. (2003). StarClass: Interactive
visual classification using star coordinates. In SDM,
pages 178–185. SIAM.
Vanhamme, L., Sundin, T., Hecke, P. V., and Huffel,
S. V. (2001). MR spectroscopy quantitation: a re-
view of time-domain methods. NMR in Biomedicine,
14(4):233–246.
Wilson, M., Reynolds, G., Kauppinen, R. A., Arvanitis,
T. N., and Peet, A. C. (2011). A constrained least-
squares approach to the automated quantitation of in
vivo
1
H magnetic resonance spectroscopy data. Mag-
netic resonance in medicine, 65(1):1–12.
Wolf, I., Vetter, M., Wegner, I., Nolden, M., Bottger, T.,
Hastenteufel, M., Schobinger, M., Kunert, T., and
Meinzer, H.-P. (2004). The Medical Imaging interac-
tion ToolKit MITK: A toolkit facilitating the creation
of interactive software by extending VTK and ITK.
volume 5367, pages 5367 – 5367 – 12.
IVAPP 2019 - 10th International Conference on Information Visualization Theory and Applications
128
