
Enhanced Deep Learning for Pathology Image Classification:
A Knowledge Transfer based Stepwise Fine-tuning Scheme
Jia Qu
1
, Nobuyuki Hiruta
2
, Kensuke Terai
2
, Hirokazu Nosato
3
,
Masahiro Murakawa
1,3
and Hidenori Sakanashi
1,3
1
Department of Intelligent Interaction Technologies, University of Tsukuba, Tsukuba, Japan
2
Department of Surgical Pathology, Toho University Sakura Medical Center, Sakura, Japan
3
Artificial Intelligence Research Center, National Institute of Advanced Industrial Science and Technology (AIST), Japan
Keywords: Pathology Image, Deep Learning, Transfer Learning, Color-Index Local Auto-Correlation (CILAC).
Abstract: Deep learning using Convolutional Neural Networks (CNN) has been demonstrated unprecedentedly
powerful for image classification. Subsequently, computer-aided diagnosis (CAD) for pathology image has
been largely facilitated due to the deep learning related approaches. However, because of extremely high
cost of pathologist's professional work, the lack of well annotated pathological image data to train deep
neural networks is currently a big problem. Aiming at further improving the performance of deep neural
networks and alleviating the lack of annotated pathology data, we propose a full-automatic knowledge
transferring based stepwise fine-tuning scheme to make deep neural networks follow pathologist’s
perception manner and understand pathology step by step. To realize this conception, we also introduce a
new type of target correlation intermediate dataset which can be yielded by using fully automated
processing. By extracting rough but stain-robust pathology-related information from unannotated pathology
images with handcrafted features, and making use of these materials to intermediately train deep neural
networks, deep neural networks are expected to acquire fundamental pathological knowledge in advance so
that boosted in the final task. In experiments, we validate the new scheme on several well-known deep
neural networks. Correspondingly, the results present solid evidence for the effectiveness and suggest
feasibility for other tasks.
1 INTRODUCTION
Cancer is one of the most terrible threats to human
health. According to the data (Ferlay J. et al., 2013),
there were approximately 14.1 million new cancer
cases and 8.2 million deaths worldwide in 2012.
Moreover, same report estimates that the number of
new cancer cases may increase to 24 million by
2035. Nowadays, we have many advanced cancer
diagnosis modalities such as Computed Tomography
(CT), Magnetic Resonance Imaging (MRI), and
Positron Emission Tomography (PET). Meanwhile,
pathology image diagnosis is still playing a key role
to assess cancer’s presence or absence. However, the
shortage of pathologists has become a conspicuous
problem in many countries. In japan, the number of
pathologists per 100,000 people is 1.95, which is
around only 1/3 of that in the United States (M.
Fukayama et al. 2015). In China, this number is even
as low as 1.35 (Cornish, 2014). The severe shortage
directly results high workload of pathologists and
increasing misjudgement in diagnosis. Although
digital pathology has widely popularized since more
than a decade ago, confirmation of a mass of large-
scale images remains heavy load to pathologists.
With respect to this issue, efforts on automatic
diagnosis of pathology images based on pattern
recognition technology are regarded as one the most
promising solution.
2 RELATED WORKS
In early periods, researchers used to adopt
conventional image classification approaches based
on pathological morphology indexes (e.g. nuclei-
cytoplasmic ratio and density) and generalized
texture descriptors to map the images to feature
spaces for further modelling. Compared to the
former approaches, the latter ones have shown more
92
Qu, J., Hiruta, N., Terai, K., Nosato, H., Murakawa, M. and Sakanashi, H.
Enhanced Deep Learning for Pathology Image Classification: A Knowledge Transfer based Stepwise Fine-tuning Scheme.
DOI: 10.5220/0007356100920099
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 92-99
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

robustness to the ever-changing cancerous
appearance. Esgiar et al. (Esgiar et al., 1998)
employed GLCM to calculate the contrast, entropy,
angular second moment, dissimilarity and
correlation from colon’s pathology images, and used
linear discriminate analysis (LDA) and k-nearest
neighbour algorithm (KNN) to distinguish normal
and cancerous images. J. Diamond et al. (J.
Diamond et al., 2004) employed an evolved version
of GLCM, called Haralick features to classify
prostate pathology images. In Masood’s study (K.
Masood and N. Rajpoot, 2009), local binary pattern
(LBP) and support vector machines (SVM) are
utilized and demonstrated effectiveness for colon
pathology images. Besides, lower-order and higher-
order histogram features, Gabor filters and
Perception-like features are involved in pathology
image classification as well. However, when using
the generalized texture features, researches have
faced a common problem: it is very hard to control
the adaptability and select the serviceable one or part
(Shen et al., 2017). Meanwhile, the non-uniform
staining quality among data resources and other
changing factors makes the classification more
challenging (R. Marée, 2017, Chen et al., 2016, B.
Bejnordi et al., 2016).
In recent years, deep learning using
convolutional neural networks (CNN) (A.
Krizhevsky et al., 2012) has shown its
unprecedented capacity to defuse these problems.
Due to more domain agnostic approach combining
both feature discovery and implementation to
maximally discriminate between the classes of
interest (Janowczyk and Madabhushi, 2016), high
hope has been placed on deep learning to accelerate
classification of pathology image (Xu et al., 2017,
Hou et al., 2016, Xu et al., 2016). When one adopts
deep learning based approaches, large datasets are
always indispensable to train more capable deep
neural networks and raise the performance.
However, unlike natural image datasets which can
be acquired based on internet and automated
categorizing techniques, building up high quality
pathology image datasets, anyhow, requires
professional observation and annotation by
pathologists. Because of the necessity of this
procedure, well-annotated data usually cost vast
financial resources and manpower. In this situation,
drawing out the maximum power of deep neural
networks with limited datasets has become a very
important practical issue.
3 STEPWISE FINE-TUNING FOR
DEEP NEURAL NETWORKS
When holding a certain amount of data, fine-tuning
the deep neural networks is one of the evidenced
techniques able to boost the performance in some
degree. Rather than training from scratch, fine-
tuning a general neural network which has been pre-
trained with large-scale image datasets (e.g.
ImageNet) to obtain a more specialized network
corresponding to target tasks can usually yield more
advantageous results (Chen et al., 2015, Shin et al.,
2016, Yosinski et al., 2014). Training a CNN
strongly depend on its initial status, thus it is
significant to obtain appropriate initialization as
much as possible in order to avoid over-fitted
learning or local minimum traps. Generally, the
forepart layers of a CNN are considered analogous
to the conventional texture features and applicable to
many of related tasks, while the later layers capture
more abstract image content by combining low-layer
features involving more specific information
corresponding to the target task (Brachmann et al.,
2017). Based on this fact, if the tasks of pre-training
and final classification are sufficiently correlated
(for instance, both of them are for color image
classification), one may only fine-tune part or all of
the pre-trained model to reach more desired results.
Actually, it is quite hard for us to understand the
correlation between these tasks. In some other
situations if target tasks possess much different
distribution compared with the pre-training datasets,
effectiveness of initialization and fine-tuning may be
largely restricted. This issue is exactly arising in
pathology image classification domain. On one
hand, in light of common human’s perception,
pathology images usually have more complicated
appearances than natural images because it is
difficult to figure out the intuitionistic difference
between benign and malignant images at a glance
due to their color uniformity of H&E (Hematoxylin
and eosin) stain and componential similarity of
tissues. On the other hand, owing to professional
knowledge, pathologists are able to distinguish
various pathological components and structures
within the image. Based on this knowledge, they can
easily tell where abnormality has occurred.
Nevertheless, natural image datasets for pre-training
rarely contain relevant information. From this
perspective, we believe that it is crucial to build a
bridge which can reasonably transfer the neural
networks from the task of pre-training classification
to the final benign/malignant judgment of the well-
annotated pathological images.
Enhanced Deep Learning for Pathology Image Classification: A Knowledge Transfer based Stepwise Fine-tuning Scheme
93
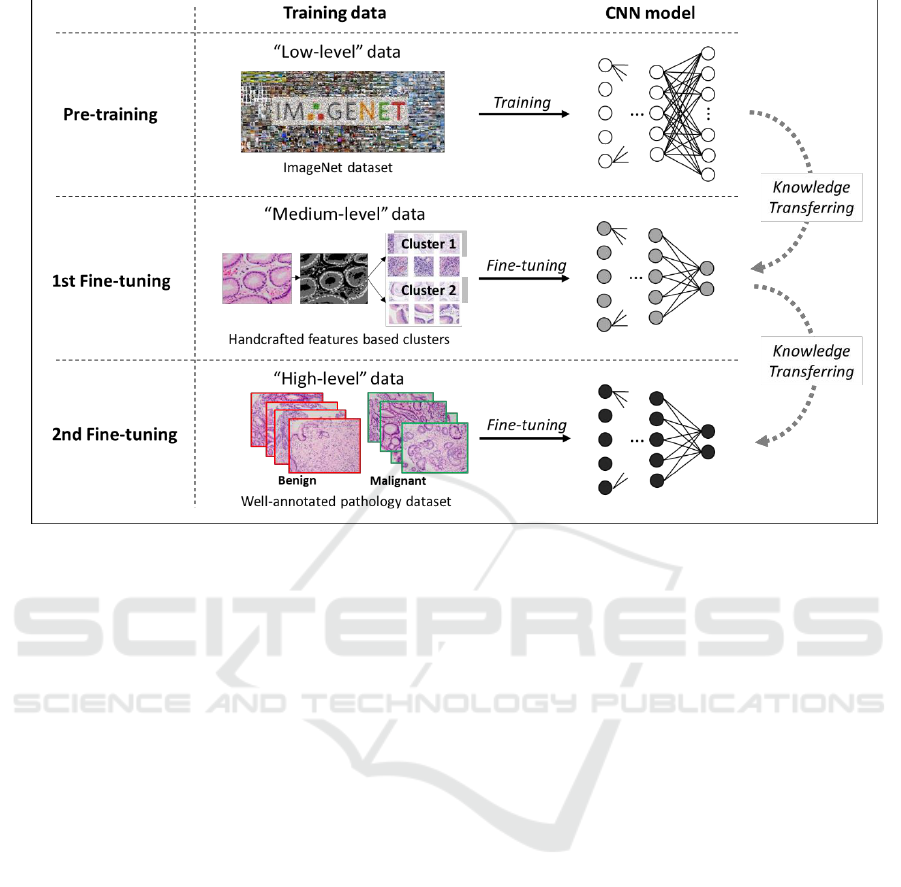
Figure 1: The proposed knowledge transferring based stepwise fine-tuning scheme. Apart from “low-level” pre-training
datasets and “high-level” well-annotated datasets, “medium-level” data are generated automatically and involved in the 1st
fine-tuning process. In CNN models corresponding to training steps along the knowledge transferring direction, darker
nodes in CNN models denote more specialized (deeper) representation which is expected for the pathology image
classification task.
3.1 Making CNNs Learn Pathology
Step by Step
In this paper, we propose a conception taking
advantage of stepwise fine-tuning to make deep
neural networks learn to understand pathology
images gradually following pathologist’s cognitive
way. Before learn to understand the differences
between benign or malignant pathology images, one
should first understand the fundamental pathological
knowledges beforehand. Such knowledges may
include but not be limited to distribution status and
density of cells, degree of nucleus distortion, nucleus
size and nuclear-cytoplasmic ratio. In the previous
section, it has been declared that specific measures
of these indexes for benign/malignant judgement
may be not reliable due to various changing factors.
Nevertheless, these morphological characteristics
can still be exploited to provide rough but task-
relative initialization to the deep neural networks
like training an unskilled pathologist.
To make deep neural networks able to pathology
in a rational way, we build up a stepwise scheme
(Suzuki et al., 2017) in which fine-tuning is adopted
to transfer several different levels of knowledge
toward the final task step by step. The scheme
consists of three main steps: pre-training, 1st fine-
tuning and 2nd fine-tuning. As shown in Figure 1, at
the beginning of the training progress, we have a
pre-trained network as initialization. The following
step of 1st fine-tuning involves a type of target-
correlative “medium-level” dataset, which is
regarded as the carrier of the fundamental
pathological knowledges. According to our
conception, rather than directly driving the deep
neural networks to learn about benign and
malignant, making it gain fundamental pathological
knowledge from the “medium-level” datasets
probably contribute to the task of higher difficulty
(Qu et al., 2018). Therefore, 1st fine-tuning with the
“medium-level” datasets is placed in the middle of
the stepwise scheme. By this step, deep neural
networks are considered more pathology-
specialized. Finally, well-annotated
benign/malignant images are used for the second
time fine-tuning. In the lower part of the figure,
corresponding to all training steps along the
knowledge transferring direction, darker nodes in
CNN models denote more specialized (deeper) rep-
resentation which is expected for the pathology
image classification task When the number of output
BIOIMAGING 2019 - 6th International Conference on Bioimaging
94
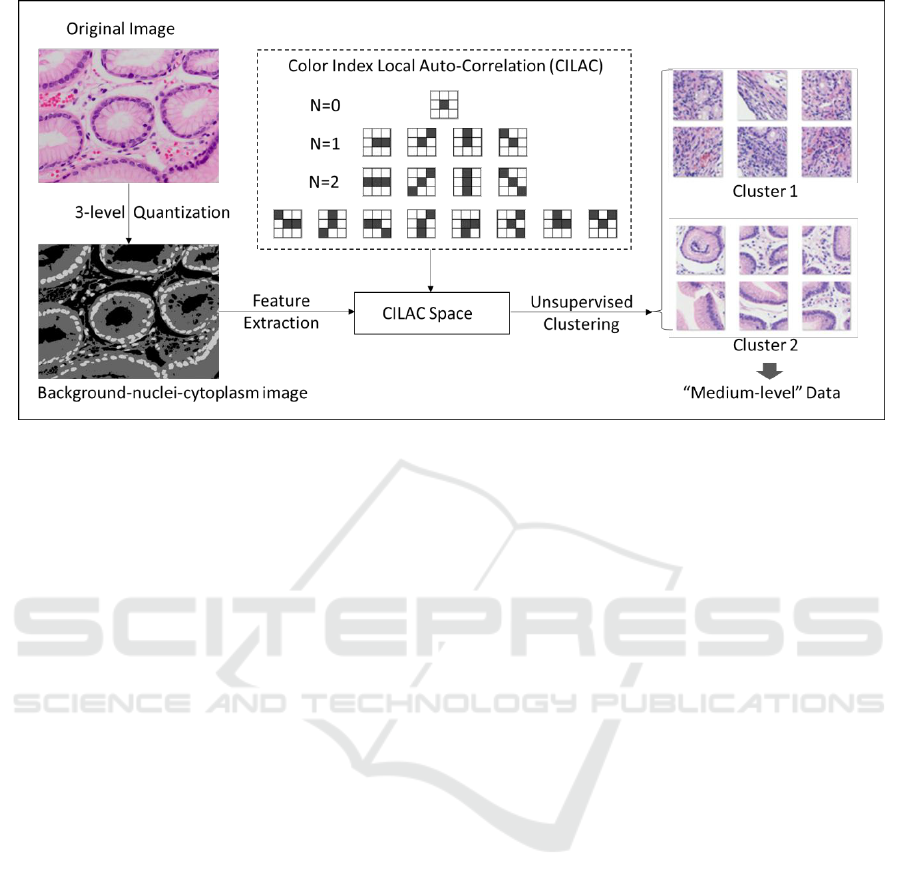
Figure 2: Procedure of generating “medium-level” dataset with color index local auto-correlation (CILAC).
classes changes, the network architecture needs to be
adjusted accordingly. As to the earlier layers, we
recommend to set all of them learnable in both of the
two fine-tuning steps in order to achieve practical
improvement.
3.2 Build “Medium-level” Dataset
using Color Index Local
Auto-Correlation
In the light of out aforesaid conception that
fundamental pathology knowledges are expected to
be involved to bridge the pre-trained model and
transfer these knowledge to the final-task-targeting
model, we consider to adopt a reliable way to
provide with rough and robust, but weakly
pathology-related information to fulfil this demand.
Meanwhile, in respect of the mission of our work, it
is a prerequisite requirement that the “medium-level”
dataset must be achievable at much lower cost than
the well-annotated datasets made by pathologists. In
order to satisfy this requirement, a full-automatic
dataset generation approach is preferentially needed.
According to our earlier study (Qu et al., 2014),
color index local auto-correlation (CILAC,
Kobayashi and Otsu, 2009) has been evidenced as an
independently competent hand-crafted feature in
pathology image classification. Notice that feature
extraction with CILAC is right the choice to
summarily evaluate the status and relation of
background, nuclei and cytoplasm based on the
three-level color indexed image. These three
components are deemed to contain most of the
crucial information for morphological analysis.
Meanwhile, because the color indexing process is
equivalent to normalizing the color space in an
extremely rough way, the color indexed images are
regarded more robust to uneven staining intensity. In
this paper, we take advantage of CILAC based
feature extraction on color-indexed images and
expect to collect sufficient anatomical pathology
information with less noise.
Specifically, CILAC feature was developed on
the basis of Higher Local Auto-Correlation. As
shown in Figure 2, CILAC consists of a set of local
patterns which are able to calculate both the local
auto-correlations of different color levels and their
statistical distribution. CILAC in order N (N = 0, 1,
2) is defined as below:
(1)
(2)
(3)
Where
denotes N-order correlation.
is a D-dimensional vector standing
for D color indexes of an color indexed image.
indicates the reference (central) pixel. a, b are
different displacements of the surrounding inspected
pixels, respectively.
,
and
denote the pixels
taken into account corresponding to all
displacements. In this paper, D is set to 3 according
to three color indexes of the 3-level image. In that
case, the 0th order CILAC (N = 0) draw out different
color indexes themselves, and the 1st and 2nd order
CILAC (N = 1 and N = 2) represent the local co-
Enhanced Deep Learning for Pathology Image Classification: A Knowledge Transfer based Stepwise Fine-tuning Scheme
95
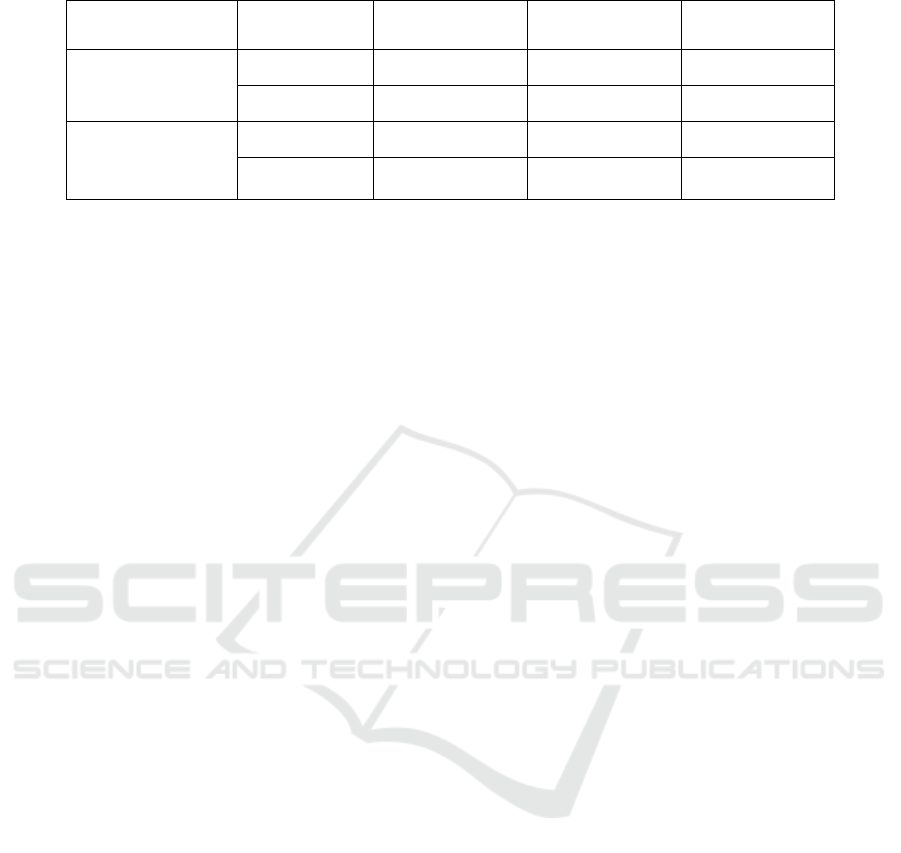
Table 1: Datasets used in experiments.
Data Type
Category
Training
Validation
Test
Medium-level Data
Cluster 1
5,016
558
-
Cluster 2
3,949
439
-
High-level
(Well-annotated)
Data
Benign
5,400
1,620
2,700
Malignant
5,400
1,620
2,700
occurrences of different color indexes. Pathological
components including nuclei, cytoplasm and
background are expected synthetically vectorized by
the CILAC patterns.
Practically, we implement a string of automatic
image pre-processing techniques including 3-level
quantization to obtain the background-nuclei-
cytoplasm images. Afterwards, CILAC feature are
extracted from these 3-level images and principal
component analysis (PCA) is also used to reduce the
dimensionality of feature vector space. Next, we
employ unsupervised K-means clustering to separate
images into several clusters within the feature vector
space. Practically, in order to obtain clusters with
large distance as possible, we set the number of
cluster k=3, and select the farthest two clusters in
line with the visualized status within the coordinate
space of finite principal components. Finally, we
pick up the most distant two clusters and assign +1
and -1 to them. Pass through the above series of
operations, the two clusters are available to be
automatically generated and employed as “medium-
level” training data for the 1st step fine-tuning.
4 EXPERIMENTS
4.1 Experimental Procedures
In order to evaluate the effectiveness of our proposed
transfer learning scheme using stepwise fine-tuning
and the automatically produced low-cost “medium-
level” datasets based on CILAC, we make use of
several well-known deep neural networks including
VGG-16 (Simonyan and Zisserman, 2015), AlexNet
and GoogLeNet (hereafter InceptionV3, Szegedy et
al., 2016). With each of the deep neural networks,
we conduct two separate procedures: (1) adopting
fine-tuning only once with high-level well-annotated
pathology images directly upon the model which has
been pre-trained by low-level large-scale datasets
(ImageNet). (2) adopting the 1st fine-tuning and 2nd
fine-tuning in sequence with the “medium-level”
data and high-level well-annotated pathology image
data, respectively. Competitions are carried out
between the two procedures based on the three deep
neural networks stated above.
4.2 Datasets
This paper employs three types of data including
“low-level”, “medium-level” and “high-level” data,
respectively used for the initialization (pre-training),
the 1st stage fine-tuning and the 2nd stage fine-
tuning. In practice, ImageNet data containing
approximately 1.2 million images in 1,000 separate
categories are customary utilized to initialize the
CNN models. As to the “medium-level” data and
high-level well-annotated pathology image data, we
make use of the gastric pathology datasets collected
by two experienced pathologists. All of the data are
illustrated in Table 1. By adopting unsupervised
clustering upon more than 10,000 patches
(256×256), we succeeded to obtain cluster 1
including 5,574 patches and 4,388 patches belong to
cluster 2. In the 1st-step fine-tuning, 90% of patches
in each cluster are used for training, remaining 10%
are used for validation. Validation data are
completely separated from training data so that well-
generalized model can be selected accordingly. As to
the well-annotated “high-level” datasets, in order to
evaluate the efficacy of the proposed two-stage
scheme, we have prepared well-annotated datasets
including 5,400 benign and 5,400 malignant patches.
All of these patches are cut off from whole
pathology images without augmentation. Except
from the former datasets, we additionally use a
validation dataset including 1,620 benign and 1,620
malignant patches to select the best model
configuration, and a test dataset of 2,700 benign and
2,700 malignant patches to finally evaluate the
performance in each optional case. It is noteworthy
BIOIMAGING 2019 - 6th International Conference on Bioimaging
96
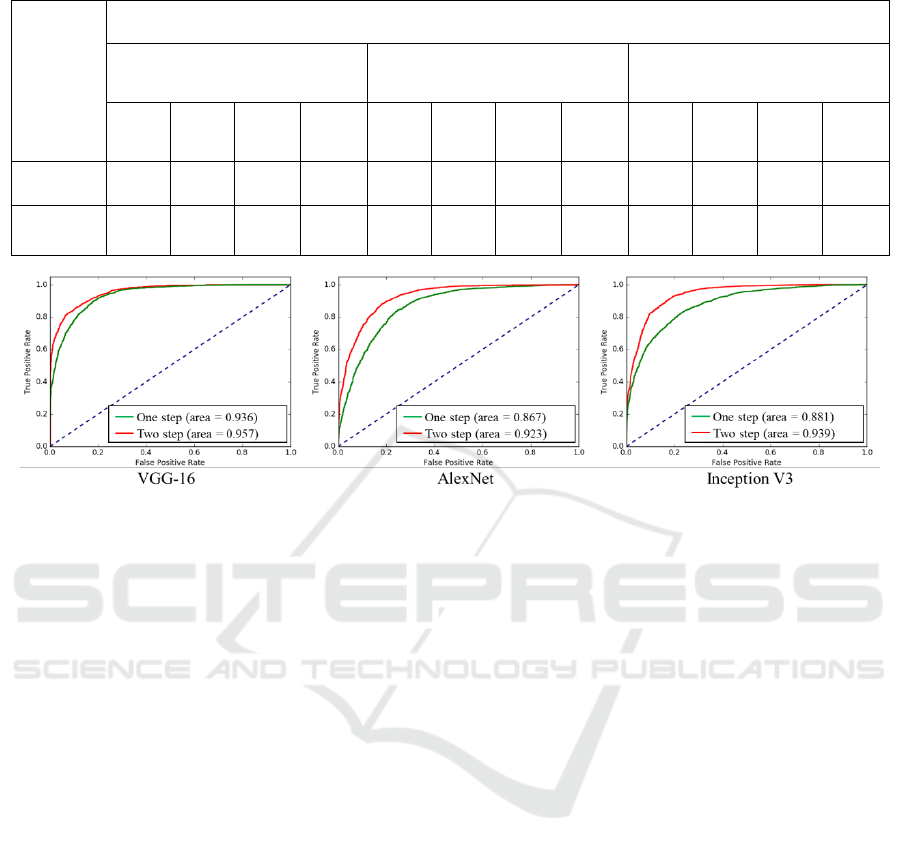
Table 2: Performances of the proposed two-stage fine-tuning using “Medium-level” data.
Scheme
CNN Architecture
VGG-16
AlexNet
GoogLeNet
(Inception V3)
AUC
ACC
Preci-
sion
Recall
AUC
ACC
Preci-
sion
Recall
AUC
ACC
Preci-
sion
Recall
One-step
0.936
0.836
0.96
0.70
0.867
0.794
0.80
0.79
0.881
0.779
0.79
0.78
Two-step
(Proposed)
0.957
0.873
0.87
0.87
0.923
0.845
0.85
0.84
0.939
0.865
0.87
0.86
Figure 3: Performances of the proposed two-stage fine-tuning presented by ROC.
that there is no overlap between the “medium-level”
datasets and the “high-level” datasets, and
meanwhile no overlap among the training, validation
and test datasets.
4.3 Results and Discussion
Next, we will present results and discuss about the
rival performances of the regular one-step fine-
tuning and our proposed stepwise fine-tuning
scheme. To be impersonal, we concurrently take
AUC (Area under the curve, which is calculated on
the class-probability output), ACC (accuracy),
Precision and Recall as the evaluation criteria
(Sokolova and Lapalme, 2009).
As denoted in Table 2, notably, in all of the
couples of competitory schemes, our proposed two-
step fine-tuning using “medium-level” dataset has
yield reasonable improvement. Specifically, AUC
value is raised by 0.021, 0.056 and 0.058, when we
adopt CNN architectures VGG-16, AlexNet and
Inception V3, respectively. Meanwhile, if we focus
on ACC values, we are aware of the fact that the
greatest improvement happens when our proposed
scheme using Inception V3 is adopted. The accuracy
has remarkably increased from 0.779 to 0.865.
Besides, precision and recall, which are commonly
used for medical image classification, are presenting
similar trend to AUC and ACC. As more intuitively
illustrated in Figure 3, three CNN architectures have
produced three separate ROC Figures. The red curve
denotes the two-stage scheme using “medium-level”
dataset, while the green curve denotes the
conventional one-stage scheme. It is clear at a
glance, in each figure, our proposed scheme
possesses overwhelming area all along both the false
positive rate axis and true positive rate axis. These
results have illustrated that our proposed scheme is
capable and rarely dependent on the deep neural
network’s architecture and the amount of well-
annotated data. To sum up, the proposed stepwise
fine-tuning scheme employing “medium-level”
dataset automatically produced based on Color-
Index Local Auto-Correlation (CILAC) has
successfully boosted the performance of the pre-
trained neural networks for gastric pathology image
classification in various situations.
5 CONCLUSION
In this paper, aiming to maximize the classification
capacity of deep neural networks and alleviate the
lack of annotated pathology data, we proposed a
stepwise fine-tuning scheme. By extracting
pathology-correlative information from unannotated
pathology images with handcrafted features, and
Enhanced Deep Learning for Pathology Image Classification: A Knowledge Transfer based Stepwise Fine-tuning Scheme
97

making use of these materials as “medium-level”
data to intermediately fine-tune deep neural
networks, we managed to make the deep neutral
networks acquire pathological knowledge step by
step following the way of pathologist’s perception.
By this mean, the initial task and the final target task
are expected to be bridged in a reasonable way. In
the experiments, our proposed scheme exerted
adequate efficacy for boosting the classification
performance and revealed high applicability for
different CNN architectures. Taking the proposed
scheme as seed, it is promising to promote such kind
of stepwise training scheme to more medical image
recognition tasks.
REFERENCES
Ferlay J, Soerjomataram I, Ervik M et al., 2013.
GLOBOCAN 2012 v1.0, Cancer Incidence and
Mortality Worldwide: IARC CancerBase, No. 11.
M. Fukayama et al., 2015. The Japanese Society of
Pathology Guideline 2015, The Japanese Society of
Pathology, pp. 6.
Toby C. Cornish, Global In-sourcing Using A Pathology
Teleconsultation Network Platform,
https://digitalpathologyassociation.org/_data/files/201
4_Pathology_Visions/PV14_Presentations/19C_In-
Sourcing_Workshop_Cornish.pdf, available on 13
Dec. 2018.
Abdelrahim N. Esgiar, R. Naguib and Bayan S. Sharif et
al., 1998, Microscopic Image Analysis for Quantitative
Measurement and Feature Identification of Normal and
Cancerous Colonic Mucosa, IEEE Transactions on
Information Technology in Biomedicine, vol. 2, no. 3,
pp. 197–203.
J. Diamond, N. H. Anderson, P. H. Bartels et al., 2004,
The Use of Morphological Characteristics and Texture
Analysis in the Identification of Tissue Composition in
Prostatic Neoplasia, Human Pathology, vol. 35, no. 9,
pp. 1121-1131.
K. Masood and N. Rajpoot, 2009, Texture Based
Classification of Hyperspectral Colon Biopsy Samples
Using CLBP, International Symposium on Biomedical
Imaging: From Nano to Macro, pp. 1011-1014.
Dinggang Shen, Guorong Wu and Heung-Il Suk, 2017,
Deep Learning in Medical Image Analysis, Annual
Review of Biomedical Engineering, no. 19, pp. 221–
248.
Raphaël Marée, 2017, The Need for Careful Data
Collection for Pattern Recognition in Digital
Pathology, Journal of Pathology Informatics. vol. 8,
no. 19.
Hao Chen, Xiaojuan Qi and Lequan Yu, 2016, DCAN:
Deep Contour-Aware Networks for Accurate Gland
Segmentation, IEEE Conference on Computer Vision
and Pattern Recognition, pp. 2487-2496.
B. Bejnordi, G. Litjens, N. Timofeeva et al., 2016, Stain
Specific Standardization of Whole-Slide
Histopathological Images, IEEE Transactions on
Medical Imaging, vol. 35, issue 2, pp. 404-415.
A. Krizhevsky, I. Sutskever and G. Hinton, 2012,
ImageNet Classification with Deep Convolutional
Neural Networks, 25th International Conference on
Neural Information Processing Systems, vol. 1, pp.
1097-1105.
Andrew Janowczyk and Anant Madabhushi, 2016, Deep
Learning for Digital Pathology Image Analysis: A
Comprehensive Tutorial With Selected Use Cases,
Journal of Pathology Informatics, vol. 7, no. 29.
Yan Xu, Zhipeng Jia, LiangBo Wang et al., 2017, Large
Scale Tissue Histopathology Image Classification,
Segmentation, and Visualization via Deep
Convolutional Activation Features, BMC
Bioinformatics, 18:281.
L. Hou, D. Samaras and TM. Kurc, 2016, Patch-based
Convolutional Neural Network for Whole Slide Tissue
Image Classification, IEEE Computer Society
Conference on Computer Vision and Pattern
Recognition, pp. 2424-2433.
Xu J, Luo X, Wang G et al., 2016, A Deep Convolutional
Neural Network for Segmenting and Classifying
Epithelial and Stromal Regions in Histopathological
Images, Neurocomputing, vol. 191, pp. 214-223.
F. Ciompi, O. Gessinnk, B. E. Bejnordi et al., 2017, The
Importance of Stain Normalization in Colorectal
Tissue Classification with Convolutional Networks,
IEEE International Symposium in Biomedical Imaging.
Manan Shah, Christopher Rubadue, David Suster et al.,
2016, Deep Learning Assessment of Tumor
Proliferation in Breast Cancer Histological Images,
arXiv:1610.03467.
H. Chen, Q. Dou, D. Ni et al., 2015, Automatic Fetal
Ultrasound Standard Plane Detectionusing Knowledge
Transferred Recurrent Neural Networks, International
Conference on Medical Image Computing and
Computer-Assisted Intervention, pp. 507-514.
Hoo Chang Shin, Holger R. Roth, Mingchen Gao et al.,
2016, Deep Convolutional Neural Networks for
Computer-Aided Detection: CNN Architectures,
Dataset Characteristics and Transfer Learning, IEEE
Transactions on Medical Imaging, vol. 35, issue 5, pp.
1285-1298.
J. Yosinski, J. Clune, Y. Bengio et al., 2014, How
Transferable Are Features in Deep Neural Networks?
Annual Conference on Neural Information Processing
Systems, pp. 3320–3328.
Anselm Brachmann, Erhardt Barth and Christoph Redies,
2017, Using CNN Features to Better Understand What
Makes Visual Artworks Special, Frontiers in
Psychology, 2017; 8: 830.
Aiga Suzuki, Satoshi Suzuki, Shoji Kido et al., 2017, A 2-
staged Transfer Learning Method with Deep
Convolutional Neural Network for Diffuse Lung
Disease Analysis, Proc. of the 2017 Intl. Forum on
Medical Imaging in Asia, pp. 160-163.
BIOIMAGING 2019 - 6th International Conference on Bioimaging
98

J. Qu, N. Hiruta, K. Terai et al., 2018, Gastric Pathology
Image Classification Using Stepwise Fine-Tuning for
Deep Neural Networks, Journal of Healthcare
Engineering, vol 2018, Article ID 8961781.
T. Kobayashi and N. Otsu, 2009, Color Image Feature
Extraction Using Color Index Local Auto-Correlations,
International Conference on Acoustics, Speech, and
Signal Processing, pp. 1057-1060.
Jia Qu, Hirokazu Nosato, Hidenori Sakanashi et al., 2014,
Computational Cancer Detection of Pathological
Images Based on An Optimization Method For Color-
Index Local Auto-Correlation Feature Extraction,
IEEE 11th International Symposium on Biomedical
Imaging, pp. 822-825.
K Simonyan and A Zisserman, 2015, Very Deep
Convolutional Networks for Large-Scale Image
Recognition, International Conference on Learning
Representations.
Christian Szegedy, Vincent Vanhoucke, Sergey Ioffe et al.,
2016, Rethinking the Inception Architecture for
Computer Vision, The IEEE Conference on Computer
Vision and Pattern Recognition, pp. 2818-2826.
M. Sokolova and G. Lapalme, 2009, A Systematic
Analysis of Performance Measures for Classification
Tasks, Information Processing & Management, vol.
45, no. 4, pp. 427-437.
Enhanced Deep Learning for Pathology Image Classification: A Knowledge Transfer based Stepwise Fine-tuning Scheme
99
