
Text-based Medical Image Retrieval using Convolutional Neural
Network and Specific Medical Features
Nada Souissi, Hajer Ayadi and Mouna Torjmen-Khemakhem
Research Laboratory on Development and Control of Distributed Applications (ReDCAD), Department of Computer
Science and Applied Mathematics, National School of Engineers of Sfax, University of Sfax, Tunisia
Keywords:
Text-based Image Retrieval, Convolutional Neural Network, Specific Medical Image Features, Word2vec.
Abstract:
With the proliferation of digital imaging data in hospitals, the amount of medical images is increasing rapidly.
Thus, the need for efficient retrieval systems, to find relevant information from large medical datasets, becomes
high. The Convolutional Neural Network (CNN)-based models have been proved to be effective in several
areas including, for example, medical image retrieval. Moreover, the Text-Based Image Retrieval (TBIR)
was successful in retrieving images with textual description. However, in TBIR, all queries and documents
are processed w ithout taking into account the influence of certain medical terminologies (Specific Medical
Features (SMF)) on the retrieval performance. In this paper, we propose a re-ranking method using the CNN
and the SMF for text-medical image retrieval. First, images (documents) and queries are indexed to specific
medical image features. Second, the Word2vec tool is used to construct feature vectors for both documents
and queries. These vectors are then integrated into a neural network process and a matching function is
used to re-rank documents obtained initially by a classical retrieval model. To evaluate our approach, several
experiments are carried out with Medical ImageCLEF datasets from 2009 to 2012. Results show that our
proposed approach significantly enhances image retrieval performance compared to several state of the art
models.
1 INTRODUCTION
The in c reasing amount of available medical images
causes a difficulty in managing and querying these
large databa ses. Thus, the need for systems provi-
ding efficient researches becomes high. However,
few works investigate the impact o f CNN-based mo-
dels on the Text-Based Image Retrieval (TBIR) p e r-
formance.
To improve the performance of the TBIR appro-
ach, authors (Ayadi et al., 2017a) and (Ayadi et al.,
2018) proposed a thesaurus which is composed of a
set of Specific Medical Features (SMF) such as image
modality, image dimensionality and image color. In
fact, the SMF have shown their effectiveness on me-
dical query classification (Ayadi et a l., 2013) and (Ay-
adi et a l., 2017b) and medical image retrieval (Ayadi
et al., 2017 a) and (Ayadi et al., 201 8). In this paper,
we propose a new re-ranking model based on CNN
and SMF (Ayadi et al., 2017b). Thus, the main con-
tribution of this paper is the exploration of SMF in a
CNN model (CSMF) for m edical image re-ranking.
In this work, we represent queries and docum ents as a
set of SMF. We propose to use the popu la r Word2Vec
model (Mikolov et al., 2013) to generate vector repre-
sentations for SMF-based doc ument and SMF-based
queries. The resulting vectors are the input of the
CSMF model, and are used to get a new semantic re-
presentation to improve the medical image retrieval
accuracy.
The remain der of this paper is organized as fol-
lows: Section 2 describes the background of our
work. Section 3 summarizes the related work. Section
4 describes the proposed CSMF model. Experiments
are presented and discussed in Section 5. Finally,
Section 6 concludes the paper and gives some per-
spectives.
2 BACKGROUND
In this section, we present the SMF set proposed in
(Ayadi et al., 20 13).
Authors in (Ayadi et al., 2017b) and (Ayadi et al.,
2013) proposed SMF to predict the best retrieval mo-
del for a given query and to retrieve images (Ayadi
et al., 2018). These features are ma nually defined by a
78
Souissi, N., Ayadi, H. and Torjmen-Khemakhem, M.
Text-based Medical Image Retrieval using Convolutional Neural Network and Specific Medical Features.
DOI: 10.5220/0007355400780087
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 78-87
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
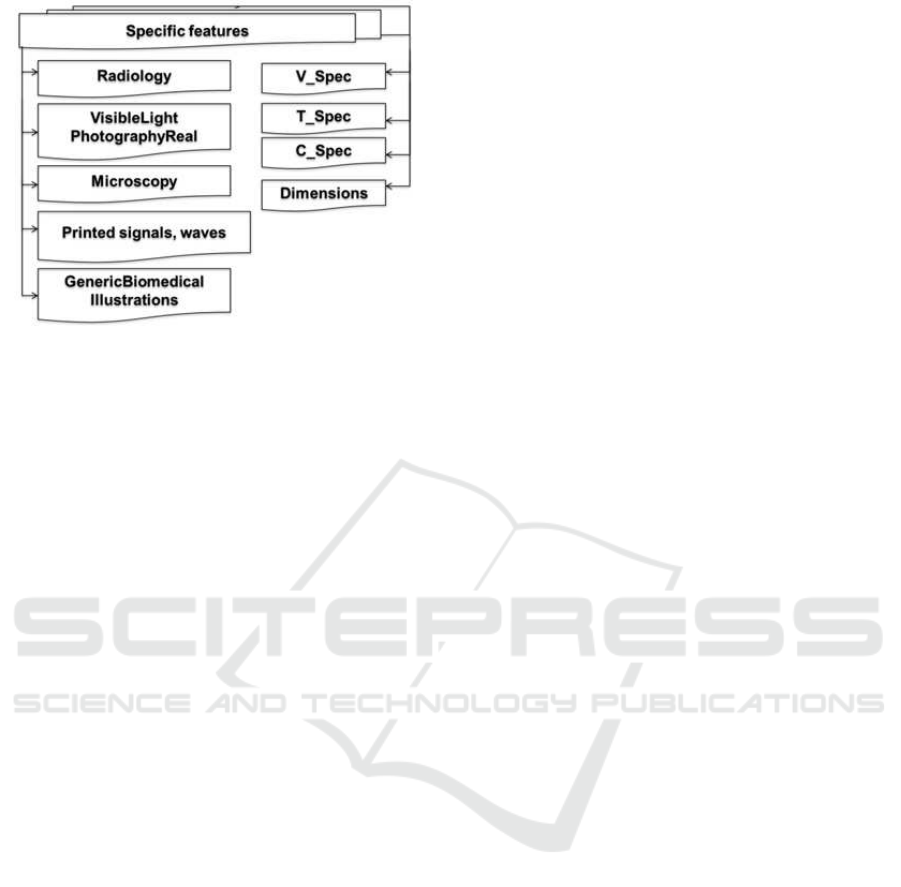
Figure 1: Specific Medical Features (Ayadi et al., 2013).
medical expe rt using imaging modalities an d medical
terminolo gy. There are 25 features that are classified
into 9 categories as illustrated in Figure 1.
• ”Radiology”: it represents the set of diag-
nostic and therapeutic modalities using radia-
tion. It denotes ”Ultraso und”, ”Computerized
Tomography”, ”Magnetic Resonance”, ”X-Ray”,
”2D Radiography”, ”PET”, ”Angiography” and
”Combined mo dalities” in one image. These mo-
dalities which en sure the provision of medical
imagery, are chosen as values for the radiology
feature.
• ”Visible light photography”: denotes the set of
modalities that use visible light inc luding ” En-
doscopy”, ”Skin”, ”Dermatology” and ”Other or-
gans”.
• ”Printed signals and waves”: combines ”elec-
tromyo graphy ”, ”electroencephalography” and
”electrocardiography”.
• ”Microscopy”: includes ”fluorescence mi-
croscopy”, ”transmission microscopy”, ” e le ctron
microscopy” an d ”light micro scopy”.
• ”Generic biomedical illustrations”: denotes, as
”modality tables and forms”, ”programs listing”,
”statistical figures”, ”graphs”, ”charts”, ”screen
shots”, ”flowcharts”, ”system overviews”, ”gene
sequences”, ”chromatography”, ”Gel”, ”chemical
structure”, ”mathematics”, ”formulae”, ”nonclini-
cal photos”, and ”hand-drawn sketches”.
• ”Dimensionality ”: using only modality features to
determine the best retrieval model is not suffcient.
A medical textual query can be expressed without
any image modality. However, in a medical query,
the user can give inform a tion about the searched
object dimension such as: ”micro”, ”gross” and
”gross-micro”.
• ”V-spec”: V-spec feature includes a fea ture rela-
ted with to the sear ched ima ge c olor. An example
of V-spec is ”colore d”.
• ”T-spec”: includes ”pathology” and ”finding”
terms.
• ”C-spec”: includes ”Histology” , which m eans a
study related to microscopic anatomy, so it inte-
resting to applied b oth image con te nt and its text
description for queries containing this term.
3 RELATED WORK
In the litterature , several studies (Qiu et al., 2017) and
(Bai et al., 2018) used CNN based model for Informa-
tion Retrieval (IR) and me dical image retrieval. This
section briefly sum marizes some of these approaches.
3.1 CNN for IR
In rec e nt literature, the CNN is increasingly used
in many disciplines such as IR (Tzelepi and Tefas,
2018), text classification (Kim, 201 4), sentiment ana-
lysis (do s Santos and Gatti, 2014), etc. Thus, it is ap-
plied to several types o f da ta such as text and images.
For textual data, the CNN has shown the ability to:
(1) automatically extract representations from input
data and (2) effectively integrate the input sentences
in vector spaces that keep the syntactic and semantic
aspects of sentences.
Authors in (Huang et a l., 2013) proposed a new
semantic mode l based on CNN to enhance the we b
search performance by extracting semantic structures
from queries or doc uments. In this model, the first
layer converts vector of terms to vector of trigrams
letters. The neuronal activities of the last layer form
a projected vector representation to a semantic space.
Finally, the CNN computes similarities of output vec-
tors to evaluate the relevance scores of documents.
In (Shen et al., 2014), a CNN-based model was
proposed. It transforms queries and documents to a
set of n -gram words. So, the n-gram is p rojected in
low-level feature vectors. Then, a max-p ooling ope-
ration is applied to select neurons with highest activa-
tion values from word featu res. Finally, a non-linear
transformation is perfor med to extract high level se-
mantic information from sequence of input words.
The parameters of the proposed mo del are learned
using click through data. In (Severyn and Mo schitti,
2015), a CNN architecture for re-ranking question-
answer pairs was presented. Additional features have
been integrated in this architecture to offer better per-
formance. This CNN model was expanded and ana-
Text-based Medical Image Retrieval using Convolutional Neural Network and Specific Medical Features
79
lyzed in (Rao e t al., 2017) and delivered reproducible
results with several implementa tions.
3.2 CNN for Medical Image Retrieval
The CNN models have been recen tly used for medical
TBIR systems. In (Rios and Kavuluru, 2015) , an ap-
proach based on bag of words was proposed. It used
CNN to index b iomedical articles by building binary
text classifiers. In this model, the input is matrix of
real numbers whic h represent the medical terms of
the input document. Then, a succession of processing
layers is do ne to classify the documen t. Another met-
hod for medical text classification that can be used for
retrieval tasks was presented in (Hughes et al., 2017).
In fact, it uses a bag of words training on a CNN to re-
present the semantics of an inpu t sentence; especially
it uses the Word2vec a lgorithm to repr esent the input
medical sentences. Also, it keeps the stop-words du-
ring the training of the CNN model which is constitu-
ted by several convolutional layers, ma x-pooling and
fully-co nnected layers. In (Soldaini et al., 2017), aut-
hors pro posed a CNN to reduce noise in clinical notes
to be used for medical literature retrieval. They used
GloVe vectors (Penn ington et al., 2014) to represent
terms of input queries.
Despite the large n umber of works using CNN,
there is a lack of studies using external semantic re-
sources such as specific features to represent que ries
and documents. Therefore, we propose a new medi-
cal image re-ranking model based o n CNN and SMF
using Wor d2vec to improve retrieval accuracy.
4 A NEW CNN MODEL FOR
TEXT-BASED MEDICA L
IMAGE RETRIEVAL: CSMF
MODEL
In this section, we explore the use of CNN for medi-
cal image retrieval. Our model, called CSMF, aims
to re-rank medical images based on their textual des-
cription. The input of the CSMF model is a set of
queries and documents indexed to a set of SMF (Ay-
adi et al., 2017b) as detailed in section II. The out-
put of our model is a set of relevant documents to a
given query. Our model is composed of several lay-
ers: (1) the input layer, which is a vector representing
the query/document, (2) the convolutional laye r, (3)
the pooling layer and (4) the Fully Connected Layer
(FCL) representing the output layer of the CSMF mo-
del. The output c ontains the scores of the similarity
between query and documents.
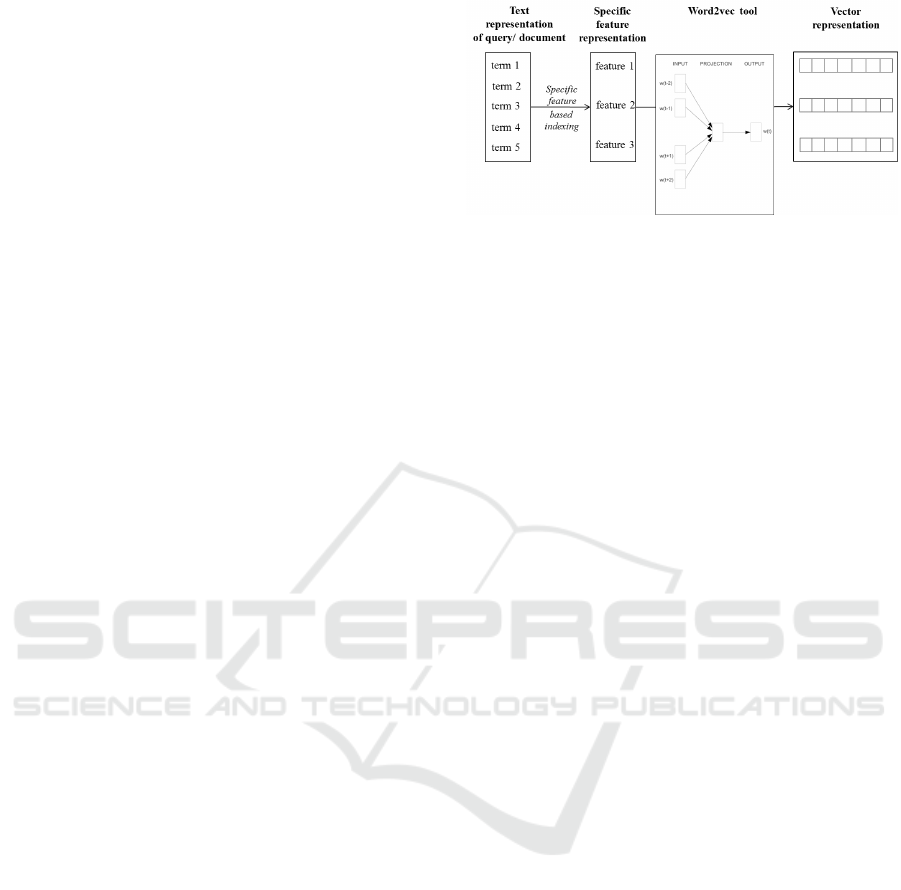
Figure 2: Transformation from text representation to vector
representation.
4.1 Vector Representation of Queries
and Documents
In this sectio n, we detail the Word2vec method (Mi-
kolov et al., 2013) for presenting queries/d ocuments
as vectors.
The input layer of the CSMF model is a
query/documen t presented by features: [ f
1
,..., f
n
],
where each feature f
i
is presented by a vector V
i
∈ R
d
using the Word2vec tool. The obtained set of vector s
are then concatenated to a matrix S ∈ R
n×d
, where n is
the number of the query (or document) features an d d
is the number of a ll fe atures (in our case d=25 as men-
tioned in section II). Each vector V
i
contains features
representation using the Word2vec tool. For e a ch in-
put query/document, the matrix S is built. Eac h row i
of S repr esents a feature f
i
at the corresponding fea-
ture position i in th e query/document.
Figure 2 shows an example of transfo rming a text
representation to a vector representation according to
the CSMF model. The que ries/documents are repre-
sented as a set of SMF in or der to extract sema ntic and
specific features fr om the text representation. Finally,
the Word2vec tool is u sed to transfor m each feature to
a vector.
To captu re semantic features in a given
query/documen t and reach high level sema ntic
informa tion, the neural network applies a series
of transformation s to the input matrix S using
convolution, non-linearity and pooling operations.
4.2 Convolutional Layer
In this layer, a set of filters F ∈ R
d
are applied to
the query/doc ument vectors representation to produce
different feature maps. Each feature map includ e s a
level of sem antic features extracted by the CNN. Each
component of the feature map c
k
∈ R is computed by
the following Equation:
c
k
=
d
∑
1
V
i
F
i
(1)
HEALTHINF 2019 - 12th International Conference on Health Informatics
80

Figure 3: Example of filter containing 25 values correspon-
ding to a feature.
where V
i
is the vector representing the
query/documen t feature, F
i
is a filter applied to
the vector V
i
, and d is the number of features. In our
work, each filter contains 25 values where each value
correspo nds to a feature semantic degree as shown in
Figure 3.
In the current work, we propose to use six filter s
initialized statistically as detailed below. In addition,
the filters applied to the queries are initialized diffe-
rently compared to the ones applied to th e documents
because the latter’s size is greater than the queries’
size. The query filter and the document filter are hen-
ceforth called (QF) and (DF), respectively.
4.2.1 Co-occurrence Filter (CoF)
(QF) The idea consists of calculating the co-
occurre nces of q uery features with all the te rminology
features.
CoF(QF) =
∑
d
0
FR(FQi)
∑
d
0
FR(FF j)
(2)
Where FR(FQ) is the frequency of query features and
FR(FF) is the frequency of features.
(DF) The document filter calculates the occur-
rence of the set of documen t features in the query.
The more the document contains query features, the
more it is relevant.
CoF(DF) =
n
∑
0
FR(FD ∈ Q) (3)
Where n is the numb er of the set of document featu-
res and FR(FD ∈ Q) is the occurrence of document
features in the query.
4.2.2 Lengt h Filter (LF)
(QF) For each query, we c ompute the documents’ size
containing query f e atures (SD). As norma liza tion, we
divide each obtained value by the highest sum of sizes
(Max (SD)).
LF(QF) =
∑
n
0
SD
Max(SD)
(4)
Where n is the n umber of the documents containing
all query features.
(DF)For docu ments, we ca lc ulate the occu rrence
of the set of document f eatures in the corresponding
query (FD) and then we divide this value b y the do-
cument length (LD). Indeed, if the document an d the
query share several fea tures and the document has a
small size, this document becomes m ore relevant.
LF(DF) =
FD
LD
(5)
4.2.3 Rank Filter (RF)
(QF) We calculate documents’ ranks (RD) containing
query features. As normalization, we divide each
obtained value by the highest rank.
RF(QF) =
∑
n
0
RD
Max(
∑
n
0
RD)
(6)
Where n is the number of documents containing all
query features.
(DF) If the organization of features in a document
is the same as in the query, the document should be
organized.
RF(DF) = FR(FQ) × Fact
org
(7)
Where FR(FQ) is the frequency of query features in
the document and Fact
org
is the o rganization factor of
query in the document: Fact
org
equals 1 if the que ry
preserves its organization in the document and 0.5 if
not.
4.2.4 Proximit y Filter (PF)
(QF) If a document contains query features, we com-
pute the distances between its feature s (DD). Then,
we divide each value by the biggest distance. In our
case, the distance between two features is the number
of features between them.
PF(QF) =
∑
n
0
DD
Max(
∑
n
0
DD)
(8)
Where n is the number of documents which contain
query features.
(DF) The more the documen t’s features existing in
the query are c loser, the more it is relevant.
PF(DF) =
1
|FD ∈ Q|
(9)
Where FD ∈ Q is the set of docume nt features exis-
ting in the query.
4.2.5 PMI Filter (PMIF)
(QF/ DF) The PMI (Pointwise Mutual Information )
(Church and Hank s, 1990) is a proposed metric to find
features with a close meaning. Indeed, the PMI of
features x an d y is defin ed usin g the occurrences of
x (FR(x)) and y (FR(y)), the co-occurrenc es FR(x,y)
Text-based Medical Image Retrieval using Convolutional Neural Network and Specific Medical Features
81
within a vector of features, and N the collection size
for QF and the document size for DF.
PMIF(QF) = log
N × FR(x,y)
FR(x) × FR(y)
(10)
This equation calculate s the semantically closest fea-
tures of the collection to the f eatures x and y.
4.2.6 Feature Difference Filter (FDF)
(QF) For each query, we co mpute the number of
its different featur es comparing to the docum ent
(Di f f D). Then, we divide this number by the max-
imum value.
FDF(QF) =
1
Di f f D
Max(
1
Di f f D
)
(11)
(DF) The more the number of the set of document
features not belonging to the query is small, the more
the document is relevant.
FDF(DF) =
1
|FD /∈ FQ|
(12)
Where FD is the set of document features and FQ is
the query features.
4.2.7 Application of Filters
Given that the input of the SemRank model is a matrix
S ∈ R
n×d
, the convolutional filters are also matrices
F ∈ R
d
. It should be noted that these filters have the
same dimensionality d as th e input matrix. Moreover,
these filters scan the vectors representation producing
a vector C ∈ R
n
at the output. Each component c
i
of
C is the result of computing the pro duct between a
vector V and the filter F, which is summed to produce
a single value.
c
i
=
d
∑
k=1
V
k
F
k
(13)
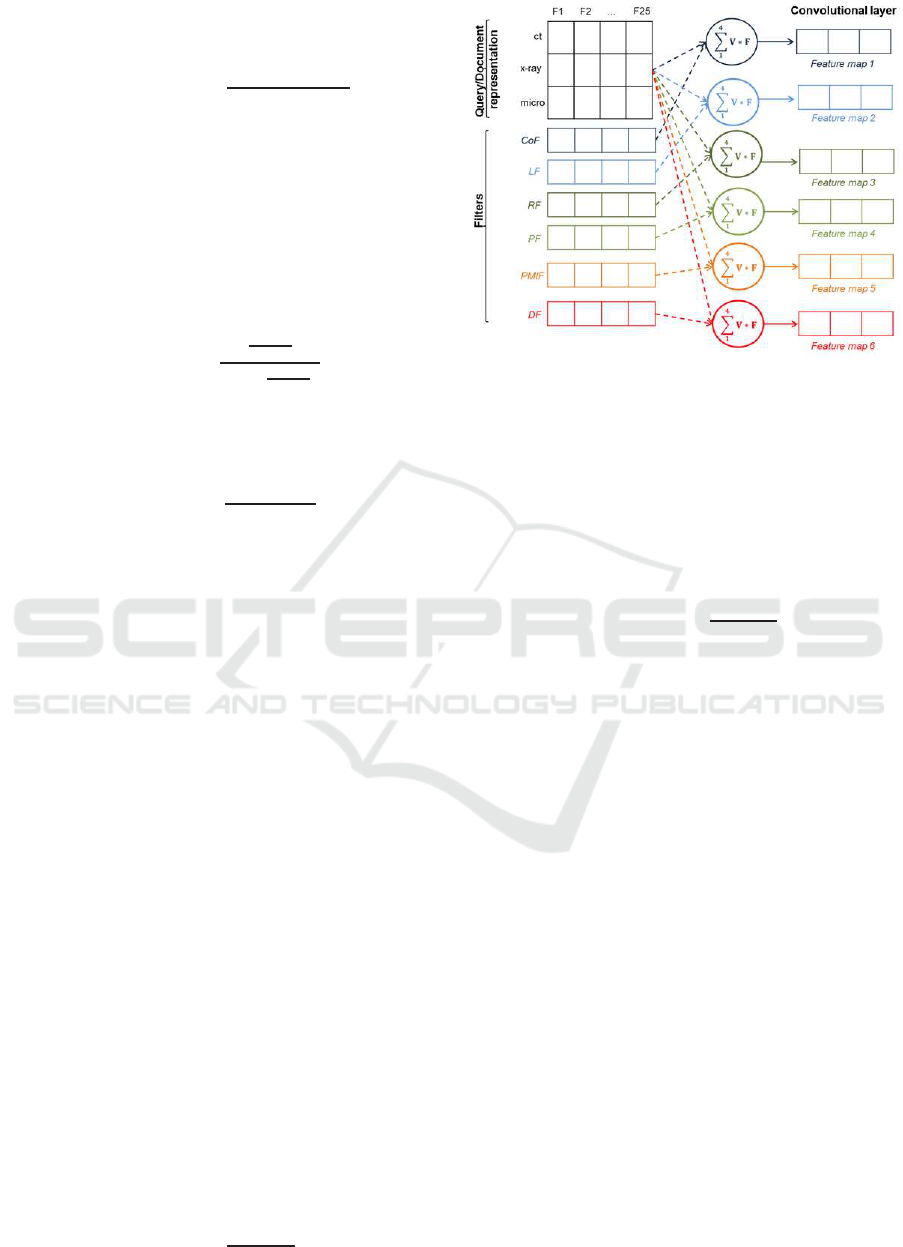
As an example, Fig. 4 shows a matrix representation
of the query ”ct x-ray micro”, as well as the six filters.
4.3 Activation Function
The convolutional layer is followed by a non-linear
activation function α applied to th e output of the pre-
ceding lay e r. This function allows transformin g the
input signal in a neuron to an output signal.
Several activation functions are proposed in the li-
terature such as:
• Sigmoid (Norouzi et al., 2009) which is defined
by:
α(x) =
1
1 − e
−λx
(14)
Figure 4: Example of convolutional layer for the query ”ct
x-ray micro”.
where x is the input of a neuron and λ a pa rame-
ter of the sigmoid function. Its name indicates in
practice an S shape . It represents the logistic dis-
tribution f unction.
• Hyperbolic tangent (tanh) ( N guyen and Widrow,
1990) is an hyperb olic function defined by:
tanh(x) =
1 − e
−2x
1 + e
−2x
(15)
where x is the input of a neuron.
• Rectified Linear Unit (ReLU) (Jarrett et al., 2009)
which is defined by:
α(x) = max(0,x) (16)
where x is the input of a neuron.
The ReLU function ensures that neural values trans-
mitted to the next layer are always positive. In fact,
authors in (Nair an d Hinton, 2010) showed that: the
ReLU function is efficient, simple and allows to re-
duce c omplexity and calculation time. Hence, we use
it as an activation fu nction in our model.
4.4 Pooling Layer
The p ooling layer aims to aggregate information, re-
duce representation and extract global features from
local ones of convolu tional layer. In the literature, two
functions have been applied:
• Average: consists of c omputing the average of
each feature map of the co nvolutional layer and
storing it in the pooling layer. However, this met-
hod suffers from a major drawback: all elements
of the input are considered even if many have low
weights (Zeiler and Fergus, 2013).
HEALTHINF 2019 - 12th International Conference on Health Informatics
82
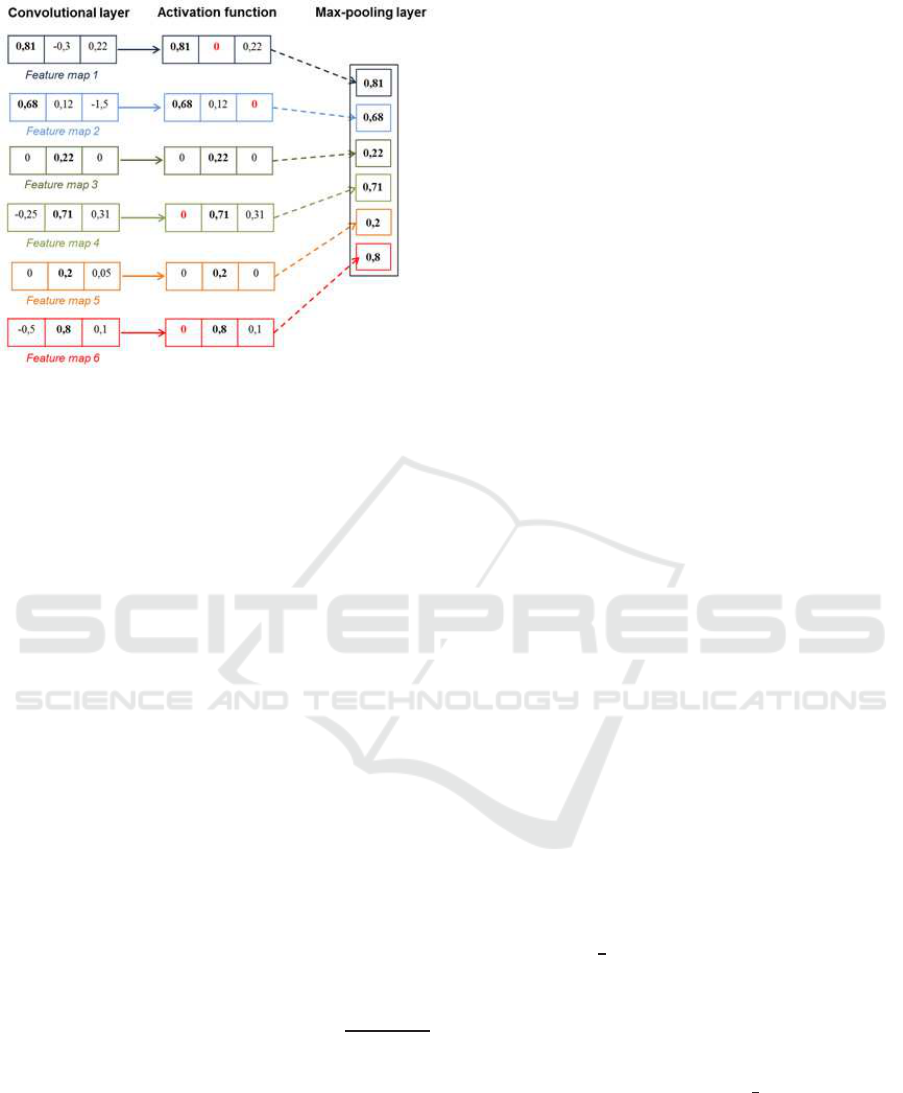
Figure 5: Example of a max-pooling layer.
• Max: c onsists of selecting the maximum value of
each feature map of the convolutional layer. Th us,
Max method only considers neur ons with h igh va-
lues of activation which can lead to poor generali-
zation of input data (Zeiler and Fergus, 201 3).
While max pooling does not suffer from this draw-
back, we chose to use it as illustrated in Figure 5.
4.5 Fully Connected Layer
A Fully Connected Layer (FCL) is, then applied to the
resulting vector, to obtain a final vector representation
of the query/document. As our objective is only to in-
terconne ct all neurons together, we propose to initia-
lize the weight vector to 1.
4.6 The Q uery/Document Matching
Function
We compute the re levance score between queries and
docume nts by c alculating the cosine similarity bet-
ween query vector representation
−→
Q and document
vector representation
−→
D . This r elevance score is defi-
ned as follows:
RSV (Q,D) = S
CSMF
(D) = cosine(
−→
Q ,
−→
D ) =
−→
Q
−→
D
−→
Q
−→
D
(17)
Finally, we combine the CSMF scores (S
CSMF
) with
Baseline model scores (S
Baseline
) using a lin ear com-
bination:
S
combination
(d
i
) = α× S
Baseline
(d
i
)+ (1 − α)× S
CSMF
(d
i
)
(18)
where α is a parameter (α ∈ [0..1]) and d
i
is a docu-
ment retrieved by the Baseline model.
As a baseline we propose to use th e well known
probablistic model BM25 model (Robertson et al,
1994).
5 EXPERIMENTS
In this section , we first d escribe the datasets and the
evaluation metrics. Then, we pr esent the baseline ap-
proach which is BM25. Finally, we discuss the ex-
perimental re sults by presenting a comparative study
with BM25, DLM and Bo1PRF models.
5.1 Datasets and Evaluation Metrics
To evaluate the proposed CSMF model, we conducted
experiments using medical ImageCLEF datasets from
2009 to 2 012 (Dimitrovski et al., 2009), (Benavent
et al., 2010), (Kalpathy-Cramer et al., 2011) and
(M¨uller et al., 2012)). Each image in the collection
has a textual descr iption presented in sem i stru ctured
format including an identifier, an URL, a captio n, a
title, e tc. These ImageCLEF collections are presen-
ted in Table 1. We note that each query is compo sed
of a text representation and few sample images. In our
work, we use only textual representations of the que-
ries. We note that ImageCLEF 2011 and 2012 data-
sets contain a greater image diversity and a lso include
charts, grap hs and other, similar, non-clinical images
(Ayadi et al., 201 3).
We no te that the size of the collection of Image-
CLEF 2011 and 2012 has been significantly increa-
sed. Indeed , these datasets contain a greater image
diversity and also include charts, graphs a nd other, si-
milar, non-clinical images (Ayadi et al., 2013).
In o ur experiments, we propose to use two me-
trics in the evaluation process: the Prec isio n at k
docume nts (P@K) and the Mean average precision
(MAP).
5.2 CSMF
BM25 Model Results
We propose to combine the scores obtained by the
CSMF model with those obtained by the BM25 mo-
del to improve medical image retr ieval accuracy. So,
we conduct a set of experiments. Consequently, we
obtain a new model called CSMF BM25 model. In
fact, α = 0 means that only the CSMF score is used
and α = 1 means th at only the BM2 5 score is used.
Figure 6 shows that the combination o f scores
obtained by the baseline mod e l and those obtained by
the CSMF model improves the results com pared to
the baseline. According to MAP measures, there a re
improvements of: 7% in the ImageCLE F 2009 when
Text-based Medical Image Retrieval using Convolutional Neural Network and Specific Medical Features
83
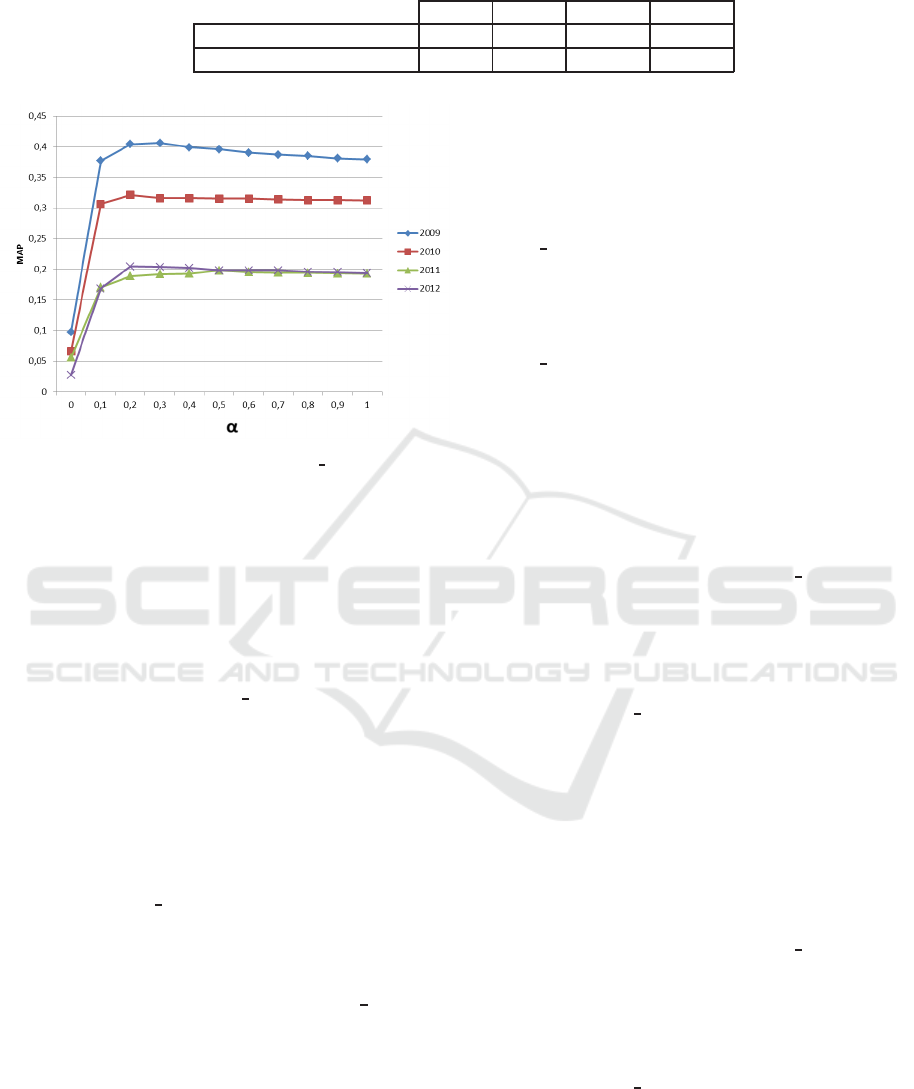
Table 1: Statistics of ImageCLEF datasets.
2009 2010 2011 2012
Total number of images 74902 77500 231000 306528
Number o f queries 25 16 30 22
Figure 6: MAP according to α of CSMF BM25 model in
ImageCLEF datasets.
α = 0.3, 2% in the ImageCLEF 2010 when α= 0.2,
2% in the ImageCLEF 2011 when α = 0.5 and 5% in
the Ima geCLEF 2012 when α = 0.3 com pared to the
baseline.
We notice that b e st results are obtained when α ∈
[0.1..0.5]. Therefore, we chose to set α = 0.3 in the
remaining experiments.
To compare the CSMF
BM25 model with the
BM25 one, we determine the im provement rate and
we conducted a statistic significanc e test. The signi-
ficance value p ∈ [0..1] estimates the probability that
the difference between two method s is due to rand-
omness. T he difference is considered statistically sig-
nificant if p < 0.05 (Hull, 1993). In this paper, the
results are followed by the * when p < 0.05. Accor-
ding to Table 2, we note that the improvement obtai-
ned by the CSMF
BM25 m odel is statistically sign ifi-
cant compared to the BM25 model for 2009 and 2012
ImageCLEF collections (p < 0.05).
5.3 Comparison between CSMF
BM25
and Some Literature Mo dels
In this section, w e propose to c ompare our proposed
model with DLM and Bo1PRF models according to
P@5, P@10 and MAP measures. The DLM (Diri-
chlet Language Model) (Yu et al., 2005) is a statistic
model that allows modeling the arrangement of words
in a language, capturing the distribution of words and
measuring the probability of observing a sequ ence of
words. The purpose of th e Bo1 PRF (Bo1 pseudo
relevance feedback) (Lioma and Ounis, 2008) is to
consider the r e levance jugement of u sers on the docu-
ments obtained initially.
Table 3 summarizes the comparison of the
CSMF
BM25 model with the DLM and the Bo1PRF
models. The best result acro ss all models and for each
metric is presented in bold. Our model o utperforms
other models significantly and reached betwee n 9%
and 24% on the 2 009 dataset. For 2010 dataset, the
CSMF
BM25 model im proves the retrieval perfor-
mance compared to DLM and Bo1PRF models. This
could be explained by the fact that 2009 and 2010 da-
tasets contain images proposed by clinicians and phy-
sicians answering the information needed.
For the 2009 and 2010 da ta sets, the combina tion
of BM25 and CSMF improves the results. For the
2011 a nd 2012 datasets, the results are reduced com-
pared to the baseline.
First, we o bserve that the CSMF
BM25 m odel
outperforms the BM25 model with a substantial ma r-
gin from 1% to 7% in MAP for the 2009, 2010 and
2012 datasets. Our model also outp e rforms D LM mo-
del with a statistically significant margin f rom 1% to
39% for different datasets. Further, compared to PRF
model, the CSMF
BM25 m odel shows a significant
improvement of 9% and 4% MAP respe c tively for
the 2009 and 2010 datasets. For the 2011 and the
2012 datasets, however, no significant ga in is ob ser-
ved. This can be exp lained that these datasets contain
a diversity of images types (ta bles, shapes, graphs ...).
Moreover, the Bo1 PRF mod e l is based on the rele-
vance feedback technique th at impr oves retrieval re-
sults.
The accuracy gain is presented in Table 4 .
First, we o bserve that the CSMF
BM25 m odel
outperforms the BM25 model with a substantial ma r-
gin from 1% to 7% in MAP for the 2009, 2010 and
2012 datasets. Our model also outp e rforms D LM mo-
del with a statistically significant margin f rom 1% to
39% for different datasets. Further, compared to PRF
model, the CSMF
BM25 m odel shows a significant
improvement of 9% and 4% MAP respe c tively for
the 2009 and 2010 datasets. For the 2011 and the
2012 datasets, however, no significant ga in is ob ser-
ved. This can be exp lained that these datasets contain
a diversity of images types (ta bles, shapes, graphs ...).
Moreover, the Bo1 PRF mod e l is based on the rele-
HEALTHINF 2019 - 12th International Conference on Health Informatics
84
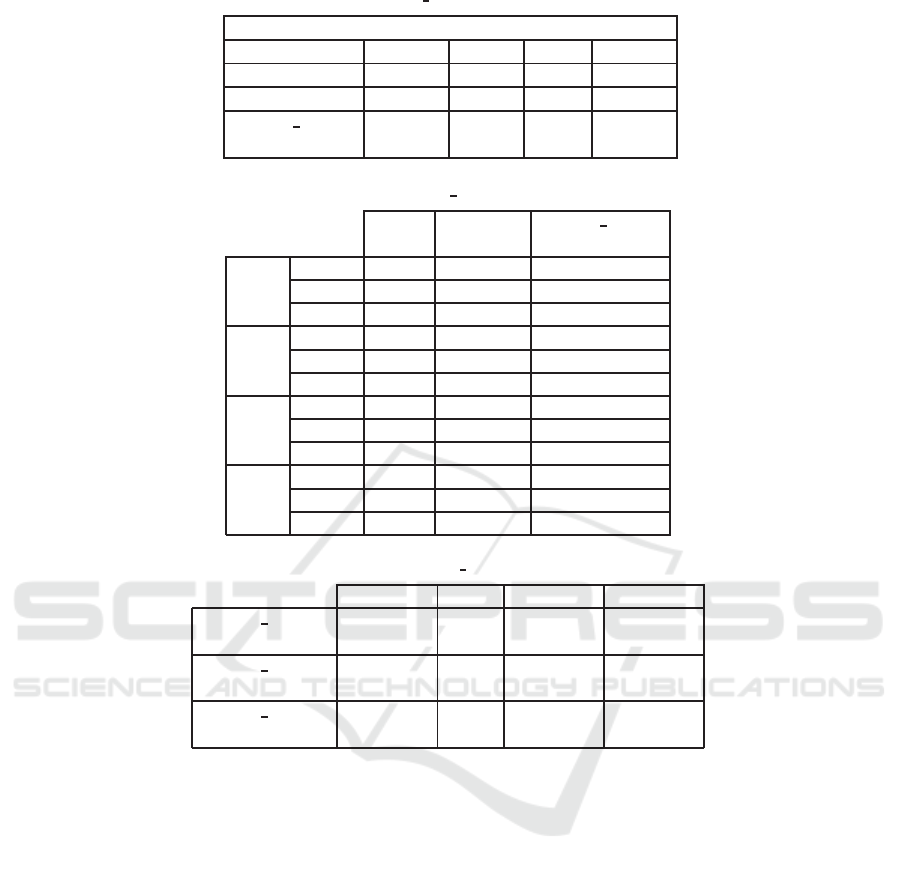
Table 2: Comparison between CSMF BM25 and BM25 according to MAP values.
ImageCLEF datasets
2009 2010 2011 2012
BM25 0.379 0.312 0.193 0.193
CSMF 0.097 0.066 0.055 0.027
CSMF BM25
(α=0.3)
0.405
(+7%*)
0.316
(+1%)
0.190
(-)
0.203
(+5%*)
Table 3: Comparative results CSMF BM25 with some literature models.
DLM Bo1PRF
CSMF BM25
(α=0.3)
2009
P@5 0.592 0.608 0.688
P@10 0.524 0.568 0.664
MAP 0.327 0.371 0.405
2010
P@5 0.436 0.361 0.413
P@10 0.375 0.330 0.460
MAP 0.313 0.305 0.316
2011
P@5 0.240 0.386 0.406
P@10 0.223 0.326 0.330
MAP 0.138 0.211 0.192
2012
P@5 0.281 0.554 0.436
P@10 0.240 0.409 0.336
MAP 0.146 0.361 0.203
Table 4: Accuracy gain of the CSMF BM25 compared to other models.
2009 2010 2011 2012
CSMF BM25/
BM25
+7% (*) +1% - +5% (*)
CSMF BM25/
DLM
+24% (*) +1% +38% (*) +39% (*)
CSMF BM25/
Bo1PRF
+9% +4% - -
vance feedback technique th at impr oves retrieval re-
sults.
To evaluate how well our pro posed app roach per-
forms compared to the state of the art approaches
(Hersh et al., 2009), (Po pescu et al., 2010), (Kalpa thy-
Cramer et a l., 2011) and (M¨uller et al., 201 2), we furt-
her compared our approach with those of the four te-
ams that achieved the best MAP using textua l runs for
the medical image retrieval tasks from 200 9 to 2012
which are:
• LIRIS (France)
• SINAI (Spain)
• YORK (Canada)
• ISSR (Egypt)
• XRCE (France)
• AUEB (Gr e ece)
• OHSU ( U SA)
• LABERINTI (Spain)
• UNED (Spain)
• IPL (Greece)
• MRIM (France)
• BIOINGENIUM (Colombia)
• BUAA AUDR (China)
• DEMIR (Turkey)
Table 5 lists th e MAP, and P@10 values of our
model and those of the state of the art app roaches.
These evaluation measures are the most c ommonly
used measures for ranking participant runs in the Ima-
geCLEFmed competition from 2009 to 2012. The re-
sults of our approach are comparab le to the state of the
art approach e s. We fir st o bserve that the CSMF BM25
model gives the best result in terms of P@10 for the
2009 dataset. For the same dataset, our model does
not outperform the highest values of MAP obtained
by existing ImageCLEFm e d approaches. However, it
was the second best approach with a MAP of 0.405.
Text-based Medical Image Retrieval using Convolutional Neural Network and Specific Medical Features
85
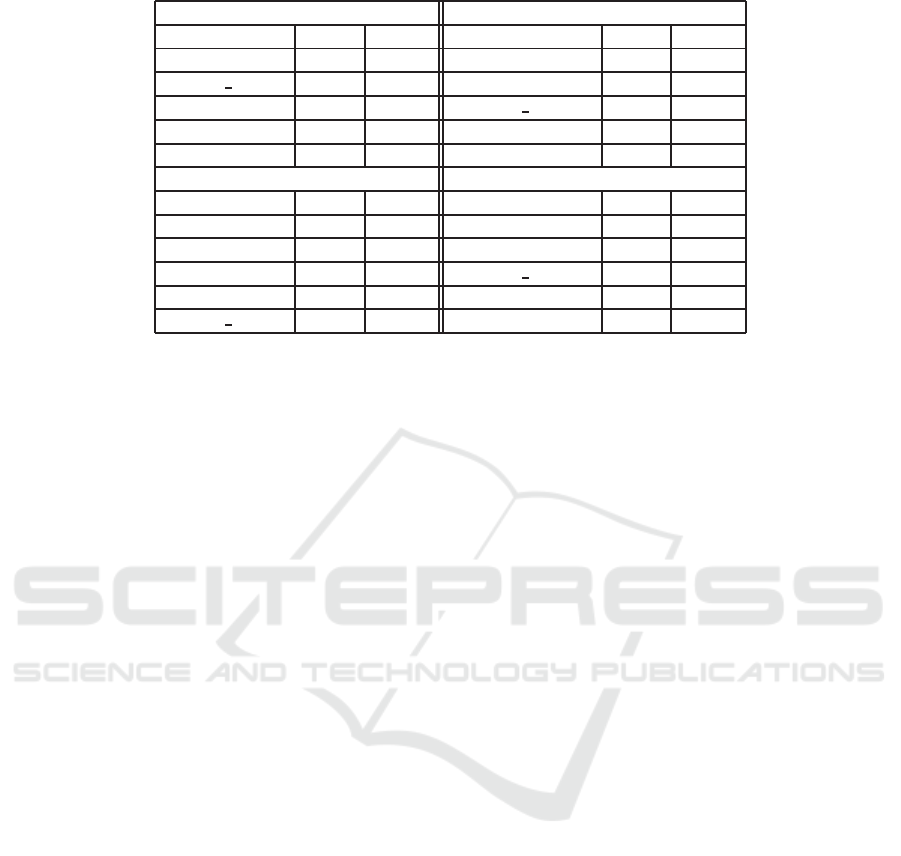
Table 5: Comparative results with the official submissions of the clef medical image retrieval track.
ImageCLEF 2009 ImageCLEF 2010
Group MAP P@10 Group MAP P@10
LIRIS 0.430 0.660 XRCE 0.338 0.506
CSMF BM25 0.405 0.664 AUE B 0.323 0.648
SINAI 0.380 0.620 CSMF BM25 0.316 0.460
YORK 0.370 0.600 OHSU 0.302 0.431
ISSR 0.350 0.560 SINAI 0.276 0.42 5
ImageCLEF 2011 ImageCLEF 2012
Group MAP P@10 Group MAP P@10
LABERINTI 0.217 0.346 BIOINGENIUM 0.218 0.340
UNED 0.215 0.353 BUAA AUDR 0.208 0.309
IPL 0.215 0.403 CSMF BM25 0.203 0.336
MRIM 0.200 0.303 IPL 0.200 0.295
CSMF BM25 0.192 0.330 DEMIR 0.190 0.331
In ImageCLEF 2011, no outperformance is shown.
We co nclude that in tegra ting SMF in a CNN im-
proves results comparing to the baseline and other
models. This could be limited to the SMF that are
purely medical.
6 CONCLUSION AND FUTURE
WORK
We proposed in this paper a novel CNN model for
re-ranking medical ima ges based on Specific Medi-
cal image Features (SMF) called CSMF. In this mo -
del, queries and do cuments are represented as a set
of SMF. The Word 2vec method is used to construct
vector representations for each query/document. The
resulting vectors are th en integrated into a CNN pro-
cess. The output is a query vector and a document
vector used to calculate new relevance scores for do-
cuments given a query. A linear combination of obtai-
ned scores with baseline scores is then used.
We carried out experiments using the Medical
ImageCLEF collections from 2009 to 2 012. The re-
sults showed that the combination of CSMF scores
and baseline scores impr oves the retrieval accuracy.
In ad dition, we compared our model with other state
of the art models and we noticed a sign ificant impro-
vement in the most of metrics’ values.
In future work, we plan to use CSMF model as a
ranking model by applying the deep lear ning techni-
que on the CNN fo r updating the filter values of this
model. Furthermore, we plan to integrate visual fe-
atures in the CSMF m odel and combine them with
textual f e atures to improve retrieval accura cy.
REFERENCES
Ayadi, H., Khemakhem, M. T., Huang, J. X., Daoud, M.,
and Jemaa, M. B. (2017a). Learning to re-rank medi-
cal images using a bayesian network-based thesaurus.
In European Conference on Information Retrieval, pa-
ges 160–172. Springer.
Ayadi, H., Torjmen, M., Daoud, M., Ben Jemaa, M.,
and Xiangji Huang, J. (2013). Correlating medical-
dependent query features with image retrieval models
using association rules. In Proceedings of the 22nd
ACM international conference on Information & Kno-
wledge Management, pages 299–308. ACM.
Ayadi, H., Torjmen-Khemakhem, M., Daoud, M., Huang,
J. X., and Ben Jemaa, M. (2017b). Mining correlati-
ons between medically dependent features and image
retrieval models for query classification. Journal of
the Association for Information Science and Techno-
logy, 68(5):1323–1334.
Ayadi, H., Torjmen-Khemakhem, M., Daoud, M., Huang,
J. X ., and Ben Jemaa, M. (2018). Mf-re-rank: A
modality feature-based re-ranking model f or medical
image retrieval. Journal of the Association for Infor-
mation Science and Technology, 69(9):1095–1108.
Bai, C., Huang, L., Pan, X. , Zheng, J., and Chen, S.
(2018). Optimization of deep convolutional neural
network for large scale image retrieval. Neurocom-
puting, 303:60–67.
Benavent, J. , Benavent, X ., de Ves, E., Granados, R., and
Garc´ıa-Serrano, A. (2010). Experiences at imageclef
2010 using cbir and tbir mixing information approa-
ches. In CLEF (Notebook Papers/LABs/Workshops).
Church, K. W. and Hanks, P. (1990). Word association
norms, mutual information, and lexicography. Com-
putational linguistics, 16(1):22–29.
Dimitrovski, I ., Kocev, D., Loskovska, S., and Dˇzeroski,
S. (2009). Imageclef 2009 medical image annotation
task: Pcts for hierarchical multi-label classification. In
Workshop of the Cross-Language Evaluation Forum
for European Languages, pages 231–238. Springer.
HEALTHINF 2019 - 12th International Conference on Health Informatics
86

dos Santos, C. and Gatti, M. (2014). Deep convolutio-
nal neural networks for sentiment analysis of short
texts. In Proceedings of COLING 2014, the 25th
International Conference on Computational Linguis-
tics: Technical Papers, pages 69–78.
Hersh, W., M¨uller, H., and Kalpathy-Cramer, J. (2009). The
imageclefmed medical image retrieval task test col-
lection. Journal of Digital Imaging, 22(6):648.
Huang, P.-S., He, X., Gao, J., Deng, L., Acero, A., and
Heck, L. (2013). Learning deep structured seman-
tic models for web search using clickthrough data.
In Proceedings of the 22nd ACM international con-
ference on Conference on information & knowledge
management, pages 2333–2338. ACM.
Hughes, M., Li, I., Kotoulas, S., and Suzumura, T. (2017).
Medical text classification using convolutional neural
networks. Stud Health Technol Inform, 235:246–50.
Hull, D. (1993). Using statistical testing in the evaluation
of retrieval experiments. In Proceedings of the 16th
annual international ACM SIGIR conference on Rese-
arch and development in information retrieval, pages
329–338. ACM.
Jarrett, K., Kavukcuoglu, K., LeCun, Y., et al. (2009). What
is t he best multi-stage architecture for object recogni-
tion? In Computer Vision, 2009 IEEE 12th Internati-
onal Conference on, pages 2146–2153. IEEE.
Kalpathy-Cramer, J., M¨uller, H., Bedrick, S., Eggel, I.,
de Herrera, A. G. S., and T sikrika, T. (2011). Over-
view of the clef 2011 medical image classifica-
tion and retrieval tasks. In CLEF (notebook pa-
pers/labs/workshop), pages 97–112.
Kim, Y. (2014). Convolutional neural networks
for sentence classification. In arXiv preprint
arXiv:1408.Conference on Empirical Methods in Na-
tural Language Processing.
Lioma, C. and Ounis, I. (2008). A syntactically-based query
reformulation t echnique for information retrieval. In-
formation processing & management, 44(1):143–162.
Mikolov, T., Chen, K., Corrado, G., and Dean, J. (2013).
Efficient estimation of word representations in vector
space. ICLR Workshop.
M¨uller, H., de Herrera, A. G. S., Kalpathy-Cramer, J.,
Demner-Fushman, D., Antani, S. K., and Eggel, I.
(2012). Overview of the imageclef 2012 medical
image retrieval and classification tasks. In CLEF (on-
line working notes/labs/workshop), pages 1–16.
Nair, V. and Hinton, G. E. (2010). Rectified linear units im-
prove restricted boltzmann machines. In Proceedings
of the 27th international conference on machine lear-
ning (ICML-10), pages 807–814.
Nguyen, D. and Widrow, B. (1990). Improving the lear-
ning speed of 2-layer neural networks by choosing
initial values of the adaptive weights. In Neural Net-
works, 1990., 1990 IJCNN International Joint Confe-
rence on, pages 21–26. IEEE.
Norouzi, M., Ranjbar, M., and Mori, G. (2009). Stacks of
convolutional r estr icted boltzmann machines for shift-
invariant feature learning. In Computer Vision and
Pattern Recognition, 2009. CVPR 2009. IEEE Con-
ference on, pages 2735–2742. IEEE.
Pennington, J. , Socher, R., and Manning, C. (2014). Glove:
Global vectors for word representation. In Procee-
dings of the 2014 conference on empirical methods in
natural language processing (EMNLP), pages 1532–
1543.
Popescu, A., Tsikrika, T., and Kludas, J. (2010). Overview
of the wi kipedia retrieval task at imageclef 2010. In
CLEF (notebook papers/LABs/workshops).
Qiu, C., Cai, Y., Gao, X., and Cui, Y. (2017). Medical
image retrieval based on the deep convolution net-
work and hash coding. In Image and Signal Proces-
sing, BioMedical Engineering and Informatics (CISP-
BMEI), 2017 10th International Congress on, pages
1–6. IEEE.
Rao, J., He, H., and Lin, J. (2017). Experiments with convo-
lutional neural network models for answer selection.
In Proceedings of the 40th International ACM SIGIR
Conference on Research and Development in Informa-
tion Retrieval, pages 1217–1220. ACM.
Rios, A. and Kavuluru, R. (2015). Convolutional neural net-
works for biomedical text classification: application
in indexing biomedical articles. In Proceedings of the
6th ACM Conference on Bioinformatics, Computatio-
nal Biology and Health Informatics, pages 258–267.
ACM.
Robertson, S. E., Walker, S. (1994). Some simple ef-
fective approximations to the 2-poisson model for pro-
babilistic weighted retrieval. In Proceedings of the
17th annual international ACM SIGIR conference on
Research and development in information retrieval.
Springer-Verlag New York, Inc., pp. 232–241.
Severyn, A. and Moschitti, A. (2015). Learning to rank
short text pairs wit h convolutional deep neural net-
works. In Proceedings of the 38th international ACM
SIGIR conference on research and development in in-
formation retrieval, pages 373–382. ACM.
Shen, Y., He, X., Gao, J., Deng, L., and Mesnil, G. (2014).
A latent semantic model with convolutional-pooling
structure for information retrieval. In Proceedings
of the 23rd ACM International Conference on Con-
ference on Information and Knowledge Management,
pages 101–110. ACM.
Soldaini, L., Yates, A., and Goharian, N. (2017). Denoising
clinical notes for medical literature retr ieval with con-
volutional neural model. In Proceedings of the 2017
ACM on Conference on Information and Knowledge
Management, pages 2307–2310. ACM.
Tzelepi, M. and Tefas, A. (2018). Deep convolutional image
retrieval: A general framework. Signal Processing:
Image Communication, 63:30–43.
Yu, G., Li, X., Bao, Y., and Wang, D. ( 2005). E valua-
ting document-to-document relevance based on docu-
ment language model: modeling, implementation and
performance evaluation. I n International Conference
on Intelligent Text Processing and Computational Lin-
guistics, pages 593–603. Springer.
Zeiler, M. D. and Fergus, R. (2013). Stochastic pooling for
regularization of deep convolutional neural networks.
Text-based Medical Image Retrieval using Convolutional Neural Network and Specific Medical Features
87
