
3D Image Deblur using Point Spread Function Modelling for
Optical Projection Tomography
Xiaoqin Tang
1
, Gerda E. M. Lamers
2
and Fons J. Verbeek
1,2
1
Leiden Institute of Advanced Computer Science, Leiden University, Niels Bohrweg 1, Leiden, Netherlands
2
Institute of Biology Leiden, Leiden University, Sylviusweg 72, Leiden, Netherlands
Keywords: 3D Imaging, Point Spread Function, Optical Projection Tomography, Magnification, Deblur.
Abstract: Optical projection tomography (OPT) is widely used to produce 3D image for specimens of size between
1mm and 10mm. However, to image large specimens a large depth of field is needed, which normally results
in blur in imaging process, i.e. compromises the image quality or resolution. Yet, it is important to obtain the
best possible quality of 3D image from OPT, thus deblurring the image is of significance. In this paper we
first model the point spread function along optical axis which varies at different depths in OPT imaging
system. The magnification is taken into account in the point spread function modelling. Afterward,
deconvolution in the coronal plane based on the modelled point spread function is implemented for the image
deblur. Experiments with the proposed approach based on 25 3D images including 4 categories of samples,
indicate the effectiveness of quality improvement assessed by image blur measures in both spatial and
frequency domain.
1 INTRODUCTION
1.1 Background: 3D Image
Deconvolution
In biomedical research, i.e. disease and drug research,
optical techniques allow efficient and high-resolution
imaging of animal cell, tissues, organs and organisms.
Compared to 2D imaging, 3D imaging provides more
structural and comprehensive information, producing
more reliable and convincing evidence for research.
Optical projection tomography (Sharpe et al.,
2002), is a typical optical 3D imaging technique for
objects at tissue-, organ- and organism-level in the
magnitude range of millimeters, thereby filling a gap
between confocal and computational tomography
imaging in the resolution range. A point object
located within the DOF of the optical system is
considered to be in focus, but not necessarily at an
optimal focus. Beyond the DOF, the object is out of
focus (Walls et al., 2007). Depth of field (DOF) is
defined as a double fan symmetric around the focal
plane. For OPT imaging and reconstruction, the DOF
is expected to be large enough to contain as much of
the sample as possible. In this manner the parts of the
sample located in the DOF will result in an image
more or less in focus. However, according to previous
studies (Walls et al., 2007, NcNally et al., 1999), large
DOF subsequently produces image blur and results in
low in-focus image quality. In this paper the image
quality is also referred to as image resolution
according to some literatures. The trade-off between
DOF and image quality should be considered when
selecting lens for an OPT imaging system. A lens
with small numerical aperture (NA) will produce
large DOF, allowing imaging of larger samples but it
results in a relatively blurred image.
One typical way to improve the image quality to
the best resolution is applying deconvolution to the
raw tomography 2D images by using a constant
theoretical or experimental point spread function
(PSF). However, this approach is not strictly suitable
for OPT images; the raw OPT images normally
integrate the sample information of different depths
within a wide field. The imaging PSF within the field
varies at different depths along the optical axis.
Considering this variation of the PSF makes
deconvolution of 3D image more feasible and
promising. A 3D image is reconstructed from the
OPT images which are obtained by rotating the
specimen and acquiring a series of wide-field images
at regular angular intervals. This is accomplished
over a full revolution of the specimen. The filtered
Tang, X., Lamers, G. and Verbeek, F.
3D Image Deblur using Point Spread Function Modelling for Optical Projection Tomography.
DOI: 10.5220/0007237700670075
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 67-75
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
67

back-projection (FBP) algorithm is typically used for
3D image reconstruction (Kak et al., 2001) in this
case. Deconvolution implemented on the
reconstructed 3D image is regarded as 3D image
deconvolution.
According to Chen et al., (2012), OPT is typically
undertaken with specimens that extend beyond the
confocal parameter i.e. the Rayleigh range, of the
imaging lens. Therefore, the tangential resolution of
the reconstructed 3D image decreases away from the
focal plane radially. When the focal plane is
coincident with the center of rotation (COR), the
tangential resolution decreases centered on the COR
in a radial-symmetrical fashion. For an imaging
system with a focal plane located away from the COR
the decrease in resolution is more complicated but the
highest resolution is still found around the focal
plane. The focal plane in the reconstructed slice
corresponds to a circle centered around the COR,
rather than a point coincident with the COR. This
subsequently appears as a cylindrical surface in the
3D image centered by the COR.
The aim of our contribution lies in deblurring the
OPT 3D image (improving the visual resolution) by
means of deconvolution, based on the modeled PSF
of the imaging system. This will, for large samples
with focal plane being at or away from the COR,
recuperate the imperfections of 3D image resulting
from the imaging system. The method in the
modelling of PSF will be explained in Section 2 and
the qualitative and quantitative image comparison
will be presented in Section 3. In section 4 we will
present our conclusions.
1.2 Related Works
Accounting for the trade-off between large DOF and
high resolution, previous studies have proposed
several methods to solve the problem. One approach
is choosing a high NA lens to acquire a high-
resolution image and combining multiple focal planes
in a simultaneous manner (Chen et al., 2013) or
scanning the focal plane through the sample (Miao et
al., 2010). These multiple focal plane approaches
solve the issue of narrow DOF, but the mechanism of
multiple measurements and scanning increases the
acquisition time and the complexity of the imaging
system. Considerably another direction is to use a
reasonable NA lens and deblur the image by
employing a deconvolution or filter on images before
or after reconstruction. Walls et al., (2007) first
applied the frequency-distance relationship (FDR)
(Xia et al., 1995) to OPT. The corresponding filter
was implemented on the sinogram before
reconstruction. The quality of the 3D image can be
further improved with weighted filtered back
projection (WFBP) (Darrell et al., 2008); this is done
by considering the intensity distribution of multiple
fluorescent spheres of known size along the optical
axis. But the implementation of evenly placing each
sphere along the optical axis is rather difficult to
achieve. Chen et al., (2012) proposed a way to
determine the modulation transfer function (MTF)
that contributed to MTF-mask filter and MTF-
deconvolution filter in the reconstruction process.
The former filter significantly reduced the artifacts
produced by sparse projection but the latter filter had
limited improvement on tangential image resolution.
Additionally, a spatial-invariant experimental PSF
was investigated by McErlean et al., (2016) in order
to improve the spatial resolution. However, spatial-
invariance of the PSF is not completely convincing
for OPT. Most recently, a new deconvolution
approach based on the reconstructed 3D image was
proposed by Horst et al., (2016). In their approaches
the PSF was modelled and as such they achieved
significant improvement on the reconstructed slice.
Nevertheless, they focused on the deconvolution of
vertically independent slices and omitted the PSF
diffractions along the optical axis that concerns the
interaction of different slices.
In this paper, we contribute by modeling the
experimental PSF of a single sphere along optical
axis, thereby considering the interaction of
contiguous slices from the reconstructed volume. At
the same time, the magnification is taken into account
in an experimental manner. As discussed in section
1.1, the tangential resolution of the OPT 3D image
slice decreases radially around the focal plane.
Theoretically the best resolution of the reconstructed
3D image can be achieved by combining all the
coronal deconvolutions of different angles. The
coronal deconvolution means deconvolving the 3D
image with the PSF slice by slice in the coronal plane
along its depth axis. This depth axis is parallel to
optical axis of the modelled PSF. We only implement
the coronal deconvolution in 2 opposite angles, i.e.
the reconstructed 3D image and its opposite sample at
180° centered by the COR, in parallel considering the
enormous time consumption of 3D matrix rotation in
N angles and the symmetry of the focal plane. When
the focal plane is off the COR during imaging
process, the shift is accounted for by a shift in PSF
modelling. This paper focuses on the presentation of
the concept of PSF modeling and coronal
deconvolution on 3D OPT data, accompanied by
some initial experimental results based on 25 3D
images including 4 categories of samples. Further
BIOIMAGING 2019 - 6th International Conference on Bioimaging
68

evaluations on a larger number of data are the topics
of our current research.
2 MATERIALS AND METHOD
2.1 The OPT Imaging System
Our homemade OPT imaging system consists of a
Leica MZ16 FA stereomicroscope with a Plan 0.5×
and 135mm working distance objective lens (Leica
10446157). Images are acquired by a 1360×1036
pixel Retiga Exi CCD with a well size of
6.45μm×6.45μm and saved as 12-bit tiff-files. A full
revolution results in a 1.13Gb tiff-file. The
acquisition is accomplished by a rotation of the
specimen driven by a stepper unit; the stepper moves
0.9°per step and it results in 400 images per
revolution. The OPT imaging system has two
modules: for bright-field imaging the specimen is
illuminated with a LED and for fluorescence imaging
a 100W mercury lamp is used in combination with a
filter-block (GFP1, Texas Red).
2.2 Sample Preparation of a Single
Fluorescence Sphere
To image the specimens in the range of several
millimeters small-valued NA (effective NA:
0.0105~0.0705) lens is used to obtain the large DOF
in our OPT imaging system. The resolution of an
optical system is defined as the minimum distance
at which two separate points can be distinguished as
individuals. According to the Rayleigh criterion
for a circular aperture where
is the emission wavelength, the minimum
size of the experimental fluorescence sphere is
supposed to be in the range between 4.40 and
29.57. To make it visible in the image the sphere
size is supposed to exceed this range. In our case, we
choose the green fluorescent protein (GFP) sphere of
size and diluted it to a concentration of 360
.
To image and model the PSF along optical axis
we have developed an injection-based protocol to
place the spheres into agarose as follows:
1% low melting point (LMP) agarose, cool down
to ~37;
Drill cylindrical agarose shapes when it is semi-
solidified in a petri dish;
Inject the diluted spheres into the outer wall of the
agarose along a line parallel to the central axis,
preferably with a small size syringe. In our case a
0.5 syringe is used with a needle length of
13mm and diameter of 0.29mm, as shown in
Figure 1;
Figure 1: The injection protocol with green spots indicating
the injection position where fluorescence spheres may
occur. The cylinder corresponds to the shape of the agarose.
Keep the agarose at 4 until it is fully solidified
(~3 hours);
Clear the sample with 70%, 80%, 90%, 96%,
100% ethanol, 100% ethanol: BABB (benzyl
alcohol: benzyl benzoate = 2: 1) = 1: 1 and BABA.
Our goal is to acquire the images of a single sphere
placed at different depths along optical axis.
Therefore, randomly sprinkling the spheres into the
agarose in a traditional way is not feasible. The main
reason is that there may be interactions and overlap
between different spheres either at the same or
different depths. This makes the extraction of the
single sphere image difficult or even impossible. The
images of each single sphere at different depths are
acquired by means of sample rotation. Each rotation
corresponds to a different depth in the OPT imaging
system. The straight-like sphere injection method in
of our protocol significantly reduces the probability
of overlapping between different spheres. In this way
the images of the same sphere in a full revolution can
be easily and efficiently acquired. The OPT imaging
system and environment is configured as explained in
(Tang et al., 2017).
2.3 PSF Modelling Concerning
Different Magnification
For our experiments a full revolution of 400 images
of the single GFP sphere are acquired. In Figure 2 the
processes of sphere image acquisition and PSF
modelling are presented. In Figure 2(a) and (b), the
green dot represents the sphere and the red arrow
indicates the sphere rotation. The excitation and
emission beams are regarded to be parallel,
demonstrated as blue and green beams in Figure 2(a).
For PSF modelling, the focal plane is set at the COR.
The 3D image whose focal plane is shifted from the
COR, requires an equal shift in the PSF. With the
protocol (cf. section 2.2) the physical rotation radius
of the sphere
can be easily measured. To this end,
we first measure the radius of the cylindrical agarose
and image it in the bright-field mode with small
3D Image Deblur using Point Spread Function Modelling for Optical Projection Tomography
69
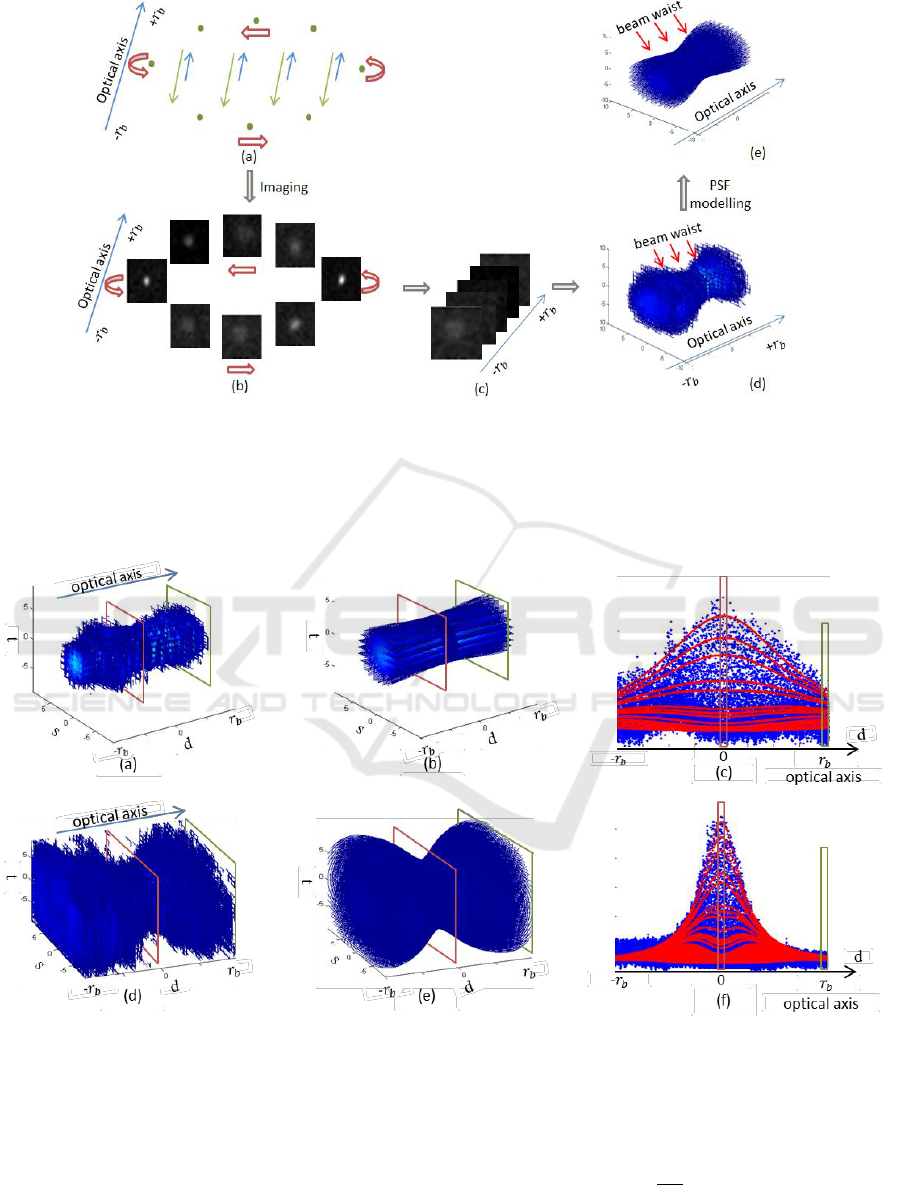
Figure 2: Image acquisition and PSF modelling of a single GFP sphere. (a) The light path of the OPT imaging system that
passes through GFP spheres (green dots). The excitation beams and emission beams are separately shown in blue and green
arrows. (b) Images of the single sphere acquired at different angles. (c) Images of the single sphere stacked according to the
defocus. Half rotation with defocus from
to
is required, in our experiment
as calculated from Eq. (1).
(d) The experimental and discrete PSF with defocus from
to
. (e) The modelled and continuous PSF with defocus
from
to
.
Figure 3: PSF modelling along the optical axis. (a), (d) Experimental PSFs acquired from images at magnification of 12.5
and 25.0. (b), (e) The corresponding modelled PSFs using Eq. (4) and Eq. (5). All the voxels of experimental data in (a)
and modelled data in (b) are respectively transformed to blue and red dots in 1D functional in (c) to visualize the modeling
performance. The vertical axis in (c) displays the intensity that corresponds to the voxel intensity in (a) and (b). Similarly,
voxels in (d) and (e) are transformed to the data in (f).
exposure time. In the same experimental
environment, the sphere is afterwards imaged in the
fluorescence mode.
is calculated by Eq. (1).
(1)
BIOIMAGING 2019 - 6th International Conference on Bioimaging
70
with
representing the rotation diameter of the
sphere in the tomogram, achieved by measuring the
distance of two opposite sphere centers that are both
in focal plane.
is the diameter of the cylindrical
agarose in the bright-field image. Dividing
by 100
rotations, the distance of each rotation along optical
axis is determined. In our case the measured
,
, producing
mm.
Therefore, the physical distance of two adjacent
rotations along optical axis is approximately.
According to convention the imaging PSF is
assumed as a focused Gaussian-like beam:
(2)
where is the beam waist (Figure 2)given by:
(3)
With
the Gaussian beam waist defined as the
value of the field amplitude in focus (van der Horst et
al., 2016), the emission wave length of fluorescence
spheres and the defocus along optical axis. For a
specific magnification,
is constant, but it varies
when imaging with different magnifications.
Additionally, in Eq. (2) and Eq. (3) the beam waist
is typically regarded as the standard deviation of
the Gaussian model in previous studies (van der Horst
et al., 2016). Different from the Gaussian model in
(van der Horst et al., 2016), we generalize the model
by employing parameter
,
and
as:
(4)
Instead of equalizing the beam waist and standard
deviation as described in (van der Horst et al., 2016)
and (Kogelnik et al., 1966), we investigate the
relationship between them by multiplying a
parameter with beam waist, considering different
magnifications.
(5)
To relate the beam waist in focus
as well as the
parameter a to the magnification, 6 magnifications
i.e. 12.5, 15.0, 17.5, 20.0, 22.5, 25.0,
are configured to acquire the images of the same
sphere. The magnifications are obtained through
zooming. The magnification of 12.5 approximately
corresponds to the minimum magnification that
renders the sphere visible in our experiment, while
25.0 approximates to the maximum magnification
that confirms that a full revolution of the sphere
remains in the field of view (FOV). The PSF of each
magnification is modelled by creating an
optimization problem and solving it with least square
curve fitting. The overall fitting error of the 6
experimental PSFs is 5.00%. The experimental PSFs
acquired from images with magnification of 12.5
and 25.0 are shown in Figure 3 (a) and (d)
respectively. The color of the voxel indicates the
intensity of PSF response. (b) and (e) represent the
modelled PSFs of the two magnifications. Voxels in
3D space are converted to a 1D space with horizontal
axis approximating the optical axis and vertical axis
displaying the intensity. The 3D voxels on the slice in
(a) and (b) match the 1D points in the box in (c)
according to the same color. The experimental PSF
differentiation between two magnifications is evident
in (a) and (d). By transforming the 3D space to 1D
functional, we can intuitively visualize the
distribution of the experimental PSF (blue dots) and
the modelled PSF (red dots), as well as showing the
differences between them.
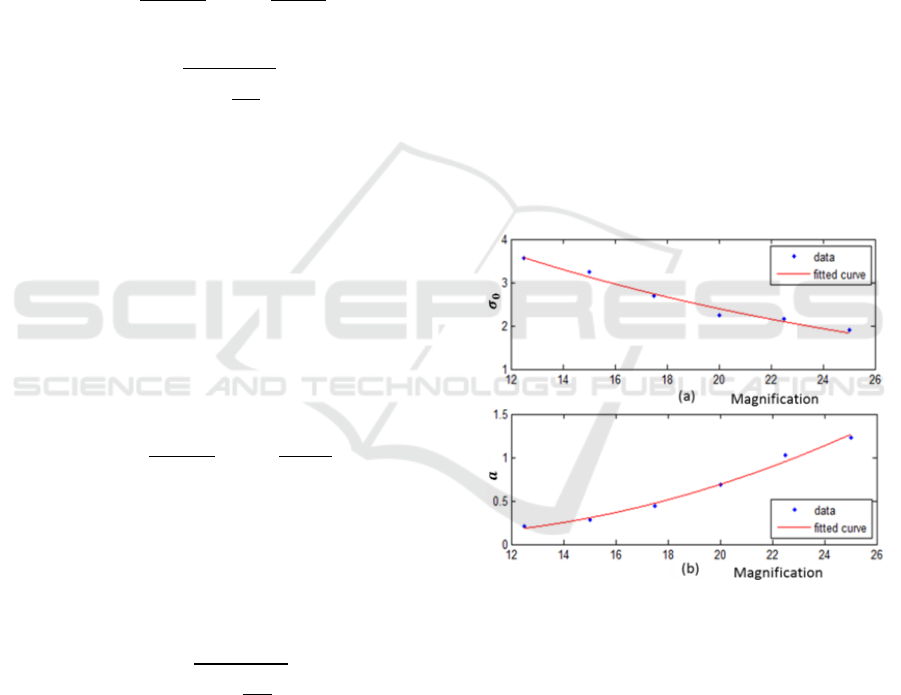
Figure 4: Fitting of
and the parameter a estimated from
6 magnifications.
is fitted by exponential function as
shown in (a) while a is fitted by quadratic function in (b).
The parameters for our modelling
and
have proved to be constant regardless of
magnifications:
,
and
. The beam waist
and parameter a
relevant to the 6 magnifications are estimated as
depicted in Figure 4. To minimize the fitting error on
the observed data, the parameters are fitted as an
exponential and a quadratic function respectively by
using Eq. (6) and Eq. (7), with representing
magnification and
to
being the parameters.
3D Image Deblur using Point Spread Function Modelling for Optical Projection Tomography
71
(6)
(7)
The PSF of any 3D image between and
along optical axis can be modelled as depicted in
section 2.3. The modeling is implemented with the
focal plane set at the COR. However, in most imaging
cases the focal plane is not in line with the COR.
Consequently, the modelled PSF will be shifted along
the optical axis by the same shift as the focal plane.
Besides, the length of the PSF along optical axis is
determined by the size of 3D image and the
resolution, because in FBP 3D reconstruction each
voxel in the 3D image corresponds to each pixel in
the 2D images. The NA is the effective value
achieved from interpolation relating to the
magnification. The relationship between effective
NA and magnification can be found in the product
manual of the Leica objective lens.
2.4 Deconvolution of 3D Images in
Coronal Plane
The modelled PSF consists of multiple 2D Gaussian
patterns along optical axis. Therefore, the 3D image
can be deconvolved slice by slice along its depth axis
that is parallel to the optical axis. As the slices are
coronal sections, the deconvolution is implemented
on the 3D image in the coronal plane as follows:
(8)
R is the reconstructed 3D image with the depth axis d
parallel to the optical axis of the PSF. stands for
the operation of deconvolution. Considering the
shifted focal plane and the reconstruction symmetry,
deconvolution of
, the opposite view of R projected
along d, is executed by applying Eq. (9).
(9)
The transform from to
is conducted by a matrix
rotation of centered by the COR. The 3D image
with the deconvolution is then achieved by combining
and back rotation of
.
3 EXPERIMENTS
3.1 Image Comparison of
Deconvolution
With respect to the magnifications, the experiments
were conducted on images at 2 different
magnifications. One is a zebra finch embryo in
fluorescence mode with magnification 13.83 and
focal plane shifted by. Considering the
resolution limit and the 3D image size, the calculated
defocus of the PSF along the optical axis ranges
from to. The deconvolution is
performed using Lucy-Richardson algorithm with 10
iterations. The result for the coronal slice is shown in
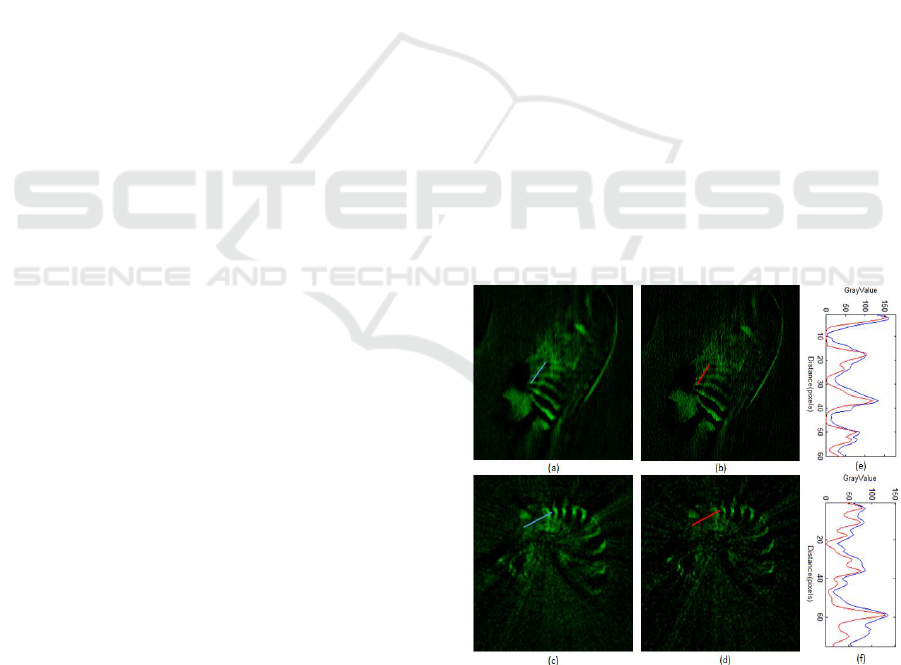
Figure 5 (b) and for the horizontal slice in Figure 5
(d). The corresponding slices without deconvolution
are displayed in Figure 5 (a) and (c). The comparisons
of intensity profile along a line with (red) and without
(blue) deconvolution are presented in (e) and (f)
respectively. In Figure 6 another sample is depicted.
This is a sample from a 3dpf batch of zebrafish larvae.
The magnification and the shifted focal plane are
separately 49.98 and -0.5mm, with the computed
defocus of PSF as between
and. Figure 6 (a) and (b) are the slices
before deconvolution in two orthogonal planes, while
(c) and (d) corresponds to the deconvolution results.
By visually comparing (c) and (d) we conclude that
the performance in horizontal plane is as good as it is
in coronal plane. This means that deconvolution in the
coronal plane simultaneously improves the quality of
the image in the horizontal plane. From a comparison
of the quantitative intensity profile for each colored
line, we state that the proposed deconvolution
sharpens and refines the 3D reconstructed images. It
enhances the strong signals and makes the intensity
profile more distinct.
Figure 5: Deconvolution results. (a) The coronal slice of the
3D zebra finch with obvious blur around the ribs. (b)
Distinct texture appears around the ribs after the
deconvolution. (e) The comparison of intensity profiles
along a line in (a) and (b). (c) and (d) The horizontal slice
comparisons with the line intensity profiles shown in (f).
BIOIMAGING 2019 - 6th International Conference on Bioimaging
72
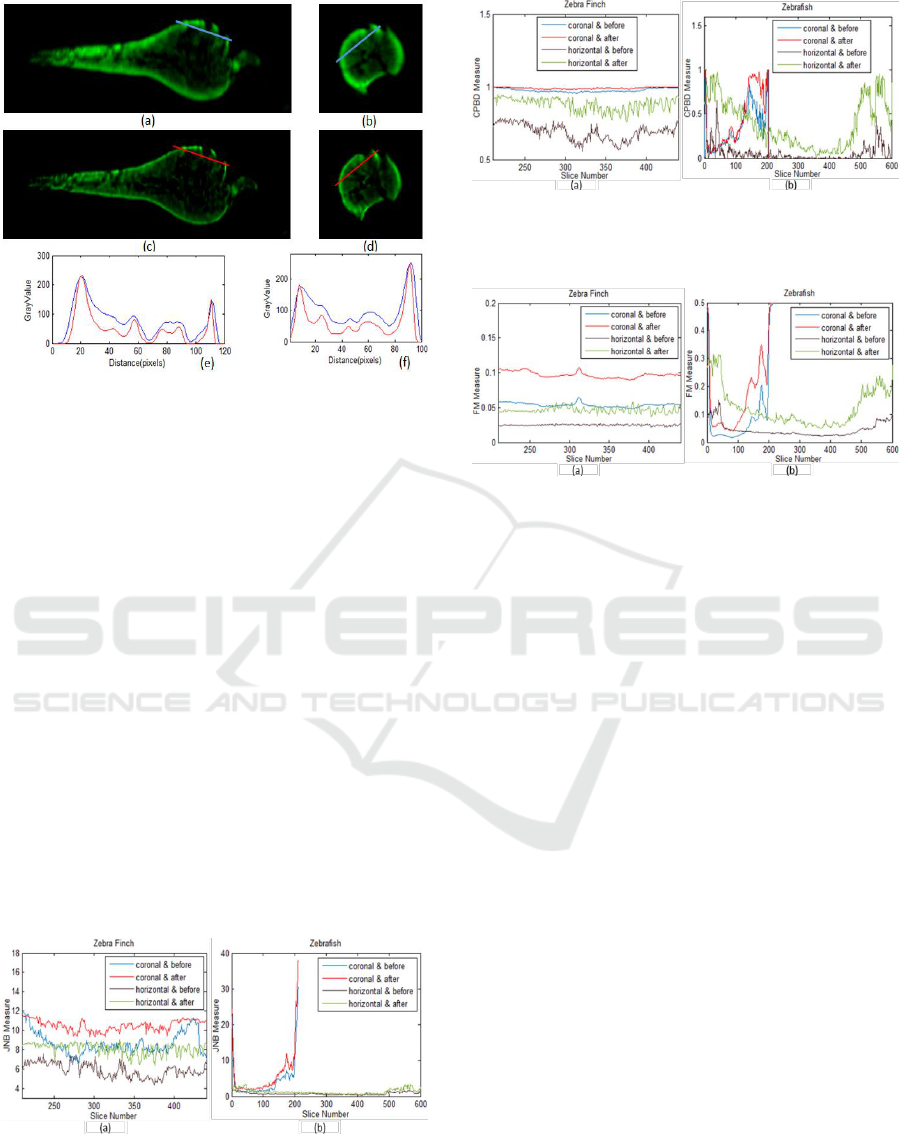
Figure 6: Coronal and horizontal slices of 3D zebrafish
before ((a) and (b)) and after ((c) and (d)) deconvolution.
The deconvolution highlights the strong signals and makes
the texture more visible. The figure below (a) and (c)
compares the intensity profile of the same line before
(labelled as blue) and after (labelled as red) deconvolution,
so does the figure below (b) and (d).
3.2 Image Blur Measurement on Slices
To quantify the image blur of each slice, 3 metrics
from literatures, i.e. the just noticeable blur (JNB)
measure (Ferzli et al., 2009), the cumulative
probability of blur detection (CPBD) measure
(Narvekar et al., 2009) and the frequency measure
(FM) (De et al., 2013) are employed to evaluate the
performance. Both the JNB and CPBD measure
acquire sharpness metric by detecting and quantifying
the blur in the spatial domain. Different from JNB and
CPBD, the FM measure quantifies the sharpness in
the frequency domain with an easier and more
efficient approach. All the three metrics characterize
the sharpness of an image, so the measure increases
at improved image quality.
Figure 7: (a) JNB measure on the zebra finch data with
magnification 13.83. (b) JNB Measure on the zebrafish
data with magnification 49.98. Coronal and horizontal
are the two orthogonal planes displaying the 3D image.
Figure 8: (a) CPBD measure on the zebra finch with
magnification 13.83. (b) CPBD Measure on the zebrafish
with magnification 49.98.
Figure 9: The FM before and after deconvolution on the two
3D image data. (a) FM measure on the zebra finch with
magnification 13.83. (b) FM Measure on the zebrafish
with magnification 49.98.
While experiments in section 3.1 give us a
qualitative comparison between the deconvolved
slice and non-deconvolved slice, in this section we
quantitatively look into all the slices in different
orthogonal planes with the three image sharpness
measurements (i.e. JNB, CPBD and FM). From
Figure 7 to Figure 9 we can see that, no matter what
measure is used, the deconvolved slices in both planes
have higher measurement values compared with the
image slices without deconvolution. This means that
on all slices, the deconvolution deblurs the images
and significantly improves the image quality.
3.3 Quantitative 3D Image Quality
Improvement of Deblur
To further quantify the deblur of the deconvolution
results on the original reconstructed 3D data across
the planes, we present the 3D image quality
improvement criterion of deblur as
in Eq. (10).
Improvement in three orthogonal individuals are
combined and encoded as a whole and each of them
are represented as Eq. (11) to Eq. (13).
3D Image Deblur using Point Spread Function Modelling for Optical Projection Tomography
73

Table 1: 3D image quality improvement of 10 zebrafish
embryos based on JNB Measure.
01
02
03
04
05
06
07
08
09
10
G
0.16
0.21
0.15
0.21
0.29
0.24
0.22
0.16
0.20
0.25
PSF
m
1.35
2.10
1.41
3.23
1.54
1.50
1.50
1.37
1.45
1.70
G
-- Gaussian-based blind deconvolution. PSF
m
-- PSF based modelling
deconvolution. 10 zebrafish embryos correspond to 01-10 with age ranging
from 3dfp to 7dfp. Magnification for 01-05 is 24.98 while for 06-10 is
22.45.
Table 2: 3D image quality improvement of 6 zebrafish brain
based on JNB Measure.
01
02
03
04
05
06
G
0.25
0.29
0.26
0.24
0.01
0.21
PSF
m
0.41
0.49
2.55
0.17
1.15
1.20
6 zebrafish embryo brains correspond to 01-06 with age ranging from
6dfp to 7dfp. Magnification for 01-02 is 20.98 while for 03-06 is
15.98.
Table 3: 3D image quality improvement of 7 chicken heart
based on JNB Measure.
01
02
03
04
05
06
07
G
0.24
0.16
0.19
0.26
0.27
0.18
0.32
PSF
m
0.93
0.49
0.29
1.15
0.49
0.38
1.07
7 chicken embryo hearts at different stages correspond to 01-07.
Magnification for 01 is 15.00 ,while for 03-06 is 11.75 and 07 is
10.00.
(10)
(11)
(12)
(13)
Where
and
are the ith deconvolved and
original reconstructed slice in x plane respectively.
By employing the image quality improvement of
deblur
, deconvolution performance of two
different methods on the same data turns to be
comparable.
We here applies our deconvolution method to 23
more 3D data, which contains 3 categories of samples
i.e. zebrafish embryo, zebrafish embryo brain and
chicken embryo heart. They are in different stages
and are acquired at different magnifications. It is
important to know that all the measurements in this
paper cannot assess the image blur across different
data, but it provides us with a comparative evaluation
of image deblur on the same data. Taking advantage
of this, we compare the presented deconvolution
method with the most commonly used Gaussian-
based blind deconvolution (Chan et al., (1998)). The
Gaussian kernel size is set to 7 for all slices. As the
most robust measurement among CPBD, JNB and
FM, JNB is employed to evaluate the image blur of
each slice. The results of the 3 categories of samples
are presented in Table 1 to Table 3. For all the 23 data,
our deconvolution approach outperforms the
Gaussian-based deconvolution, thereby indicating the
success of the method.
4 CONCLUSIONS
In this paper we have focused on 3D image deblur and
quality improvement, under the condition of the
limitation of small NA for imaging of large samples.
We investigated and modeled the PSF along the
optical axis, exploring the influence of magnification
on PSF. The sample of a single GFP sphere is
prepared with the protocol in section 2.2. The
experimental PSF is then modelled to deconvolve the
3D image in a coronal plane. A number of measures
for image blur are employed to convincingly evaluate
the performance of the deconvolution. They provide
quantitative information about how much
improvement is achieved. The overall
improvement
gives us a criterion to compare
image quality improvement regardless of different
data. All the experimental results including the image
comparisons and quantitative measures sustain the
effectiveness of the proposed PSF modelling and
deconvolution methodology.
The deconvolution results presented in this paper
represent a proof of concept. The datasets used in the
experiments are composed of 25 samples (i.e. 4
categories: zebrafish embryo, zebra finch embryo,
zebrafish brain and chicken heart). Regarding the
evaluation of performance on a large scale of dataset,
our data are far from perfect in terms of ‘large
dataset’. However, it presents a clear idea that our
model is not constrained by several samples, it also
works on many other types of objects or images. This
will help to explain its potential capability of
improving image quality on more 3D data, including
those from other OPT imaging systems, which is a
part of our current work. In the future we will take
BIOIMAGING 2019 - 6th International Conference on Bioimaging
74

more effort on further generalizing the model to other
imaging set-ups. In addition, the fluorescent sphere
used in the experiments is fix-sized. The effect of
sphere size on PSF modelling and deblur performance
will be investigated afterwards.
REFERENCES
Sharpe, J. et al., 2002. Optical projection tomography as a
tool for 3D microscopy and gene expression studies.
Science (80-. ), vol. 296, no. 5567, pp. 541–545, 2002.
Walls, J. R., Sled, J. G., Sharpe, J. and Henkelman, R. M.,
2007. Resolution improvement in emission optical
projection tomography. Phys. Med. Biol., vol. 52, no.
10, p. 2775.
McNally, J. G., Karpova,T., Cooper, J. and Conchello, J.
A., 1999. Three-dimensional imaging by deconvolution
microscopy. Methods, vol. 19, no. 3, pp. 373–385.
Kak, A. C. and Slaney, M., 2001. Principles of
computerized tomographic imaging. SIAM.
Chen, L. et al., 2012. Incorporation of an experimentally
determined MTF for spatial frequency filtering and
deconvolution during optical projection tomography
reconstruction. Opt. Express.
Chen, L., Andrews, N., Kumar, S., Frankel, P., McGinty, J.
and French, P. M. W., 2013. Simultaneous angular
multiplexing optical projection tomography at shifted
focal planes. Opt. Lett., vol. 38, no. 6.
Miao, Q., Hayenga, J., Meyer, M. G., Neumann, T., Nelson,
A. C. and Seibel, E. J., 2010. Resolution improvement
in optical projection tomography by the focal scanning
method. Opt. Lett., vol. 35, no. 20, pp. 3363–5.
Xia, W., Lewitt, R. M. and Edholm, P. R., 1995. Fourier
correction for spatially variant collimator blurring in
SPECT. IEEE Trans. Med. Imaging, vol. 14, no. 1, pp.
100–115.
Darrell, H. Meyer, K. Marias, M. Brady, and J. Ripoll.,
2008. Weighted filtered backprojection for quantitative
fluorescence optical projection tomography. Phys.
Med. Biol., vol. 53, no. 14, pp. 3863–81.
McErlean, C. M., Bräuer-Krisch, E., Adamovics,J. and
Doran, S. J., 2016. Assessment of optical CT as a future
QA tool for synchrotron x-ray microbeam therapy.
Phys. Med. Biol., vol. 61, no. 1, pp. 320–37.
van der Horst, J., and Kalkman, J., 2016. Image resolution
and deconvolution in optical tomography. Opt.
Express, vol. 24, no. 21, pp. 24460–24472.
Tang, X., van der Zwaan, D., Zammit,M., Rietveld, A., K.
F. D. and Verbeek, F. J., 2017. Fast Post-Processing
Pipeline for Optical Projection Tomography. IEEE
Trans. Nanobioscience, vol. 16, no. 5, pp. 367–374, Jul.
Kogelnik, H. and Li, T., 1966. Laser Beams and
Resonators. Proc. IEEE, vol. 54, no. 10, pp. 1312–
1329.
Ferzli, R. and Karam, L. J., 2009. A no-reference objective
image sharpness metric based on the notion of Just
Noticeable Blur (JNB). IEEE Trans. Image Process.,
vol. 18, no. 4, pp. 717–728.
Narvekar, N. D. and Jaram, L. J., 2009. An improved no-
reference sharpness metric based on the probability of
blur detection. IEEE Int. Work. Qual. Multimed. Exp.,
vol. 20, no. 1, pp. 1–5.
De, K. and Masilamani, V., 2013. Image Sharpness
Measure for Blurred Images in Frequency Domain.
Procedia Eng., vol. 64, no. Complete, pp. 149–158.
Chan, T.F. and Wong, C.K., 1998. Total variation blind
deconvolution. IEEE Transactions on Image
Processing, vol. 7, no. 3, pp. 370—375.
3D Image Deblur using Point Spread Function Modelling for Optical Projection Tomography
75
