
Features Selection and k-NN Parameters Optimization based on
Genetic Algorithm for Medical Datasets Classification
Rizki Tri Prasetio
1
, Ali Akbar Rismayadi
1
, Nana Suryana
2
, Rochmanijar Setiady
2
1
Department of Computer Science, Universitas BSI, Bandung, Indonesia
2
Department of Computer Science, Universitas Kebangsaan, Bandung, Indonesia
Keywords: Classification, Genetic Algorithm, Features Selection, Parameters Optimization, k-Nearest Neighbors
Abstract: Medical dataset classification is a major data mining problem being researched about for a decade. Most
classifiers are designed to learn from the data itself through training process, because expert knowledge to
determine classifier parameters is difficult. This research proposes a methodology based on data mining
paradigm. This paradigm integrates the search heuristic that is inspired by natural evolution called genetic
algorithm with the simplest and the most used learning algorithm, k-nearest Neighbors. The genetic algorithm
is used for feature selection and parameter optimization while k-nearest Neighbors is used as a classifier. The
proposed method is experimented on five medical datasets of the UCI Machine Learning Repository and
compared with original k-NN and other feature selection algorithm i.e., forward selection, backward
elimination and greedy feature selection. Experiment results show that the proposed method is able to achieve
good performance with significant improvement with p value of t-Test is 0.0011.
1 INTRODUCTION
Recently, the application of machine learning in
medical diagnosis is a major trend for medical data
applications. Most of the diagnosis techniques in
medical field are systematized as intelligent data
classification approaches (Subbulakhsmi & Deepa,
2015). Use of Computer-Aided Diagnosis (CAD)
systems can assist doctors to diagnose patient
illnesses (Unal & Kocer, 2013), among the various
assignments performed by a CAD system,
classification is most common. Medical dataset
classification problem may be categorized as a class
of complex optimization problem with an objective to
guarantee the diagnosis aid accurately (Subbulakhsmi
& Deepa, 2015).
Various researchers have attempted to apply
numerous techniques to improve the accuracy of data
classification for identify the potential patients (Babu
& Suresh, 2013). In the recent studies, metaheuristic
algorithms such as particle swarm optimizations
(Subbulakhsmi & Deepa, 2015) (Inbarani, et al.,
2014) (Chang, et al., 2012) or genetic algorithms
(Raymer, et al., 2000) (Yang & Honavar, 1998) (Shah
& Kusiak, 2007) and also data mining techniques
such as neural networks (Mazurowski, et al., 2008)
(Brameier & Banzhaf, 2001) (Amato, et al., 2013) or
k-nearest neighbors (Prasetio & Pratiwi, 2015)
(Suguna & Thanushkodi, 2010) (Jabbar, et al., 2013)
were applied for classification of medical data and
obtained with remarkably meaningful results.
k-Nearest Neighbors (k-NN) algorithm is a
method that uses a supervised Algorithm (Wu, et al.,
2008). Which is a classification technique that is easy
to understand and implement (Wu & Kumar, 2009)
and simplest amongst of all machine learning
algorithms (Gorunescu, 2011). k-NN method
represents the technique to classify an object based on
the closest (k) objects in its neighborhood
(Harrington, 2012). k-NN is particularly well-suited
for multimodal classes as well as applications in
which an object can have many class labels (Wu &
Kumar, 2009).
There are several key issues that affect the
performance of k-NN. One is the choice of k (Wu &
Kumar, 2009). If k is too small, then the result can be
sensitive to noise points that may lead the algorithm
toward overfitting (Larose, 2005). On the other hand,
if k is too large, then the neighborhood may include
too many points from other classes (Wu, et al., 2008).
The best choice of k depends upon the data
(Gorunescu, 2011).
Another key issue is the accuracy of the k-NN
algorithm can be severely degraded by the presence
of noisy or irrelevant features (Han, et al., 2012), or if
3080
Tri Prasetio, R., Akbar Rismayadi, A., Suryana, N. and Setiady, R.
Features Selection and k-NN Parameters Optimization based on Genetic Algorithm for Medical Datasets Classification.
DOI: 10.5220/0009947130803086
In Proceedings of the 1st International Conference on Recent Innovations (ICRI 2018), pages 3080-3086
ISBN: 978-989-758-458-9
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

the feature scales are not consistent with their
importance (Gorunescu, 2011).
The medical data involved in diagnostic models
are usually high dimensional. High-dimensional
datasets increase the complexity of classification and
reduce the effect of models (Bharti & Singh, 2014), a
serious obstacle to the efficiency of most data mining
algorithms. This obstacle is sometimes known as the
“curse of dimensionality” (Maimon & Rokach,
2010). It is necessary to reduce the data dimension
while retaining essential information. Feature
extraction (Liu, et al., 2015) and feature selection
(Jirapech-Umpai & Aitken, 2005) are the main
methods in dimensionality reduction. The data
mining process requires high computational cost
when dealing with large data sets. Reducing
dimensionality can effectively cut this cost (Maimon
& Rokach, 2010), reduces time and memory
(Shilaskar & Ghatol, 2013).
The main purpose of feature selection is to reduce
the number of features used in classification while
maintaining an acceptable classification accuracy
(Raymer, et al., 2000). The selection of features can
have a considerable impact on the effectiveness of the
resulting classification algorithm (Jain & Zongker,
1997), in some cases, as a result of feature selection,
accuracy on future classification can be improved
(Maimon & Rokach, 2010).
The problem of feature selection is defined as
given a set of candidate features and optimized a
subset that performs the best under some
classification system (Jain & Zongker, 1997). Genetic
algorithms are often used to perform optimization.
Genetic algorithms have less of a tendency to become
stuck in local minima (Gorunescu, 2011), and more
sophisticated optimization (Witten, et al., 2011).
Genetic algorithms are a wide range of
optimization depending on the objective function
(fitness) (Prasetio & Riana, 2015), easily
parallelizable and have been used for classification as
well as other optimization problems. In data mining,
they may be used to evaluate the fitness of other
algorithms (Han, et al., 2012).
In this research, we integrates genetic algorithm
for features selection and parameters optimized k-NN
applies to classify five benchmarked medical
datasets, namely, Wisconsin breast cancer diagnostic
and prognostic (Mangasarian, et al., 1995), diabetic
retinopathy Debrecen (Antal & Hajdu, 2014),
cardiotocography (Ayres-de-campos, et al., 2000)
and SPECTF image of heart disease (Kurgan, et al.,
2001). Main objectives of this research are to improve
accuracy of five benchmarked medical datasets
classification by applying genetic algorithm as
feature selection and to improve performance of k-
NN classifier algorithm by optimizing k value using
genetic algorithm.
2 DATASETS
This research is experimented on five medical
datasets obtained from UCI Machine Learning
(https://archive.ics.uci.edu/ml/datasets.html). The
details of these medical datasets is listed in Table 1
that contains number of instances, features and
classes. The training and testing datasets are
randomly generated.
Table 1: Description of datasets
Dataset
Number
of
instances
Number
of
features
Number
of
classes
Wisconsin Breast Cancer
(Diagnostic)
569 32 2
Wisconsin Breast Cancer
(Prognostic)
198 34 2
Diabetic Retinopathy
Debrecen
1151 20 2
Cardiotocography (CTGs) 2126 23 3
Heart Disease (SPECTF) 267 44 2
Wisconsin Breast Cancer (Diagnostic), the dataset
is available at the University of Wisconsin. It contains
569 instances with 32 features which are used to
predict benign or malignant growths (Mangasarian, et
al., 1995).s
Wisconsin Breast Cancer (Prognostic), the dataset
is obtained from University of Wisconsin. There are
198 instances with 20 features which are used to
predict recurrent and nonrecurrent (Mangasarian, et
al., 1995).
Diabetic Retinopathy, this dataset was collected
from University of Debrecen and contains about 1151
instances with 20 features which are used to predict
whether it is contain diabetic retinopathy or not
(Antal & Hajdu, 2014).
Cardiotocography (CTGs), this dataset was
created by Diogo Ayres-de-campos at the University
of Porto. It contains 2126 instances with 23 features
which are used to predict fetal state (Ayres-de-
campos, et al., 2000).
Heart Disease (SPECTF), this dataset is based on
data from University of Colorado. It contains 45
features with 267 instances which are used to identify
whether patients are normal or not (Kurgan, et al.,
2001).
Features Selection and k-NN Parameters Optimization based on Genetic Algorithm for Medical Datasets Classification
3081

3 METHODS
This research proposes a methodology based on data
mining paradigm. This paradigm integrates the search
heuristic that is inspired by natural evolution called
genetic algorithm with the simplest and the most used
learning algorithm, k-nearest Neighbors.
3.1 Genetic Algorithm
Genetic algorithm (GA) is a stochastic, parallel,
heuristic search algorithm inspired by basic principle
of natural selection introduced by Charles Darwin
(Nowe, 2014). The basic principles of GA were first
proposed by Holland (Holland, 1975). Genetic
algorithms (GAs) attempt to mimic computationally
the processes by which natural selection a biological
process in which stronger individuals are likely be the
winners in a competing environment (Man, et al.,
1996) operates and apply them to solve business and
research problems (Larose, 2006).
Genetic algorithms provide a framework for
studying the effects of such biologically inspired
factors as mate selection, reproduction, mutation, and
crossover of genetic information. Three operators are
used by genetic algorithms: (Gorunescu, 2011)
1. Selection. The selection operator refers to the
method used for selecting which chromosomes
will be reproducing. The fitness function evaluates
each of the chromosomes (candidate solutions),
and the fitter the chromosome, the more likely it
will be selected to reproduce.
2. Crossover. The crossover operator performs
recombination, creating two new offspring by
randomly selecting a locus and exchanging
subsequences to the left and right of that locus
between two chromosomes chosen during
selection. For example, in binary representation,
two strings 11111111 and 00000000 could be
crossed over at the sixth locus in each to generate
the two new offspring 11111000 and 00000111.
3. Mutation. The mutation operator randomly
changes the bits or digits at a particular locus in a
chromosome: usually, however, with very small
probability. For example, after crossover, the
11111000 child string could be mutated at locus
two to become 10111000. Mutation introduces
new information to the genetic pool and protects
against converging too quickly to a local
optimum.
Algorithm 1: Genetic Algorithms
Begin
INITIALISE population with random candidate
solutions;
EVALUATE each candidate;
REPEAT UNTIL (termination condition is
satisfied) DO
1. SELECT parents;
2. RECOMBINE pairs of parents;
3. MUTATE the resulting offspring;
4. EVALUATE new candidates;
5. SELECT individuals for the next generation;
End
3.2 k-Nearest Neighbours
k-Nearest Neighbors (k-NN) algorithm is a method
that uses a supervised Algorithm (Wu, et al., 2008).
Which is simplest (Gorunescu, 2011), often used for
classification, although it can also be used for
estimation and prediction. k-nearest neighbors is an
example of instance-based learning, in which the
training data set is stored, so that a classification for a
new unclassified record may be found simply by
comparing it to the most similar records in the
training set (Larose, 2005).
k-NN method represents the technique to classify
an object based on the closest (k) objects in its
neighborhood (Gorunescu, 2011) and bases the
assignment of a label on the predominance of a
particular class in this neighborhood (Wu & Kumar,
2009). We look at the top k most similar pieces of data
from our known dataset; this is where the k comes
from (Harrington, 2012). To conclude, the k-nearest
neighbor algorithm is amongst the simplest of all
machine learning algorithms, since it simply consists
in classifying an object by the majority vote of its
neighbors (Gorunescu, 2011) from the k most similar
pieces of data (Harrington, 2012).
“Closeness” between object with its neighborhood
is defined in terms of a distance metric, such as
Euclidean distance or Manhattan distance (Han, et al.,
2012). To construct the algorithm, we need the
following items (algorithm input):
1. A set of labeled stored records (Gorunescu, 2011)
to be used for evaluating a test object’s class (Wu
& Kumar, 2009) (training dataset);
2. A distance (metric) to compute the similarity
between objects (Gorunescu, 2011) that can be
used to compute the closeness of objects (Wu &
Kumar, 2009);
3. The value of k, the number of nearest neighbors
(Wu & Kumar, 2009) (records) belonging to the
training dataset, based on which we will achieve
the classification of a new object (Gorunescu,
2011);
ICRI 2018 - International Conference Recent Innovation
3082
4. The method used to determine the class of the
target object based on the classes and distances of
the k nearest neighbors (Wu & Kumar, 2009).
Based on these three requirements, a new (not yet
classified) object will be classified by performing the
following steps: (Gorunescu, 2011)
1. Compute the distance (similarity) between all the
training records and the new object (naive
approach);
2. Identify the k nearest objects (most similar k
neighbors), by ordering the training objects taking
into account the computed distances in the first
step;
3. Assign the label which is most frequent among the
k training records nearest to that object (majority
voting).
Algorithm 2: Basic k-NN Algorithm
Input: D, the set of training objects, the test object, z,
which is a vector of attribute values, and L, the set of
classes used to label the objects
Output: c
z
∈ L, the class of z
foreach object y ∈ D do
| Compute d(z, y), the distance between z and y;
end
Select N ⊆ D, the set (neighborhood) of k closest
training objects for z;
c
z
= argmax ∑
y
∈
N
I(v = class(c
y
));
where I is an indicator function that returns the value 1 if
its argument is true and 0 otherwise
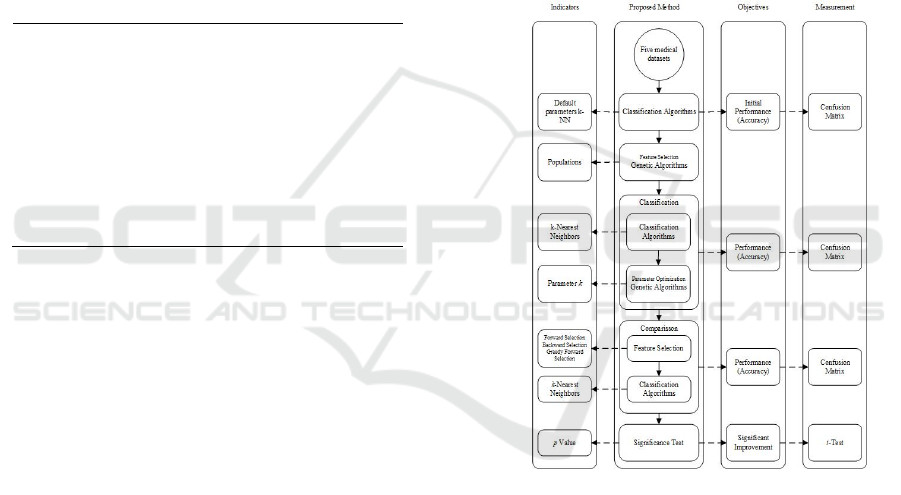
3.3. Proposed Method
The proposed method integrates genetic algorithm for
features selection and parameters optimized k-NN
applies to classify five benchmarked medical dataset
that explained in Table 1. The proposed method can
be seen in Figure 1. An early data processing begins
by dividing five datasets into training and testing data
using split validation, respectively. k-NN with default
parameters is applied for each training data to results
initial performance.
Genetic algorithms are applied for each training
data for features selection. Features selection is used
to find the features that best represents the class on
that dataset. Parameters optimized k-NN then applied
to training data that has been feature selected. After
that, validate the models which are produced by k-
NN, calculate how much accuracy generated by the
model tested in testing data. If the desired accuracy
has not been reached, repeat the process of feature
selection using genetic algorithms. This iteration will
continue until optimal feature is resulted.
The results obtained from the performance of the
features selection by genetic algorithm are then
compared with other algorithms that can be used for
feature selection i.e., backward elimination (Guyon &
Elisseeff, 2003) (Abe, 2005) (Derksen & Keselman,
1992), forward selection (Blanchet, et al., 2008)
(Abe, 2010) (Jain & Zongker, 1997) and greedy
feature selection (Dyer, et al., 2013) (Vafaie & Imam,
1994) (Farahat, et al., 2013). This comparison is to
determine whether performance of genetic algorithms
is better than any other algorithms in performing
feature selection.
The results obtained from proposed method then
tested with results obtained from k-NN with default
parameters to determine whether the proposed
method performance results improved the accuracy of
the five datasets significantly using a t-test (Prasetio
& Pratiwi, 2015) (Setiyorini & Wahono, 2015)
(Prasetio & Riana, 2015) significance test.
Figure 1. Proposed Method
4 RESULT AND DISCUSSION
This research conducted several experiments,
experiments using the k-NN algorithm with
unoptimized parameters of the five unselected
features datasets, experiments using the k-NN
algorithm with optimized parameters of the five
datasets in Table 1 that have not been selected
features dan experiments using the k-NN algorithm
with optimized parameters of five datasets in Table 1
which have been selected feature using genetic
algorithm, backward elimination, forward selection
dan greedy feature selection.
Features Selection and k-NN Parameters Optimization based on Genetic Algorithm for Medical Datasets Classification
3083
All experiments use split validation to split the
datasets randomly. The experiment using default
parameters configuration for genetic algorithm,
backward elimination, forward selection and greedy
forward selection.
Table 2: Experiment Results of Proposed Method and k-
NN
Datasets
Proposed
Method
k-NN
breast-cancer (D) 99.2.% 94.15%
breast-cancer (P) 86.44% 78.75%
diabetic-retinopathy 71.69% 61.16%
cardiotocography 98.59% 90.91%
heart (SPECTF) 87.5% 77.5%
The experimental results set forth in Table 2 stated
that the proposed method can improve the accuracy
of the five benchmarked datasets with a 5% - 10%
increase in comparison with the k-NN algorithm
without optimization and features selection.
Highest improved performance was obtained
from the classification of the Diabetic Retinopathy
dataset with an increase of 10.53% of 61.16% with
the most optimal k is 86. Meanwhile, the lowest
improved performance was obtained from the
classification of Breast Cancer Diagnostic dataset
with only 5.05% increase from 94.15% with the
optimal k is 8.
Improved performance on Breast Cancer
Prognostic dataset is 7.69% from 78.75% with
optimal k is 57, Cardiotocography datasets increased
by 7.68% from the original 90.91% with optimal k is
23 dan SPECTF Heart dataset increased by 10% from
77.5% with the most optimal k is 23.
Based on experiment results in this research, to
determine whether the proposed method can improve
performance in the classification of medical datasets
significantly. Testing using significance test was
done, t-Test Paired Two Sample for Means were used
in results between before and after using proposed
method.
The test results of t-Test generate that the
proposed method can improve the performance of k-
NN in terms of accuracy significantly in all datasets
marked with p value of t-Test < 0.05. t-Test results
can be seen in Table 4.
Table 3: Experiment Results of Featured Selection k-NN
Datasets
Proposed
Method
Forward
Selection
Backwar
d
Selection
Greed
y
Selecti
on
breast-
cancer (D)
99.2% 98.83% 97.08% 92.4%
breast-
cancer (P)
86.44% 84.75% 83.05%
79.66
%
diabetic-
retinopathy
71.69% 68.99% 69.28%
68.12
%
cardio-
tocography
98.59% 91.22% 92.63%
79.78
%
heart
(SPECTF)
87.5% 86.25% 85% 82.5%
The results of the experiments described in Table
3 stated that the proposed method is superior when
compared to other feature selection algorithms across
all benchmarked datasets. The results on backward
elimination and forward selection were slightly lower
0.37% - 5.96% when compared to genetic algorithm,
and the lowest results obtained by greedy feature
selection. Based on experiment results, to determine
whether feature selection can improve performance in
the classification of medical datasets significantly. t-
Test Paired Two Sample for Means were used in
results obtained from all features selection
algorithms.
Table 4: t-Test Results of Proposed Method compared
with k-NN
Proposed
Method
Normal
Mean 88.684 80.494
Variance 125.98713 170.19713
Observations 5 5
Pearson
Correlation
0.995007081
df 4
t Stat 8.376046049
P(T<=t) one-tail 0.000555628
t Critical one-tail 2.131846786
P(T<=t) two-tail 0.001111256
t Critical two-tail 2.776445105
The test results of t-Test generate that the feature
selection can improve the performance of k-NN in
terms of accuracy significantly in all datasets except
greedy feature selection marked with p value of t-Test
< 0.05. t-Test results for significance of using features
selection can be seen in Table 5.
Table 5: t-Test Results of Featured Selection Classifier
compared with k-NN
Algorithms
p Value of
t-Test
Results
Genetic Algorithms 0.0011 Sig. (p<0.05)
Forward Selection 0.02 Sig. (p<0.05)
Backward Elimination 0.01 Sig. (p<0.05)
Greedy Feature
Selection
0.99 Not Sig. (p>0.05)
k-nearest neighbors algorithm is easy to
implement (Gorunescu, 2011) and high accuracy
(Harrington, 2012) for a variety of applications
because excess k-NN is considered comparable to the
ICRI 2018 - International Conference Recent Innovation
3084

much more complex algorithms such as neural
networks or support vector machine. From the results
of this research, it can be concluded that parameter
optimized k-NN combine with genetic algorithms as
feature selection is superior when compared to other
feature selection algorithms on five benchmarked
medical datasets.
5 CONCLUSIONS
Genetic algorithms are applied to select features and
optimizing k parameter for k-nearest neighbors to
improve accuracy of five benchmarked medical
datasets. Proposed method is proven effective to be
able improve accuracy, and furthermore the different
test results among five datasets produce significant
difference.
Comparison of the feature selection algorithms are
proposed to compare the accuracy of the results
among genetic algorithms, forward selection,
backward elimination and greedy feature selection.
Genetic algorithms are proven to have the highest
accuracy compared with any others feature selection
algorithms.
In this research, in general, genetic algorithms
applied to select features and optimizing parameters
to improve accuracy of five benchmarked medical
datasets. In further research, some things can be
applied to enhance the research, which uses other
algorithms for parameter optimizing or other methods
to reduce dimensionality of medical datasets.
ACKNOWLEDGEMENTS
This research is supported by The Ministries of
Research, Technology, And Higher Education of
Republic Indonesia
REFERENCES
Abe, S., 2005. Modified Backward Feature Selection by
Cross Validation. Bruges, European Symposium on
Artificial Neural Networks, pp. 163-168.
Abe, S., 2010. Support Vector Machine for Pattern
Classification. Second Edition ed. New York: Springer
London.
Amato, F. et al., 2013. Artificial neural networks in medical
diagnosis. Journal of Applied Biomedicine, 11(2), pp.
47-58.
Antal, B. & Hajdu, A., 2014. An ensemble-based system
for automatic screening of diabetic retinopathy.
Knowledge-Based Systems, Volume 60, pp. 20-27.
Ayres-de-campos, D. et al., 2000. SisPorto 2.0: A Program
for Automated Analysis of Cardiotocograms. The
Journal of Maternal-Fetal Medicine, Volume 9, pp.
311-318.
Babu, G. S. & Suresh, S., 2013. Meta-cognitive RBF
network and its projection based learning algorithm for
classification problems. Applied Soft Computing
Journal, 13(1), pp. 654-666.
Bharti, K. K. & Singh, P. K., 2014. A three-stage
unsupervised dimension reduction method for text
clustering. Journal of Computational Science, 5(2), pp.
156-169.
Blanchet, F. G., Legendre, P. & Borcard, D., 2008. Forward
Selection of Explanatory Variables. Ecology, 89(9), pp.
2623-2632.
Brameier, M. & Banzhaf, W., 2001. A comparison of linear
genetic programming and neural networks in medical
data mining. IEEE Transactions on Evolutionary
Computation , 5(1), pp. 17-26.
Chang, P.-C., Lin, J.-J. & Liu, C.-H., 2012. An attribute
weight assignment and particle swarm optimization
algorithm for medical database classifications.
Computer Methods and Programs in Biomedicine,
107(3), pp. 382-392.
Derksen, S. & Keselman, H. J., 1992. Backward, Forward
and Stepwise Automated Subset Selection Algorithms.
British Journal of Mathematical and Statistical
Psychology, Volume 45, pp. 265-282.
Dyer, E. L., Sankaranarayanan, A. C. & Baraniuk, R. G.,
2013. Greedy Feature Selection for Subspace
Clustering. Journal of Machine Learning Research,
Volume 14, pp. 2487-2517.
Farahat, A. K., Ghodsi, A. & Kamel, M. S., 2013. Efficient
Greedy Feature Selection for Unsupervised Learning.
Knowledge Information System, Volume 35, pp. 285-
310.
Gorunescu, F., 2011. Data Mining. Berlin: Springer.
Gorunescu, F., 2011. Data Mining: Concepts, Models, and
Techniques. Verlag Berlin Heidelberg: Springer.
Guyon, I. & Elisseeff, A., 2003. An Introduction to
Variable and Feature Selection. Journal of Machine
Learning Research, Volume 3, pp. 1157-1182.
Han, J. & Kamber, M., 2007. Data Mining: Concepts and
Techniques: 2nd (second) Edition. Amsterdam:
Elsevier Science.
Han, J., Kamber, M. & Pei, J., 2012. Data Mining Concepts
and Techniques. San Fransisco: Morgan Kauffman.
Harrington, P., 2012. Machine Learning in Action. New
York: Manning Publication.
Holland, J. H., 1975. Adaption in Natural and Artificial
Systems. Cambridge: MIT Press.
Inbarani, H. H., Azar, A. T. & Jothi, G., 2014. Supervised
hybrid feature selection based on PSO and rough sets
for medical diagnosis. Computer Methods and
Programs in Biomedicine, 113(1), pp. 175-185.
Jabbar, M. A., Deekshatulu, B. L. & Chandra, P., 2013.
Classification of Heart Disease Using K- Nearest
Features Selection and k-NN Parameters Optimization based on Genetic Algorithm for Medical Datasets Classification
3085

Neighbor and Genetic Algorithm. Procedia
Technology, Volume 10, pp. 85-94.
Jain, A. & Zongker, D., 1997. Feature Selection:
Evaluation, Application and Small Sample
Performance. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 19(2), pp. 153-158.
Jirapech-Umpai, T. & Aitken, S., 2005. Feature selection
and classification for microarray data analysis:
evolutionary methods for identifying predictive genes.
BMC Bioinformatics, Volume 6, p. 148.
Kurgan, L. A. et al., 2001. Knowledge discovery approach
to automated Cardiac SPECT Diagnosis. Artificial
Intelligence in Medicine, Volume 23, pp. 149-169.
Larose, D. T., 2005. Discovering Knowledge in Data: An
Introduction to Data Mining. New Jersey: John Wiley
& Sons, Inc..
Larose, D. T., 2006. Data Mining Methods and Models.
New Jersey: John Wiley & Sons, Inc..
Liu, Z., Chai, T. & Tang, J., 2015. Multi-frequency signal
modeling using empirical mode decomposition and
PCA with application to mill load estimation.
Neurocomputing, Volume 169, pp. 392-402.
Maimon, O. & Rokach, L., 2010. Data Mining and
Knowledge Discovery Handbook. Second Edition ed.
New York: Springer.
Mangasarian, O. L., Street, W. N. & Wolberg, W. H., 1995.
Breast cancer diagnosis and prognosis via linear
programming. Operations Research, 43(4), pp. 570-
577.
Man, K. F., Tang, K. S. & Kwong, S., 1996. Genetic
Algorithms: Concepts and Applications. IEEE
Transactions on Industrial Electronics, 43(5), pp. 519-
534.
Mazurowski, M. A. et al., 2008. Training neural network
classifiers for medical decision making: The effects of
imbalanced datasets on classification performance.
Neural Networks, 21(2), pp. 427-436.
Nowe, A., 2014. Genetic Algorithms. Encyclopedia of
Astrobiology ed. Berlin: Springer.
Prasetio, R. T. & Pratiwi, 2015. Penerapan Teknik Bagging
pada Algoritma Klasifikasi untuk Mengatasi
Ketidakseimbangan Kelas pada Dataset Medis.
Informatika, 2(2), pp. 395-403.
Prasetio, R. T. & Riana, D., 2015. A Comparison of
Classification Methods in Vertebral Column Disorder
with the Application of Genetic Algorithm and
Bagging. Bandung, IEEE.
Raymer, M. L. et al., 2000. Dimensionality reduction using
genetic algorithms. IEEE Transactions on
Evolutionary Computation, 4(2), pp. 164-171.
Setiyorini, T. & Wahono, R. S., 2015. Penerapan Metode
Bagging untuk Mengurangi Data Noise pada Neural
Network untuk Estimasi Kuat Tekan Beton. Journal of
Intelligent Systems, 1(1), pp. 37-42.
Shah, S. & Kusiak, A., 2007. Cancer gene search with data-
mining and genetic algorithms. Computers in Biology
and Medicine, 37(2), pp. 251-261.
Shilaskar, S. & Ghatol, A., 2013. Dimensionality Reduction
Techniques for Improved Diagnosis of Heart Disease.
International Journal of Computer Applications ,
61(5), pp. 1-8.
Subbulakhsmi, C. V. & Deepa, S. N., 2015. Medical
Dataset Classification: A Machine Learning Paradigm
Integrating Particle Swarm Optimization with Extreme
Learning Machine Classifier. The Scientific World
Journal, Volume 2015, pp. 1-12.
Suguna, N. & Thanushkodi, K., 2010. An Improved k-
Nearest Neigbor Classification Using Genetic
Algorithm. IJCSI International Journal of Computer
Science, 7(2), pp. 18-44.
Unal, Y. & Kocer, E., 2013. Diagnosis of Pathology on the
Vertebral Column with Backpropagation and Naive
Bayes Classifier. Turkey, IEEE, pp. 278-281.
Vafaie, H. & Imam, I. F., 1994. Feature Selection Method:
Genetic Algorithms vs Greedy-like Search. Louisville,
Proceedings of the 3rd International Fuzzy Systems and
Intelligent Control Conference.
Witten, I. H., Frank, E. & Hall, M. A., 2011. Data Mining:
Practical Machine Learning Tools and Technique.
Third Edition ed. Amsterdam: Elsevier Inc..
Wu, X. & Kumar, V., 2009. The Top Ten Algorithms in
Data Mining. Boca Raton: Taylor & Francis Group,
LLC.
Wu, X. et al., 2008. Top 10 Algorithms in Data Mining,
London: Springer-Verlag.
Yang, J. & Honavar, V., 1998. Feature Subset Selection
Using a Genetic Algorithm. Feature Extraction,
Construction and Selection, pp. 117-136.
ICRI 2018 - International Conference Recent Innovation
3086
