
Study of the Modification of Loop-mediated Isothermal Amplification
(LAMP) using Taq Polymerase for Halal Testing
Rosy Hutami
1
, Mira Suprayatmi
1
, Nida Idzni
1
, Raafqi Ranasasmita
2
, Henny Nuraini
3
and Joko Hermanianto
4
1
Department of Food Technology and Nutrition, Djuanda University, Bogor, Indonesia
2
Halal Laboratory, The Assessment Institute for Foods, Drugs And Cosmetics, Bogor, Indonesia
3
Department of Animal Production and Technology, Bogor Agricultural University Bogor, Indonesia
4
Department of Food Science and Technology, Bogor Agricultural University Bogor, Indonesia
Keywords: LAMP, Taq Polymerase, Enzyme, Bst, Betaine, Halal.
Abstract: Loop-Mediated Isothermal Amplification (LAMP) is a new method in nucleic acid analysis that amplifies
target in isothermal or constant conditions. This method is suitable for analysis with low-resource condition.
Commonly, this method needs a polymerase enzyme that has strand displacement activity such as Bacillus
stearothermopilus (Bst) polymerase. But, this enzyme was rarely available in the laboratory. Taq polymerase
is an polymerase enzyme that usualy available in molecular laboratory, easier to be accessed, and has a lower
cost comparing to the Bst polymerase. This research conducted with Taq polymerase in order to study the
compatibility of taq polymerase in amplificating the deoxyribonucleic acid (DNA) with LAMP method. We
used cytochrome b (cyt b) from pork as terget gene for halal detection need. For conducting the strand
displacement activity in DNA amplification, we used denaturation process (95
o
C), following by annealing
(65
o
C), and enzyme inactivation (80
o
C). The result showed there was no band appeared on the agarose after
electrophoresis. It was suggested that the Taq polymerase was not suitable for LAMP analysis although it has
been combined with denaturation process. The LAMP should be conducted with a suitable polymerase that
has strand displacement activity and some supporting reagents such as betaine and LAMP buffer that can
strengthen the reaction under isothermal condition.
1 INTRODUCTION
Halal food is currently one of the centers of attention
in the global food industries. Muslims are obliged to
consume only halal and thayyib foods and drinks. In
Islamic perspective, basically everything on earth is
allowed to be consumed except those that are
prohibited, and among those that are forbidden are
pigs and their derivatives (Al-Baqarah: 168, Al-
Baqarah: 173, Al An'am: 145, Al-Maidah: 3, An
Nahl: 115).
Today, various technologies for testing the
authenticity of a product are progressing rapidly.
Many analytical methods are developed and offer fast
and authentic results, one of which is a DNA-based
method. Methods of analysis using DNA have several
advantages, including DNA can be found in all cell
types in each individual with identical genetic
information, DNA is a stable molecule in the
extraction process, and DNA analysis is very likely to
be done from several different types of samples (Jain,
2004).
One of the most widely used nucleic acid
amplification technologies (DNA and RNA) is
Polymerase Chain Reaction (PCR) technology. PCR
is one of the most widely used methods of DNA
propagation in authentication and can also copy
nucleic acids millions of times from the initial
reaction. However, conventional PCR has the
disadvantage that this method requires the separation
of post-PCR products using electrophoresis gel which
is time-consuming and only semi-quantitative
(Kumar, 2007). These weaknesses can be overcome
by real-time PCR using a fluorescence system so that
the amplification results can be seen directly. Real-
time PCR has a disadvantage that is the expensive
price so that not many laboratories in Indonesia have
it. Because the methods commonly used for DNA
detection have disadvantages, alternative methods
that can detect DNA with the same high sensitivity as
Real-time PCR are needed but use simple and real-
Hutami, R., Suprayatmi, M., Idzni, N., Ranasasmita, R., Nuraini, H. and Hermanianto, J.
Study of the Modification of Loop-mediated Isothermal Amplification (LAMP) using Taq Polymerase for Halal Testing.
DOI: 10.5220/0009940721912198
In Proceedings of the 1st International Conference on Recent Innovations (ICRI 2018), pages 2191-2198
ISBN: 978-989-758-458-9
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
2191

time equipment, so they can be applied in the field or
in laboratories with limited resources.
The study of identification of pork protein
contamination in food products such as meat can be
tested using several methods, namely the PCR
method, PDK (Porcine Detection Kit), and ELISA
(Enzyme-Linked Immunosorbent Assay). Ardi
(2012) has analyzed the presence of pig
contamination in meatball products sold in the market
by PCR method, and found a minimum level of pig
contamination that can be identified by PCR
technique of 0.5%. Rasyid (2015) has analyzed the
contamination of pork in beef products using real-
time PCR with the hydrolysis probe method, and
stated that by using real-time PCR and using the
hydrolysis probe method can amplify DNA from
meatballs using specific primers of cattle and pork in
mitochondrial cytochrome B area with 120 bp and
131 bp amplicons using annealing temperature in
61oC and 60oC.
In this study, the inspection method of a halal
product to be developed is the Loop-Mediated
Isothermal Amplification (LAMP) method. The
LAMP method is one of the molecular diagnostic
techniques that has been developed from 1999 in
Japan. The LAMP technique uses DNA amplification
at a fixed temperature, so that the use of expensive
thermocycler devices is not needed.
In the study using the LAMP method, Wilopo et
al. (2015) stated that the LAMP method can be used
as an alternative examination in detecting blaTEM
genes, especially in areas with limited laboratory
infrastructure. As for, Soleha (2015) states that the
LAMP method is very specific and has a high
sensitivity, fast, and economical. LAMP has a high
selectivity because it recognizes the target 6 different
sequences at the beginning of the reaction. Because
of the advantages of the LAMP method, it is
necessary to explore the method of detection of pork
DNA using the LAMP method by using the Taq
Polymerase enzyme, because the Taq Polymerase
enzyme is easily access, more economical prices, and
commonly used in DNA-based sample testing.
2 MATERIALS AND METHOD
The materials used in this study were pork, beef,
chicken, goat, fish, pork DNA primers for gene
fragments Cytochrome B, Sure Food DNA extraction
kits (R Biopharm AG, from buffer lysis, proteinase,
buffer bindings, pre wash buffer, wash buffer, elution
buffer), PCR GoTaq green master mix (Promega)
PCR reagent, DNA free aquades, agarose powder,
Tris-Borat-EDTA (TBE), FlouroSafe, loading dye
and DNA ladder 100bp.
The tools used in this research are analytical
balance (Precisa XB220A), Sorvall ST 16R (Thermo
Scientific) centrifuge, Genova Nano (Jenway)
spectrophotometer, thermoshaker heating block
Dipabis MHR 13, coolant, eppendorf 0.1-2.5μl
micropipette, micropipette eppendorf 0,5-10μl,
Eppendorf 100μl micropipette, Eppendorf 200μl
microphone, Eppendorf 1000μl micropipette, Gilson
20μl micropipette, vortex, PCR Gene Amp AB
system 9700, PCR (Esco), glassware, microwave,
magenetic stirrer, electrophoresis (Mupid- exu), UV-
Transilluminator (Alphalmager EP), plastic bags,
latex gloves, and spatula.
2.1 DNA Isolation
Isolation or DNA extraction was carried out using
extraction techniques with extraction kits issued by R
Biopharm AG (SureFood® PREP Basic Art kit. No.
S1052). Fresh meat to be extracted is pork, beef,
chicken, goat and fish. Then, the concentration and
purity of the DNA extract were analyzed using a
spectrophotometer and visualized by gel
electrophoresis.
Testing the quality of DNA extracts is done by
looking at the results of DNA visualization, DNA
concentration, and DNA purity resulting from
extraction. DNA visualization from extraction results
was carried out by electrophoresis on 1% gel. The gel
is made from 0.45 grams of agarose and 30 ml of
buffer solution (0.5 x TBE) is heated. The agarose
solution is left to cool a little while stirring with the
stirrer magnet, then adding 1.8 µl of Flouro Safe dye.
A total of 5 µl of DNA samples were dissolved in 1
µl of loading dye. Added 100 μl of 100 bp DNA
ladder as a tape measure on the gel. Electrophoresis
is carried out for 40 minutes at a constant voltage of
100 volts. After electrophoresis is complete, the gel is
taken to take photos using UV-Transilluminator.
Evaluating the quality of DNA extract was done
by testing the concentration and purity of DNA by
spectrophotometric analysis. The elution buffer
solution was used as blank as much as 2 µl of the
solution was dripped into a spectrophotometer, then a
sample of 2 µl of the solution was dripped into a
spectrophotometer and tested at a wavelength of 260
nm and 280 nm.
The obtained DNA extracts were diluted into
uniform DNA concentration. Dilution was aimed to
ensure that the amount of DNA to be amplified at the
next stage were uniform in each samples. The final
DNA concentration of all samples was diluted to 30
ng / µl and 50 ng / µl.
ICRI 2018 - International Conference Recent Innovation
2192

2.2 Primer Design
Primer design was carried out with the NCBI program
to examine primer specificity, then specific DNA
segments were chosen as targets. The complete
sequence of this DNA segment was then downloaded.
Primer was made by using the Primer Explorer
version 5 program and produced 4 primer sets for
LAMP. Each primer set was checked for specificity
with BLAST alignment; examination of melting
temperature, self-complimentary and secondary
structure with Oligo Analyzer 3.1; and primer-dimer
checks between primers with the Thermo Multiple
Primary Analyzer.
2.3 DNA Amplification
This research stage is the stage of DNA amplification
by using 4 primers through denaturation (95 ° C),
annealing (65 ° C), and final extension (80 ° C)
stages. The purpose of this stage was to determine the
success of the LAMP method with modifications to 4
primers F3, B3, FIP, BIP with Cytochrome B DNA
template.
3. RESULT AND DISCUSSION
3.1 DNA Isolation
DNA concentrations ranged from 46.15 to 455.76 ng
/ μl. The amount of DNA produced was influenced by
several factors which are at the time of extraction and
also the condition of the sample. Komalasari (2009)
states that the concentration of DNA extraction was
influenced by two factors, namely the extraction
speed and the composition of buffer lysis adition.
The DNA extract concentration was not uniform.
Therefore, then dilution was conducted to uniform the
sample concentration. In this study, DNA extracts
were diluted up to of 30 ng / μl and 50 ng / μl
concentration. This concentration range was
commonly used in the PCR process. The use of non-
dense DNA concentrations can also avoid primer
attachment errors when the target DNA amplification
occurs.
DNA purity with absorbance ratio λ260 / λ280 has
a good result between 1.822 to 2.099. According to
Sambrook et al., (1989) and Muladno (2010) DNA
isolates can be said as pure and have met the
requirements required for molecular analysis when
the ratio λ260 / λ280 ranges between 1.8-2.0.
Extraction results with a ratio of 1.8 to 2.0 are high
purity DNA and are not contaminated with protein
residues. A value of 260 nm is the maximum value of
DNA that can absorb the light. This value can be used
to estimate the concentration of DNA. The value of
280 is the maximum value of protein residues that can
absorb light.
The extraction method used refers to the
extraction method issued by the Biopharm AG R
(SureFood® PREP Basic Art kit. No. S1052) by
making several modifications. This method of DNA
isolation or extraction used an extraction buffer that
was ready to use. In SureFood kit, the filtration
equipment used was spin filter, there were two types
of spin filters, namely clear spin filter and yellow spin
filter. Cells were lysed by using a lysis buffer. The
principle of lysis method was the destruction of cell
walls without having to damage the desired DNA.
Therefore, cell wall destruction was generally done
by breaking the cell wall using a lysis buffer
(Handoyo and Rudiretna, 2000). Then the cell
components, especially proteins, were destroyed by
using Proteinase-K (protease enzyme). Then, binding
the nucleic acids on spin filters using binding buffer,
filtered and bound of nucleic acids, and washed
nucleic acid using washing buffer (pre-wash buffer
and wash buffer). In the final stage the DNA was
dissolved in the elution buffer.
According to Sunarno et al., (2014), SureFood's
commercial extraction kit has the same principles as
other commercial extractions that use the principle of
mini column or DNA filtration. DNA extraction using
the mini column principle is the most common
extraction method because the results obtained are
very good with a process that is not too long when
compared to the phenol-chloroform method and a
very cheaper cost compared to the magnetic beads
method.
3.2 Primer Design
The primer design steps obtained several primer
pairs. Each primer candidates were tested for
specificity in pork DNA using the BLAST alignment
program from NCBI. The specificity test result by
BLAST alignment program can be seen in Table 1 to
Table 4.
Table 1. Primer Candidate 1
Primer Sequence Primer Specificity
F3 CCCTGAAT
CACCCGTA
TC
Sus scrofa, Sus scrofa
breed, Sus
domesticus,
Prionailurus
bengalensis, Neofelis
nebulosa
mitochondrion, Lutra
lutra, Felis nigripes.
Study of the Modification of Loop-mediated Isothermal Amplification (LAMP) using Taq Polymerase for Halal Testing
2193
Primer Sequence Primer Specificity
B3 TGGTTTTT
GGTTATAC
TACTGC
Sus scrofa breed,
Podomys floridanus,
Sus Scrofa, Priodontes
maximus,
Chaetophractus
vellerosus, Sus
Barbatus,
F2 AAATTACT
CAATCCCC
AAGC
Sus scrofa breed, Sus
domesticus, Sus
Scrofa, Sus cebifrons.
F1C TATGCATT
GAAGGAA
GAGGAAG
TAG
Sus scrofa breed, Sus
Scrofa
B2 CATGGCTA
CTGAGATG
TACC
Sus scrofa breed, Sus
Scrofa, Diplodia
corticola, Sus
verrucosus.
B1c CAGAAAC
AAATGCTC
CAAAAAC
AGT
Sus scrofa breed, Sus
domesticus, Sus scrofa
breed, Sus Scrofa
Table 2. Primer Candidate 2 (Hutami et al. 2017)
Primer Sequence Primer
Specificity
F3 GTCTTATTAG
AAACTCAAAC
CTCA
Sus scrofa breed,
Sus domesticus,
Sus Scrofa
B3 TTTTCTTCTAA
ACCCTCTCCTA
Sus scrofa breed,
Sus domesticus,
Sus Scrofa
F2 GGGTACATCT
CAGTAGCCAT
Sus scrofa breed,
Sus domesticus,
Sus Scrofa
F1C TGGTGTTTTTG
ATTTATTTGGG
GGG
Sus scrofa breed,
Sus domesticus,
Sus Scrofa
B2 TGGACTTGGG
TTGATTGT
Sus scrofa breed,
Sus domesticus,
Sus Scrofa, Sus
cebifrons, Sus
verrucosus,
Tursiops
truncates.
B1c CCTAAAAAAG
ACCCACCAAA
ATTCA
Sus scrofa breed,
Sus domesticus,
Sus Scrofa,
Capra hircus,
ovis aries,
Tayassu pecari,
Pecari tajacu,
Catagonus
wagneri,
Bootherium
bombifrons,
Capra aegagrus,
Ovis ammon.
Table 3. Primer Candidate 3
Primer Sequence Primer Specificity
F3 TCAACTACAA
GAACCTTAAT
GAC
Sus scrofa breed,
Sus domesticus,
Sus Scrofa,
Delphinapterus
leucas, Ursus
arctos, Ursus
spelaeus, Ursus
ingressus,
Nannoperca
variegate.
B3 AGCTGTTGTT
GTGTCTGA
Sus scrofa breed,
Sus Scrofa,
Acanthochromis
polyacanthus,
Calomyscus
bailwardi,
Caenorhabditis
elegans.
F2 AACATCCGA
AAATCACACC
Sus scrofa breed,
Sus domesticus,
Sus scrofa, Lagopus
lagopus,
Myonycteris sp.,
Eonycteris spelaea
Sphaerias blanfordi,
Epomophorus
minimus, Epomops
buettikoferi.
F1C TGGGAGGTC
AATGAATGC
GT
Sus scrofa breed,
Sus scrofa, Rusa
unicolor hainana,
Nanger dama
mhorr, Eudorcas
rufifrons, Gazella
dorcas, Hippotragus
leucophaeus,
Tragelaphus
strepsiceros, Ovis
vignei blanfordi,
Rupicapra
rupicapra, Lemmus
trimucronatus,
Capricornis sp.
B2 TGTGTAATGT
ATTGCTAAGA
ACA
Sus scrofa breed,
Sus scrofa,
Phlebotomus
perniciosus,
B1c AACTTCGGTT
CCCTCTTAGG
C
Sus scrofa breed,
Sus domesticus,
Sus scrofa,
Macrognathus
semiocellatus,
Phlebotomus
perniciosus.
ICRI 2018 - International Conference Recent Innovation
2194
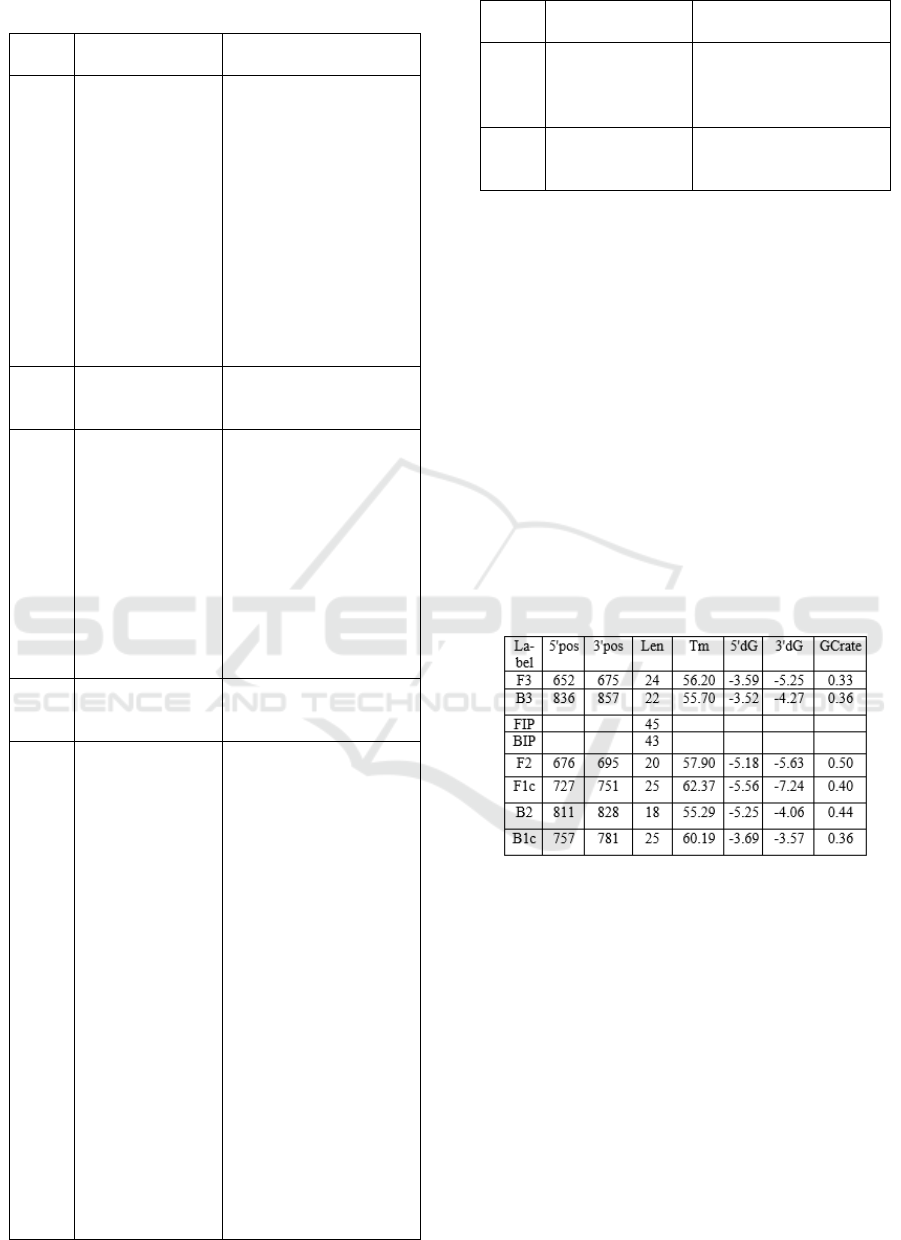
Table 4. Primer Candidate 4
Prime
r
Sequence Primer Specificity
F3 ATTCATTGAC
CTCCCAGC
Sus scrofa breed, Sus
domesticus, Sus scrofa,
Hubei orthoptera virus,
Microtus arvalis, Canis
lupus, Chodsigoa
hoffmanni, Canis
himalayensis, Rusa
unicolor hainana,
Nanger dama mhorr,
Eudorcas rufifrons,
Gazella dorcas,
Tragelaphus buxtoni,
Ovis vignei blanfordi,
Rupicapra rupicapra
B3 TGTAGGTAGC
GAATAACTCA
T
Sus scrofa breed, Sus
scrofa, Phacochoerus
africanus.
F2 CATCTCATCA
TGATGAAACT
TCG
Sus scrofa breed, Sus
scrofa breed, Sus
domesticus, Sus scrofa,
Microtus transcaspicus,
Microtus arvalis
mystacinus, Alophoixus
finschii, Bullimus
luzonicus, Habromys
ixtlani, Oecomys
catherinae, Blarinomys
breviceps, Tragelaphus
buxtoni,
F1C AGAACAGGC
CTGTTAGGAT
TTGC
Sus scrofa
B2 CGTAATTTAC
GTCTCGACAG
AT
Sus scrofa breed, Sus
scrofa, Potos flavus,
Phyllotis definitus,
Planigale ingrami,
Blarinomys breviceps,
Halichoeres
maculipinna,
Acrossocheilus
beijiangensis,
Gerbilliscus
nigricaudus, Myotis
bechsteinii,
Neophocaena
phocaenoides sunameri,
Proechimys roberti,
Peromyscus furvus,
Aphyosemion georgiae,
Myodes glareolus,
Necromys lilloi,
Ichthyoelephas
longirostris, Cribroheros
robertsoni, Neoromicia
robertsi, Embiotoca
jacksoni, Ronquilus
Prime
r
Sequence Primer Specificity
jordani, Psammomys
obesus.
B1c ACATTACACA
TCAGACACAA
CAACA
Sus scrofa breed, Sus
scrofa, Grammomys sp.
Primer selection was based on the level of primer
specificity of pork DNA. Thus, Primer Candidate 2
was chosen with the consideration that, compared to
the other primer pairs, Primer Candidate 2 was more
specific for each primer with the DNA Sus scrofa
(Wild boar). Although primer B2 cross-reacts with
dolphins, it can be ignored because this organism was
not as common as food. Meanwhile, B1c primers
cross react with goats, sheep, Bootherium bombifrons
(variants of the extinct bull) and wild variations of
pigs (Tayassu pecari, Tajacu and Catagonus
wagneri). B1c primer cross reaction can also be
ignored because there were still 6 other primers which
prevent cross-linking with other species. The
characteristic of Primer Candidate 2 showed at Table
5.
Table 5. Characteristic of Primer Candidate 2
When compared with the requirements of primer
design according to Viljoen et al., (2005) Primer
Candidate 2 have fairly good feasibility, including
primers having a nucleotide length of 18-28 bp, Tm
ranging from 55 ° C-72 ° C, hairpin and dimers would
not occur, primers (F3, B3, FIP, F2, F1C, B2, B1C)
were free from self-annealing, whereas in BIP
primers they had the possibility of self-annealing,
because they had ΔG value of -9.43 kcal / mol. But
there were some conditions that are not fulfilled,
including the composition of G and C not 50-60% and
at the primary end -3 'dominated by bases A and T.
However, the primer can still be used in research.
This was evidenced by the results of amplification of
pork DNA with conventional PCR methods using
selected primers that was success. Visualization of
Study of the Modification of Loop-mediated Isothermal Amplification (LAMP) using Taq Polymerase for Halal Testing
2195
pork DNA amplification results with selected primers
is shown in Figure 1 and Figure 2.
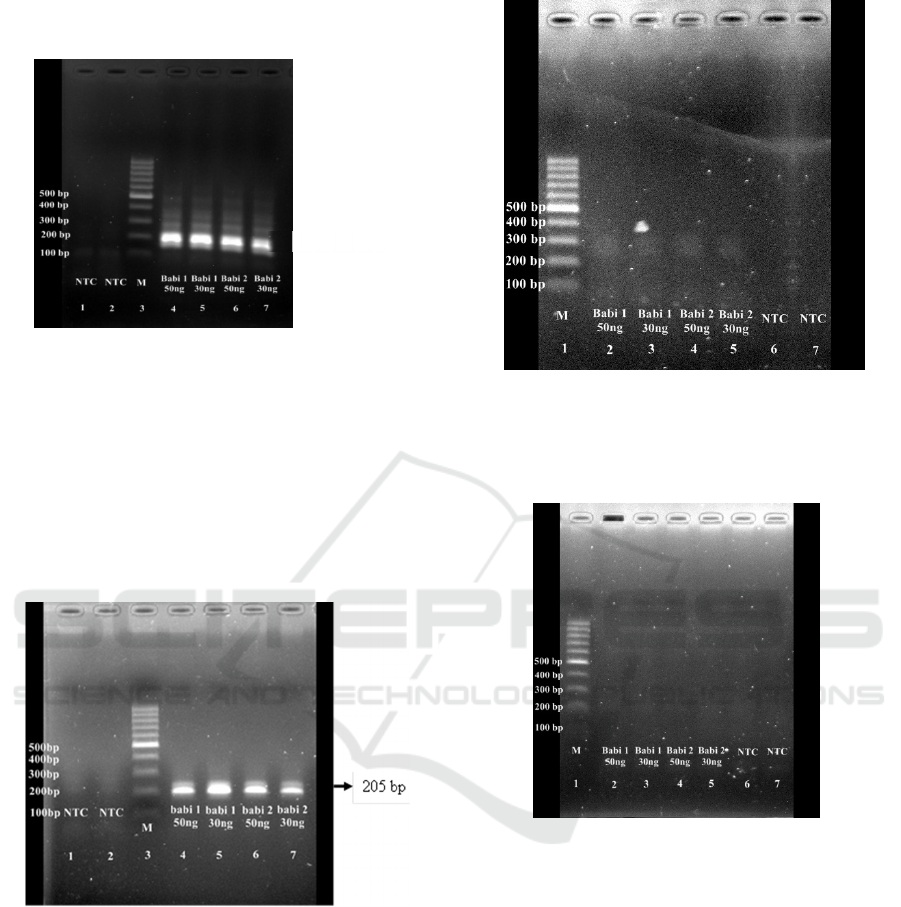
Figure 1: Visualization of Amplified DNA by PCR
(replication 1), 1: Non Template Control, 2 : Non Template
Control, 3 : Marker, 4 : Pork DNA 50 ng/µl, 5 : Pork DNA
30 ng/µl, 6 : Pork DNA 50 ng/µl, 7: Pork DNA 30 ng/µl.
Appeared bands showed that the primers and the
reagent including Taq polymerase enzyme were
suitable for DNA amplification using PCR protocol
(Fig. 1 and Fig 2). Then, the modified LAMP was
performed to study whether the primers and Taq
polymerase will also work in the LAMP protocol.
Figure 2: Visualization of Amplified DNA by PCR
(replication 2), 1: Non Template Control, 2: Non Template
Control, 3 : Marker, 4 : Pork DNA 50 ng/µl, 5 : Pork DNA
30 ng/µl, 6 : Pork DNA 50 ng/µl, 7: Pork DNA 30 ng/µl.
The LAMP procedure that used in this research
was modified by the procedure that developed by
Kanchanaphum et al. (2014). As modification, we
used denaturation process in 95
o
C. The result of
modified LAMP was showed in Figure 3 and Figure
4.
Figure 3: Visualization of Amplified DNA by LAMP with
Modification (replication 1); 1: marker, 2 : Pork DNA 50
ng/µl, 3 : Pork DNA 30 ng/µl, 4 : Pork DNA 50 ng/µl, 5:
Pork DNA 30 ng/µl, 6 : Non Template Control, 7 : Non
Template Control.
In
Figure 4: Visualization of Amplified DNA by LAMP with
Modification (replication 2); 1 : marker, 2 : Pork DNA 50
ng/µl, 3 : Pork DNA 30 ng/µl, 4 : Pork DNA 50 ng/µl, 5:
Pork DNA 30 ng/µl, 6 : Non Template Control, 7 : Non
Template Control.
According to the results, there was no bands
appeared in DNA visualization after DNA
amplification by modified LAMP method (Fig. 3 and
Fig. 4).
This was allegedly caused by several factors, due
to differences in the used procedure compared with
The LAMP procedure used by Kanchanaphum et
al. (2014). The first factor was the type of polymerase
used. The function of polymerase functions is to
catalyze the formation of a phosphodiester bond
between OH at the end of the 3' carbon with the
phosphate group from the added dNTP (Notomi et al.,
205 b
p
ICRI 2018 - International Conference Recent Innovation
2196
2000). In the Kanchanaphum’s procedure, the
polymerase used is Bst polymerase that has strand
displacement activity (Notomi et al., 2000), while in
this modified LAMP procedure, the polymerase used
(Taq polymerase) does not have any strand
displacement activity. Enzymes used in PCR (e.g.,
Taq DNA polymerase) possess high thermostability
and robust polymerase activity but do not exhibit a
strong strand displacement activity and are therefore
not suitable for isothermal amplification methods
such as LAMP (Konstantin et al. 2018). The second
factor was the temperature used. In the LAMP
procedure (Kanchanaphum et al. 2014), DNA
denaturation temperature was not used. In this
modified LAMP procedure, denaturation temperature
was available in order to displace the DNA stand.
However, the duration for denaturating was less than
common denaturation duration in PCR while using
Taq polymerase (5 minutes of pradenaturation
following by 2 seconds of denaturation). In this study,
denaturation was only carried out for 2 minutes. So
that the heating stage at 95
o
C (denaturation) was
considered unsuccessful in doing strand
displacement. The third factor was the used of
betaine. In the LAMP protocol (Kanchanaphum et al.
2014), betaine was used, when in the modified LAMP
protocol was not. Betaine has an important role in
strand displacement activity, because the presence of
betaine (N, N, N-trimethylglyine or L-proline) can
help destabilize the double helix structure in DNA
(Notomi et al. 2000). The fourth factor was the used
of LAMP buffer. In this LAMP modified protocol,
buffer was not used, while in the LAMP protocol
(Kanchanaphum et al. 2014), it was used. LAMP
reaction needs a certain pH, and LAMP buffer will
provide it (Yang et al. 2006). The summary of method
used in LAMP (Kanchanaphum et al. 2014) and
modified LAMP in this research was showed in Table
6.
Table 6: The Summary of Method between LAMP
(Kanchanaphum et al. 2014) and Modified LAMP
Components
Method
LAMP
Method
(Kanchanaphu
m et al. 2014)
Modified LAMP
Primer
4 Primer :
F3, B3, FIP,
BIP
4 Primer :
F3, B3, FIP, BIP
Reagents
- Bst
polymerase
- MgSO
4
- Betaine
- dNTP
- Taq
polymerase
- MgCl
2
- dNTP
- Aquades
bebas DNA
- LAMP
Buffer
- Aquades bebas
DNA
Temperatur
e and Time
Annealing :
T: 65
°
C; t: 45
min
Enzyme
Inactivation :
T: 80
°
C; t: 5
min
Denaturation :
T: 95
°
C; t: 2 min
Annealing :
T: 65
°
C; t: 45 min
Enzyme
Inactivation :
T: 80
°
C; t: 5 min
4 CONCLUSIONS
Modification of the LAMP method was carried out by
changing the type of polymerase used, eliminating the
use of betaine and LAMP buffer and adding
denaturation temperature to the amplification
process. Based on the results, Taq polymerase was
not suitable for LAMP method because it does not
have the strand displacement activity of DNA
structure, although it was combined with denaturation
process (95
o
C, 2 min). The absent of betaine and
LAMP buffer were suspected giving impact in the
failure of reaction, because betaine plays role in
destabilizing the double helical structure of DNA and
LAMP buffer plays role in giving the suitable pH for
LAMP reaction. Thus, Taq polymerase and
modification of some reagents were not suitable to be
applied in LAMP method.
ACKNOWLEDGEMENTS
We would like to thanks for the support of Ministry
of Research Technology and Higher Education of
Indonesia for Gave the Grant of This Research in
accordance with the Research Contract No.
1598/K4/KM/2017. We would like to thanks also for
Department of Animal Production and Technology,
Faculty of Animal Science, Bogor Agricultural
University, Indonesia and The Assesment Institute for
Foods, Drugs and Cosmetics, The Indonesian Council
of Ulama, Indonesia.
REFERENCES
Ardi, A. 2012. Validasi Metode Ekstraksi DNA pada
Analisis DNA Babi dalam Produk Bakso [skripsi].
Fakultas MIPA, Institut Pertanian Bogor, Bogor.
Study of the Modification of Loop-mediated Isothermal Amplification (LAMP) using Taq Polymerase for Halal Testing
2197

Gunimaladevi I., Kono T., Venugopal M.N., Sakai M.
2004. Detection of koi herpesvirus in common carp,
Cyprinus carpio L., by loop-mediated isothermal
amplification. J Fish Dis 27:583–589.
Gunimaladevi I., Kono T., LaPatra S.E., Sakai M. 2005. A
loop mediated isothermal amplification (LAMP)
method for detection of infectious hematopoietic
necrosis virus (IHNV) in rainbow trout (Oncorhynchus
mykiss). Arch Virol 150: 899–909.
Hafner, G. J., Yang I. C., Wolter L. C., Stafford M. R.,
Giffard P. M.. 2001. Isothermal amplification and
multimerization of DNA by Bst DNA polymerase.
BioTechniques 30: 852-867
Handoyo, D dan Rudiretna, A. 2000. Prinsip Umum dan
Pelaksanaan Polymerase Chain Reaction (PCR). Pusat
Studi Bioteknologi, Universitas Surabaya, Surabaya.
Hutami R, Ranasasmita R, Suprayatmi M, Idzni N, Nuraini
H, Hermanianto J. 2017b. Perancangan dan optimasi
primer loop-amplification mediated polymorphism
untuk deteksi kehalalan pangan. Prosiding Seminar
Nasioal PATPI 2017. 2: 1005-1008.
Kanchanaphum, P., Maneenin, S., Chaiyana, W. 2014.
Analysis of Pork Meat Using LAMP to Confirm Halal
Status. Int J Biosci 4(9): 62-68.
Komalasari, K. 2009. Pengaruh perbandingan volume
darah dan lisis buffer serta kecepatan sentrifugasi
terhadap kualitas produk DNA pada sapi Frensian
Holstein (FH) [Skripsi]. Fakultas Peternakan, Institut
Pertanian Bogor, Bogor.
Konstantin B. I., Ekaterina V. B., Arkady F. F., Konstantin
A. B., Tatiana V. K., Vladimir M. K. 2018. A Strong
Stand Displacement Activity of Thermostable DNA
Polymerase Margedly Improves the Results of DNA
Amplification. Biotechniques 57 (2) : 81-87.
Kumar Y., Bansal S., Jaiswal P. 2017. Loop-Mediated
Isothermal Amplification (LAMP): A Rapid and
Sensitive Tool for Quality Assessment of Meat
Products. Comprehensive Reviews in Food Science and
Food Safety 16 : 1359-1378. doi: 10.1111/1541-
4337.12309.
Muladno. 2010. Teknologi Rekayasa Genetika. IPB Press,
Bogor.
Notomi T, Okayama H, Masubuchi H, Yonekawa T,
Watanabe K, Amino N, Hase T. 2000. Loop-mediated
isothermal amplification of DNA. Nucleic Acids
Research. 28(12): 63.
Rasyid, S. 2015. Analisis Cemaran Daging Babi pada
Produk Daging Sapi Menggunakan Real-time PCR
dengan Metode Hydrolisis Probe [skripsi]. Fakultas
Kedokteran dan Ilmu Kesehatan, UIN Syarif
Hidayatullah, Tanggerang Selatan.
Sambrook J, Fritsch F, Miniatis T. 1989. Molecular Cloning
Laboratory Manual. 3rd Edition. Cold Spring Harbor
Laboratory Pr., New York (US).
Savan R., Igarashi A., Matsuoka S., Sakai M. 2004.
Sensitive and rapid detection of edwardsiellosis in fish
by a loop mediated isothermal amplification method.
Appl Environ Microbiol 70:621–624.
Soleha, T.U. 2015. Teknik Pemeriksaan DNA dengan
Metode Loop Mediated Isothermal Amplification
(LAMP). Fakultas Kedokteran, Universitas Lampung,
Lampung.
Sunarno, S., Muna, F., Fitri, N., Malik, A., Karuniawati, A.,
Soebandrio, A. 2014. Metode Cepat ekstraksi DNA
Corynebacterium diphteriae untuk pemeriksaan PCR.
Peneliti Kesehat. 42(2):85-92.
Thai H.T.C., Le M.Q., Vuong C.D., Parida M., et al. 2004.
Development and evaluation of a novel loop-mediated
isothermal amplification method for rapid detection of
severe acute respiratory syndrome Coronavirus. J Gen
Virol 36: 93–109.
Viljoen, G.J., Nel, L.H., Crowther, J.R. 2005. Molecular
Diagnostik PCR Handbook. Springer Netherlands.
Wahyudi, T.H. 2007. Pengaruh Suhu Annealing dan
Jumlah Siklus yang Berbeda Pada Program PCR
Terhadap Keberhailan Isolasi dan Amplifikasi mtDNA
Ikan Patin (Pangasius hypothalmus) [Skripsi]. Fakultas
Perikanan dan Ilmu Kelautan, Institut Pertanian Bogor,
Bogor.
Wilopo, Bayu A.P., Sudigdoadi, S., Sahiratmadja, E., Dewi,
Intan M.W. 2015. Loop-Mediated Isothermal
Amplification untuk Mendeteksi Gen blaTEM Sebagai
Penyandi Extended-Spectrum Beta-Lactamase pada
Isolat Enterobacteriaceae. Fakultas Kedokteran,
Universitas Padjajaran, Bandung.
Yang W, Lee JY, Nowotny M. 2006. Making and breaking
nucleic acids: Two-Mg
2+
-ion catalysis and substrate
specificity. Molecular Cell. 22:5-13.
ICRI 2018 - International Conference Recent Innovation
2198
