
Metabolite Profiling of 96 % Ethanol Extract Marsilea Crenata Presl.
Leaves Using Uplc-Qtof-Ms/Ms
Agnis Pondinekaria Aditama*, Mangestuti Agil
Keywords Marsilea crenata Presl., metabolite profiling, UPLC-QTOF-MS/MS, 96% ethanol.
Abstract Marsilea crenata Presl. plants grow in east java area, usually consumed by local people, and was known
having medical purposes. Some researches were conducted toward to the plant and showed that the plant
having potential treatment to some diseases. The aim of research is to know the contain of Marsilea crenata
Presl. compound by using UPLC MS/MS methode. Marsilea crenata Presl. M crenata was ekstracted using
etanol 96% by using Ustrasonic Assisted Extraction methode. The first step was prepare 100 extrac ppm,
and then were injected 5 µL to UPLC MS/MS. The next step, the data obtained was total ion chromatogram
(TIC), and the last step, data was analyzed by using soffware Masslynx 4.1. Which shown in each
equipment dichloromethane (DCM) blank 47 compound and methanol blank 50 compound. This is the first
report of the application of non-targeted metabolomics in Marsilea crenata Presl.
1 INTRODUCTION
Marsilea crenata Presl. Contains of different
phytochemical which having medical purposes. Tthe
benefit explained above is the effect of metabolit
secunder that was obtained in Marsilea crenata
Presl. Secondary metabolism is chemical material
that was resulted from the plant metabolism process
that is useful to the plant. Secondary metabolism is
classified according to chemical structured
functional characteristic such as alkaloid, flavonoid,
saponin, tannin, poliphenole, antraquinone and
volatile oil (Manitto, 1992; Jacoeb et al., 2010).
Some research had been done to know the activity of
Marsilea crenata Presl. Some of them are, Marsilea
crenata Presl. Leaves had been observed by using
Radio Immuno Assay (RIA) and activity observation
in vivo in female mice. The result showed that 96 %
ethanol extract Marsilea crenata Presl. Leaves
enable to inhibit osteoporosis to pascamenopouse
woman by increasing bone remodelling process
mechanism especially in the bone forming (Putra
and Laswati, 2011).
The research that had been done was Gas
Chromatography-Mass Spectrometry (GC-MS)
analysis where the result showed that some
compound such as monoterpenoid, diterpenoid, fatty
acid, and other compound have not been known in
n-hexane extract of Marsilea crenata Presl. Leaves.
and Palmitat contain was assumed enable to increase
the bone forming process with induction mechanism
in osteoblast cell so that it can be used as
phytoestrogen (Ma’arif et al., 2016).
According to the previous research GC-MS
instrument was used in order to know Marsilea
crenata Presl. Metabolit secondary contain, but not
all secondary metabolit chemical compound can be
analysed because lack of instrument, so only volatile
compound can be analysed. Periodic and update
library is needed because there are some compound
having similar m/z model, so it is known as
similarityindex (SI). Therefore metabolit profiling
must be done by using Ultra Performance Liquid
Chromatography-Mass Spectrometer (UPLC-MS)
Instrument. UPLC-MS instrument is liquid
chromatography technique with mass spectrometer
detector. Bio analysis research use UPLC-MS. The
instrument is specific and having wide application as
well as practical method. The application of this
instrument is not restricted only for volatile
molecule, high flecsibility and limited time (K
Naresh et al., 2014; Chawla and Ranjan, 2016). The
using of UPLC-MS can give scientific data that is
benefical for the user of the plant drug.
Metabolite Profiling of 96 .
DOI: 10.5220/0009841600002406
In Proceedings of BROMO Conference (BROMO 2018) - Symposium on Natural Product and Biodiversity, page 1
ISBN: 978-989-758-347-6
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
1

2 MATERIAL AND METHOD
2.1 Material
We performed UPLC-QTOF-MS/MS (Waters),
Oasis C18 Cartridge (Waters), Sonicator (Sonica),
Moisture Analyzer (Mettler Toledo), Vacuum
Rotary Evaporator (Heidolph), TLC (Camag), TLC
Visualizer (Camag), analytical scales (XX), flasks,
beaker glass, measuring cups, petri dishes, stirrer
bars, spatulas, dropper pipes, funnel, filter paper,
eppendorf, and computers.
Marsilea crenata Presl. Leaves were obtained
from Benowo village at Surabaya, ethanol 96%
(Merck), aquadest, dichloromethane (Merck),
acetonitrile (Merck) and formic acid (Merck).
2.2 Methods
2.2.1 Sample Preparation
The extract preparation was done by simplicia of
Marsilea crenata Presl. Leaves powder weighed
30 g and put into the Erlenmeyer flask, then
dissolved with 500 ml ethanol solvent with
replication 3 times (200 ml, 150 ml, 150 ml). Further
extraction is done with the help of ultrasonic waves
(> 20 kHz) for 6 minutes with 3 pauses every 2
minutes. Ethanol 96% extraction was performed by
single extraction. The extract was evaporated using a
Rotary evaporator, then stored in an oven with a
temperature of 40
0
C.
2.2.2 Extract Preparation to UPLC-QTOF-
MS/MS Analysis
Sample was injected to instrument UPLC MS/MS
5µl, and than chromathogram was obtained and the
data was processed by using software Masslynx so
that peak area, retention time, spectra m/z dan
elemental composition was obtained from each peak
area was detected. The next step, data interpretation
was done by using website Chemspider to get the
level of data similarity from chromagram and
spectra, so that the similarity explained above, we
can get the suitable IUPAC name and it can be
concluded that metabolit contain was in M.crenata
extract.
3 RESULT AND DISCUSSION
The extraction method used by ultrasonic assisted
extraction (UAE) which has advantages, among
others, accelerating the extraction process
(compared with conventional extraction eg
maceration), more time efficient, and can increase
the crude rendement rate of the extract. In addition,
ultrasonic extraction may also be used in the
extraction of heat resistant materials (Handayani et
al., 2016).
Fourty seven compounds in DCM blank and
Fifty compounds in methanol blank were obtained
from UPLC MS/MS analysis. Data obtained was
total ion of kromatogram (TIC) and 96 % ethanol
extract from Marsilea crenata Presl. leaves that was
processed by using software Masslynx so that peak
area, retention time, spectra m/z dan elemental
composition was obtained from each peak area was
detected. The next step, data interpretation was done
by using website Chemspider to get the level of data
similarity from chromagram and spectra, so that the
similarity explained above, we can get the suitable
IUPAC name.
Fifty major contain were tentatively assigned
based on their accurate masses, MS/MS
fragmentation patterns in methanol blank and Forty-
seven major contain in dichloromethane (DCM)
blank, in comparison to standard compounds and
references (Table 1 and 2).
The largest compound in 96% ethanol extract
leaves Marsilea crenata Presl. on methanol blank
with % area 23,3199 %; 11.9297% and 10.3549%
are unknown compounds where the chemspider
application does not recognize it or has never been
published. Whereas in the DCM blank on % area
37.6384 % is C
36
H
36
N
5
O
6
SCl after data
interpretation was done by using website
Chemspider and software Chemdraw so that
compound similarity 4-[(N-{2-[(6-Chloro-2-methyl-
4-quinolinyl)amino] ethyl}-N-[(4-methoxyphenyl)
sulfonyl] -β-alanyl) amino] -3-methoxy-N-
phenylbenzamide was obtained ; peak area 26.3455
% is C
38
H
38
N
5
O
11
Cl and suitable with compound
(1R, 13S, 16S, 17R, 28R) -28-Amino-20-chloro-
17,25-dihydroxy-5,8,10,24-tetramethoxy-N-methyl-
15,29, 31-trioxo-22-oxa-14,30,32-triazahexacyclo
14.14.2.218,21.12,6.123,27,07,12] hexatriaconta-2
(36), 3,5, 7,9,11,18,20,23 (33), 24,26,34-dodecaene-
13-carboxamide and we did not obtaine the
compound name that was not suitable with the
compound name reference. So that we catagorized
as unknown compound.
The activity of the major compound explained
above had non been obtained yet before. According
2

to the research was done, it need to analyzed deeply
in order to get the data about unknown compound.
4 CONCLUSIONS
From the analysis data, we can conclude that there
are some phytochemical compound in Marsilea
crenata Presl. leaves that was known having major
unknown compound.
REFERENCES
Afriastini, J. J. 2003. Marsilea Crenata Presl. De Winter
WP, Amoroso VB. Edited by Cryptograms. Bogor :
LIPI: Ferns and fern allies.
Chawla, Gita, and Chanda Ranjan. 2016. Principle,
Instrumentation, and Applications of UPLC: A Novel
Technique of Liquid Chromatography.Open
Chemistry Journal 3 (1): 1–16.
https://doi.org/10.2174/18748422016 03010001.
Fiehn, O, J Kopka, P Dormann, T Altmann, R N
Trethewey, and L Willmitzer. 2000. Metabolite
Profiling for Plant Functional Genomics. Nature
Biotechnology 18: 1157–61.https://doi.org/10.1038/
81137.
Handayani Hana, Feronika Heppy Sriherfyna, and
Yunianta. 2016. Ekstraksi Antioksidan Daun Sirsak
Metode Ultrasonic Bath (Kajian Rasio Bahan : Pelarut
Dan Lama Ekstraksi. Jurnal Pangan Dan Agroindustri
4 (1): 262–72.
Jacoeb AM, Nurjanah, Arifin M, Sulistiono W, Kristiono
SS. 2010. Deskripsi Histologi Dan Perubahan
Komposisi Kimia Daun Dan Tangkai Semanggi
(Marsilea Ccrenata Presl., Marsileaceae) Akibat
Perubahan. Pengolahan Hasil Perikanan Indonesia 13
(2).
K Naresh, S Bhawani and T Maneesh Kumar. 2014.
Ultra Performance Liquid. 3 (3): 84–94.
Ma’arif, Burhan, Mangestuti Agil1 and Hening Laswati.
2016. Phytochemical Assessment On N-Hexane
Extract And Fractions Of Marsilea Crenata Presl .
Leaves Through GC-MS” 21 (August): 77–85.
Ma’arif, Burhan. 2015. Aktivitas Ekstrak N-Heksana Dan
Fraksi Hasil Pemisahan Daun Marsilea Crenata Presl.
Terhadap Diferensiasi Sel Preosteoblas MC3T3-E1
Melalui Pengukuran Alkaline Phosphatase In Vitro.
Manitto, P. 1992. Biosintesis Metabolit Sekunder. Edited
by Koensomardiyah dan B. Sudarto. semarang: IKIP
Semarang Press.
Putra, Hening Laswati. 2011. Green Clover
Potentiates Delaying the Increment of Imbalance
Bone Remodeling Process in Postmenopausal Women.
Folia Medica Indonesiana 47 (2): 112–17.
Singh, Sheelendra Pratap, Nistha Dwivedi, Kanumuri Siva
Rama Raju, Isha Taneja, and Mohammad Wahajuddin.
2016. Validation of a Rapid and Sensitive UPLC-MS-
MS Method Coupled with Protein Precipitation for the
Simultaneous Determination of Seven Pyrethroids in
100 μL of Rat Plasma by Using Ammonium Adduct as
Precursor Ion. Journal of Analytical Toxicology 40
(3): 213–21. https://doi.org/10.1093/jat/bkw002.
Waters. 2008.
Metabolite Profiling of 96
3
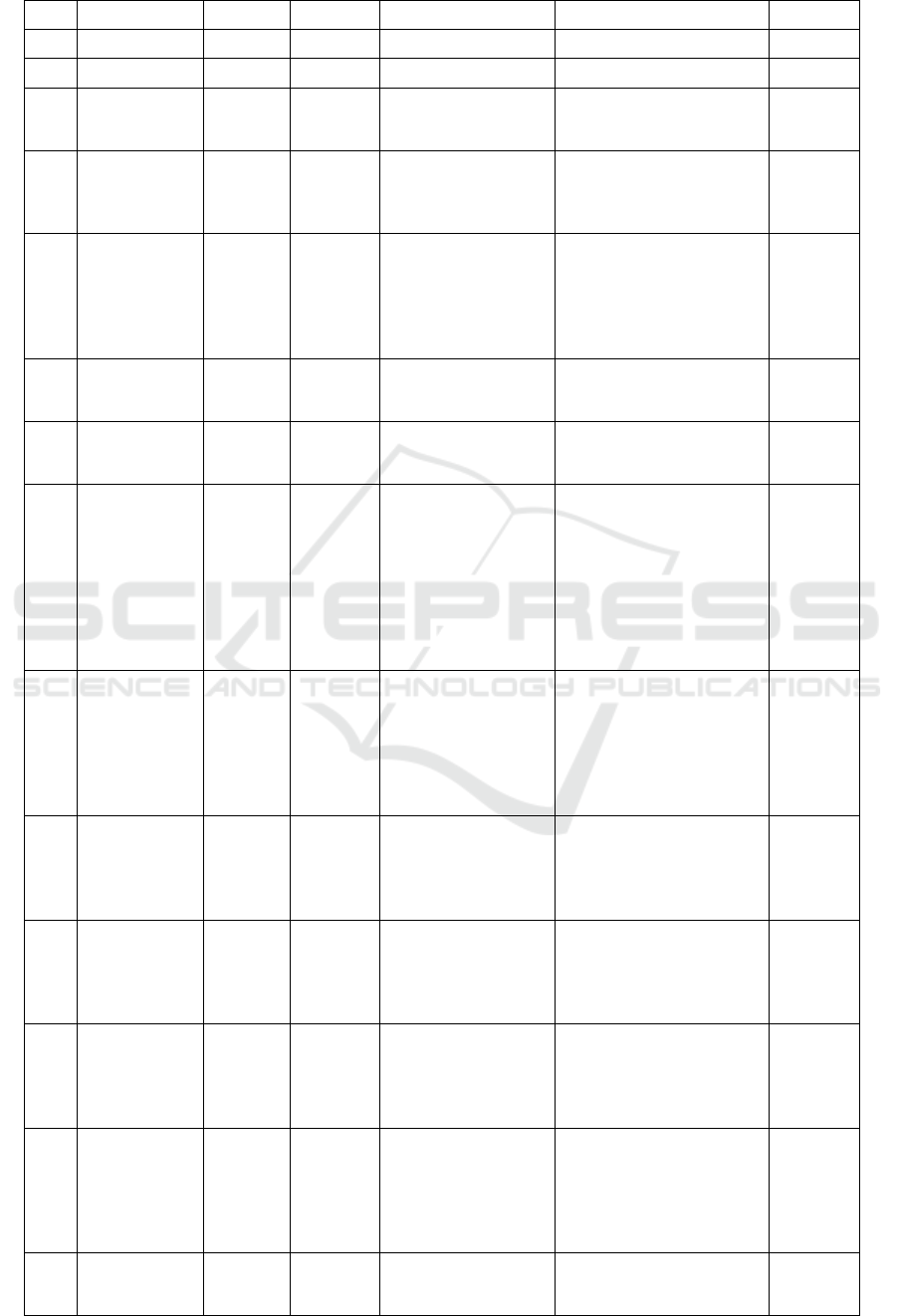
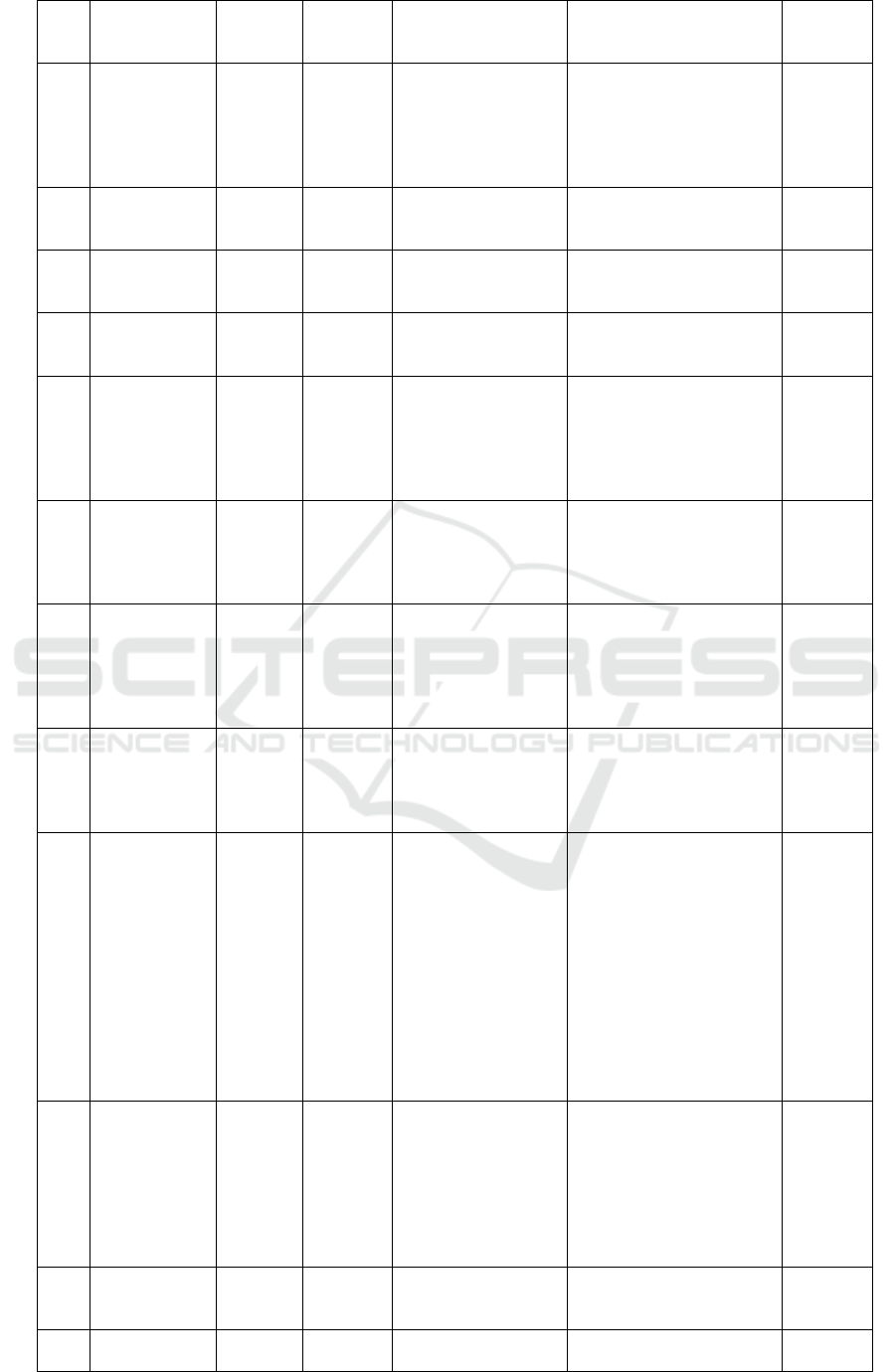
Table 1. Metabolite profiling Marsilea crenata Presl.in methanol blank by UPLC-QTOF-MS/MS.
No
RT
% Area
formula
Trivial name
IUPAC name
Activity
1
0,200694
0,0039%
-
-
-
-
2
0,478472
0,0014%
-
-
-
-
3
1.535
2,4313%
C10H21
NO5
4-(3-
Hydroxypropyl)-4-
nitro-1,7-heptanediol
4-(3-Hydroxypropyl)-4-
nitro-1,7-heptanediol
-
4
2.232
0,1510%
C11H21
NO7
2-[(tert-
Butoxycarbonyl)ami
no]-2-deoxy-D-
glucopyranose
2-Deoxy-2-({[(2-methyl-
2-
propanyl)oxy]carbonyl}a
mino)-D-glucopyranose
-
5
2.518
1,5144%
C12H23
NO7
Methyl 4,6-dideoxy-
4-{[(2R)-2,4-
dihydroxybutanoyl]a
mino}-2-O-methyl-
α-D-
mannopyranoside
Methyl 4,6-dideoxy-4-
{[(2R)-2,4-
dihydroxybutanoyl]amino
}-2-O-methyl-α-D-
mannopyranoside
-
6
3.799
1,4856%
C15H21
NO7
Methyl (3,4,5-
triethoxy-2-
nitrophenyl)acetate
Methyl (3,4,5-triethoxy-2-
nitrophenyl)acetate
-
7
4.427
1,4055%
C5H15N
3Cl2
4-
Hydrazinopiperidine
dihydrochloride
4-Hydrazinopiperidine
dihydrochloride
-
4.610
0,3629%
C9H6O3
3 hydroxycoumarin
3-Hydroxy-2H-chromen-
2-one
Pengham
batan
kompetiti
f DAAO
rekombin
an
manusia
(Molla,
2017).
8
4.896
0,1836%
C20H24
N3SCl
Prochlorperazine
2-Chloro-10-[3-(4-methyl-
1-piperazinyl)propyl]-
10H-phenothiazine
Analgesi
k (callan,
2008),
amtiemet
ik
(roberge,
2006)
9
5.228
0,9215%
C13H18
N5O5Cl
Ethyl 4-[3-(4-chloro-
3-nitro-1H-pyrazol-
1-yl)propanoyl]-1-
piperazinecarboxylat
e
Ethyl 4-[3-(4-chloro-3-
nitro-1H-pyrazol-1-
yl)propanoyl]-1-
piperazinecarboxylate
-
10
5.445
0,0257%
C33H37
N3
4-{Bis[4-(1-
pyrrolidinyl)phenyl]
methyl}-N,N-
dimethyl-1-
naphthalenamine
4-{Bis[4-(1-
pyrrolidinyl)phenyl]methy
l}-N,N-dimethyl-1-
naphthalenamine
-
11
5.628
0,9906%
C10H21
N3O8S
1-Azido-1-deoxy-
2,3-bis-O-
(methoxymethyl)-5-
O-(methylsulfonyl)-
D-ribitol
1-Azido-1-deoxy-2,3-bis-
O-(methoxymethyl)-5-O-
(methylsulfonyl)-D-ribitol
-
12
5.845
0,6908%
C29H18
N4O6S
2-(2-{(E)-2-Cyano-
2-[4-(2-oxo-2H-
chromen-3-yl)-1,3-
thiazol-2-yl]vinyl}-
4-nitrophenoxy)-N-
phenylacetamide
2-(2-{(E)-2-Cyano-2-[4-
(2-oxo-2H-chromen-3-yl)-
1,3-thiazol-2-yl]vinyl}-4-
nitrophenoxy)-N-
phenylacetamide
-
13
6.177
1,0895%
C25H22
O11
4-(1,3-Benzodioxol-
5-yl)-6-hydroxy-1-
oxo-1,3-
4-(1,3-Benzodioxol-5-yl)-
6-hydroxy-1-oxo-1,3-
dihydronaphtho[2,3-
-
4
dihydronaphtho[2,3-
c]furan-5-yl
hexopyranoside
c]furan-5-
yl hexopyranoside
14
6.577
0,3205%
C24H22
O14
2-(3,4-
Dihydroxyphenyl)-5-
hydroxy-4-oxo-4H-
chromen-7-yl 6-O-
(carboxyacetyl)-β-D-
glucopyranoside
2-(3,4-Dihydroxyphenyl)-
5-hydroxy-4-oxo-4H-
chromen-7-yl 6-O-
(carboxyacetyl)-β-D-
glucopyranoside
-
15
6.908
0,2713%
C14H21
NO
1-[1-(4-
Methoxyphenyl)cycl
ohexyl]methanamine
1-[1-(4-
Methoxyphenyl)cyclohex
yl]methanamine
-
16
7.206
2,0878%
C11H16
O3
1-carboxy-3-
hydroxyadamantane
3-Hydroxy-1-
adamantanecarboxylic
acid
-
17
7.423
0,6567%
C16H23
NO2
UNII:891H89GFT4
1-(7-Ethyl-1-benzofuran-
2-yl)-2-[(2-methyl-2-
propanyl)amino]ethanol
-
18
7.640
0,2325%
C11H24
N5Cl
1-Hexyl-6,6-
dimethyl-1,6-
dihydro-1,3,5-
triazine-2,4-
diamine hydrochlori
de (1:1)
1-Hexyl-6,6-dimethyl-1,6-
dihydro-1,3,5-triazine-2,4-
diamine hydrochloride
(1:1)
-
19
7.903
0,3096%
C14H22
N5Cl
1-methyl-2-[(4-
methylpiperazin-1-
yl)methyl]benzimida
zol-5-amine
hydrochloride
1-Methyl-2-[(4-methyl-1-
piperazinyl)methyl]-1H-
benzimidazol-5-amine
hydrochloride (1:1)
-
20
8.406
1,4141%
C36H46
N4O
Manzamine J
(1R,2R,12R,13S,16Z)-25-
(9H-β-Carbolin-1-yl)-
11,22-
diazatetracyclo[11.11.2.12
,22.02,12]heptacosa-
5,16,25-trien-13-ol
-
21
8.886
0,0560%
C17H31
NO9
6-O-(N-{[(2-Methyl-
2-
propanyl)oxy]carbon
yl}-D-leucyl)-α-D-
allopyranose
6-O-(N-{[(2-Methyl-2-
propanyl)oxy]carbonyl}-
D-leucyl)-α-D-
allopyranose
-
22
9.321
0,1071%
C18H27
NO2
dyclonine
1-(4-Butoxyphenyl)-3-(1-
piperidinyl)-1-propanone
Inhibitor
Aldehyd
e
Dehydro
genase 1
(ALDH1
A1)
(Collard,
2007).
Antimicr
oba
(Floresta
no,1956)
23
9.584
0,1649%
C13H29
N3O4S
(3R,4R)-3-{[(2-
Hydroxyethyl)(meth
yl)amino]methyl}-4-
(hydroxymethyl)-N-
isopropyl-N-methyl-
1-
pyrrolidinesulfonami
de
(3R,4R)-3-{[(2-
Hydroxyethyl)(methyl)am
ino]methyl}-4-
(hydroxymethyl)-N-
isopropyl-N-methyl-1-
pyrrolidinesulfonamide
-
24
10.601
0,6568%
C12H18
NO
N,N,N-Trimethyl-3-
oxo-3-phenyl-1-
propanaminium
N,N,N-Trimethyl-3-oxo-
3-phenyl-1-
propanaminium
-
25
10.830
0,3341%
C47H61
N3O8S
2-
({(3β,7β,8ξ,9ξ,10α,1
2-
({(3β,7β,8ξ,9ξ,10α,12β,13
-
Metabolite Profiling of 96
5
2β,13α,14ξ,17α,20S)
-3-[(2-{[(3-Acetyl-2-
methyl-4-
quinolinyl)amino]me
thyl}phenyl)ethynyl]
-3,7,12-trihydroxy-
24-oxocholan-24-
yl}amino)ethanesulf
onic acid
α,14ξ,17α,20S)-3-[(2-
{[(3-Acetyl-2-methyl-4-
quinolinyl)amino]methyl}
phenyl)ethynyl]-3,7,12-
trihydroxy-24-oxocholan-
24-
yl}amino)ethanesulfonic
acid
26
11.082
0,4582%
-
-
-
-
27
11.379
0,8714%
C37H47
N9OS
-
-
-
28
11.562
1,7782%
C14H19
N4O2Cl
Lintopride
4-Amino-5-chloro-N-[(1-
ethyl-4,5-dihydro-1H-
imidazol-2-yl)methyl]-2-
methoxybenzamide
-
29
11.928
0,4325%
C28H49
NO12
2-Methyl-2-propanyl
2-cyano-3-[(4S,5R)-
5-{(5S,6R)-6-[(4R)-
2,2-dimethyl-1,3-
dioxolan-4-yl]-
2,4,7,9-
tetraoxadecan-5-yl}-
2,2-dimethyl-1,3-
dioxolan-4-yl]-2-(1-
ethoxyethoxy)propan
oate
2-Methyl-2-propanyl 2-
cyano-3-[(4S,5R)-5-
{(5S,6R)-6-[(4R)-2,2-
dimethyl-1,3-dioxolan-4-
yl]-2,4,7,9-tetraoxadecan-
5-yl}-2,2-dimethyl-1,3-
dioxolan-4-yl]-2-(1-
ethoxyethoxy)propanoate
-
30
12.179
0,3815%
C27H49
NOS2
2-[(Bis{2-[(2-
methyl-2-
propanyl)sulfanyl]et
hyl}amino)methyl]-
4,6-bis(2-methyl-2-
propanyl)phenol
2-[(Bis{2-[(2-methyl-2-
propanyl)sulfanyl]ethyl}a
mino)methyl]-4,6-bis(2-
methyl-2-propanyl)phenol
-
31
12.397
1,5741%
C25H45
NO9
Pederin
(2S)-N-[(S)-{(2S,4R,6R)-
6-[(2S)-2,3-
Dimethoxypropyl]-4-
hydroxy-5,5-
dimethyltetrahydro-2H-
pyran-2-
yl}(methoxy)methyl]-2-
hydroxy-2-[(2R,5R,6R)-2-
methoxy-5,6-dimethyl-4-
methylenetetrahydro-2H-
pyran-2-yl] acetamide
Anticanc
er
(ghoneim
, 2013)
32
12.614
1,9858%
C33H59
NO14
2-(aziridin-1-
yl)ethanol;
decanedioic acid;
2,2-
dimethylpropane-
1,3-diol; 2-ethyl-2-
(hydroxymethyl)pro
pane-1,3-diol;
isophthalic acid
-
-
33
12.797
2,5108%
C29H39
N7O2
1-(2-Methylalanyl-5-
phenyl-D-norvalyl)-
4-{2-[2-(2H-tetrazol-
5-
yl)ethyl]phenyl}pipe
ridine
1-(2-Methylalanyl-5-
phenyl-D-norvalyl)-4-{2-
[2-(2H-tetrazol-5-
yl)ethyl]phenyl}piperidine
-
34
13.208
0,9465%
C30H53
NO12
(3S)-16-{[(1S)-1-
Carboxyethyl]amino
}-2-methyl-16-oxo-
3-hexadecanyl 6-O-
(3-
carboxypropanoyl)-
β-D-glucopyranoside
(3S)-16-{[(1S)-1-
Carboxyethyl]amino}-2-
methyl-16-oxo-3-
hexadecanyl 6-O-(3-
carboxypropanoyl)-β-D-
glucopyranoside
-
6
35
13.460
2,6423%
C29H45
N5O2
8-(Benzylamino)-7-
hexadecyl-3-methyl-
3,7-dihydro-1H-
purine-2,6-dione
8-(Benzylamino)-7-
hexadecyl-3-methyl-3,7-
dihydro-1H-purine-2,6-
dione
-
36
13.677
2,4722%
C28H46
N5O2Cl
N4-(5-Chloro-2,4-
dimethoxyphenyl)-
N6-hexadecyl-4,5,6-
pyrimidinetriamine
N4-(5-Chloro-2,4-
dimethoxyphenyl)-N6-
hexadecyl-4,5,6-
pyrimidinetriamine
-
37
14.409
10,3549
%
C25H50
NO6Cl
-
-
-
38
14.740
2,0423%
C22H48
N9Cl
N2-[3-({12-[(3-
Aminopropyl)amino]
dodecyl}amino)prop
yl]-N4-methyl-1,3,5-
triazine-2,4,6-
triamine
hydrochloride (1:1)
N2-[3-({12-[(3-
Aminopropyl)amino]dode
cyl}amino)propyl]-N4-
methyl-1,3,5-triazine-
2,4,6-
triamine hydrochloride
(1:1)
-
39
15.106
23,3199
%
C8H39N
23O
-
-
-
40
15.404
4,7166%
C24H50
N9Cl
-
-
-
41
15.769
1,1138%
C8NO15
S6Br2
-
-
-
42
15.952
0,6060%
C8NO15
S6Br2
-
-
-
43
16.718
5,9510%
C36H36
N5O6SC
l
4-[(N-{2-[(6-Chloro-
2-methyl-4-
quinolinyl)amino]eth
yl}-N-[(4-
methoxyphenyl)sulfo
nyl]-β-
alanyl)amino]-3-
methoxy-N-
phenylbenzamide
4-[(N-{2-[(6-Chloro-2-
methyl-4-
quinolinyl)amino]ethyl}-
N-[(4-
methoxyphenyl)sulfonyl]-
β-alanyl)amino]-3-
methoxy-N-
phenylbenzamide
-
44
17.004
1,3681%
C7H24N
19O9Cl
-
-
-
45
17.999
4,6577%
C46H48
N5OS4C
l
-
-
-
46
18.330
11,9297
%
C8NO15
S6Br2
-
-
-
47
21.509
0,0036%
-
-
-
-
48
21.726
0,0049%
-
-
-
-
49
22.389
0,0047%
-
-
-
-
50
22.755
0,0043%
-
-
-
-
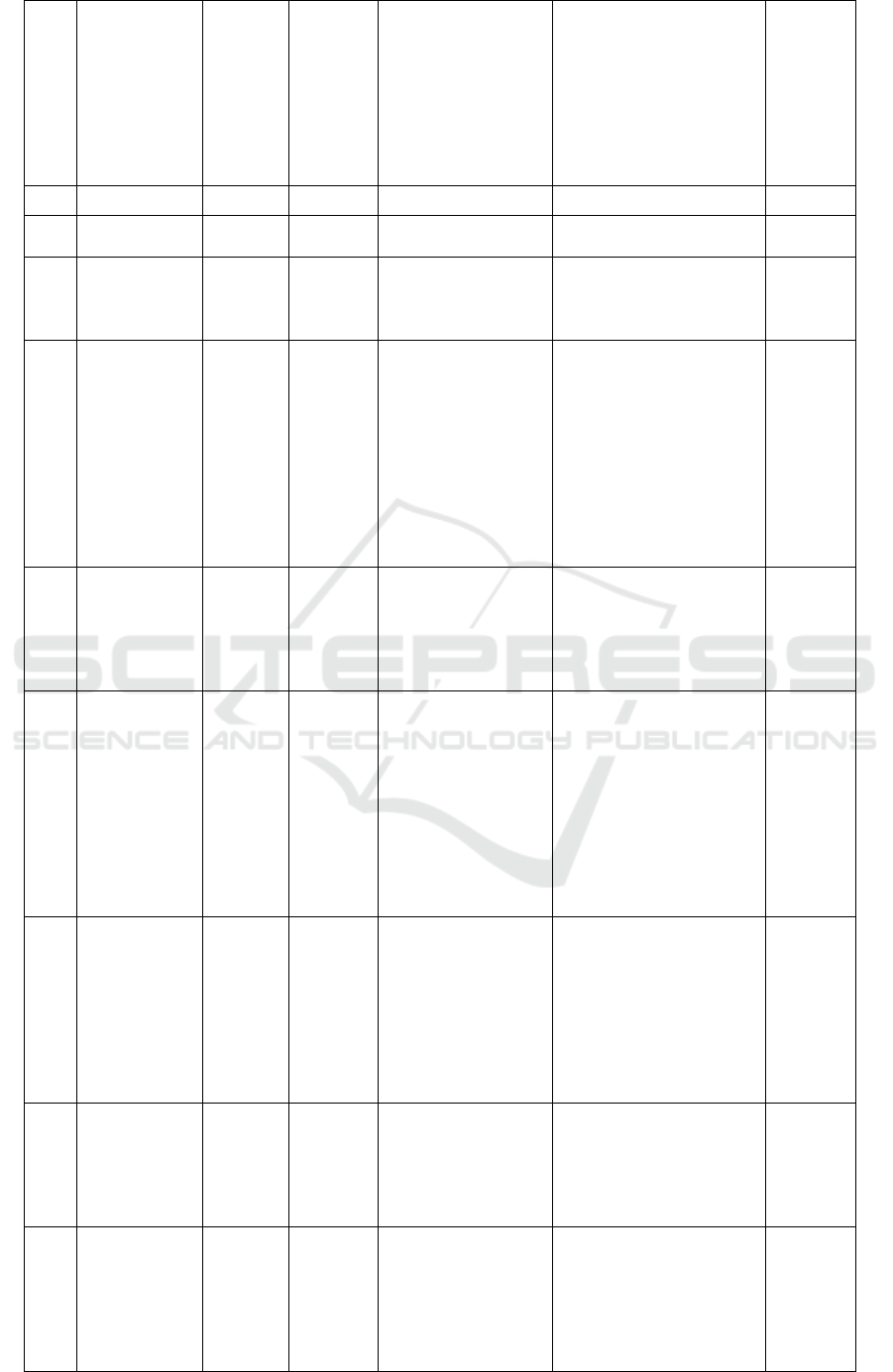
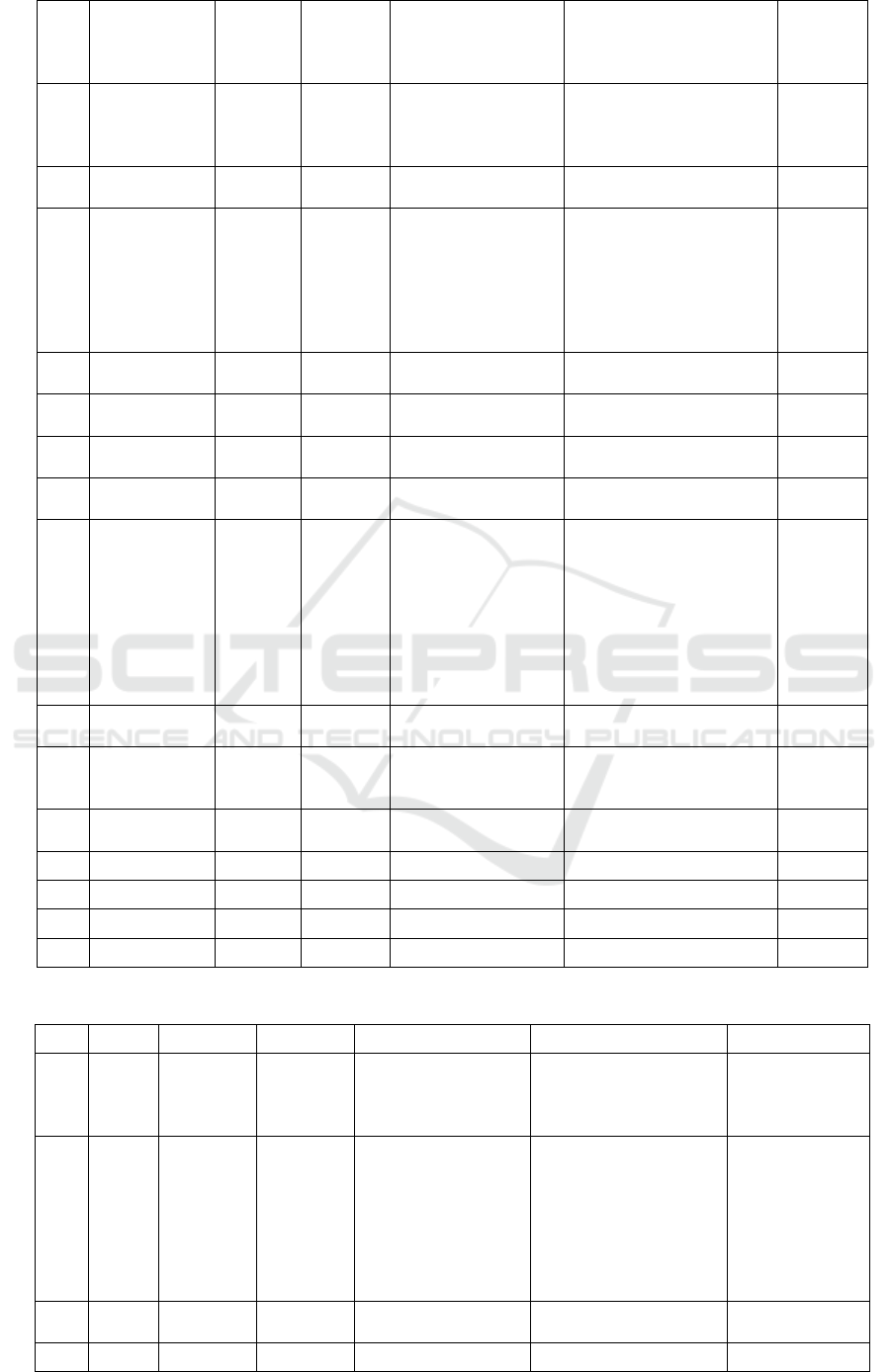
Table 2. Metabolite profiling Marsilea crenata Presl.in DCM blank by UPLC-QTOF-MS/MS.
No
Rt
%Area
Formula
Trivial name
IUPAC name
Activity
1
0.289
0,0032%
C11H23N
4O2Cl
Tert-Butyl 4-
carbamimidamidopip
eridine-1-carboxylate
hydrochloride (1:1)
2-Methyl-2-propanyl 4-
carbamimidamido-1-
piperidinecarboxylate
hydrochloride (1:1)
-
2
0.540
0,0278%
C16H22O
4
Dibutyl phthalate
Dibutyl phthalate
Antibacteri
(Khatiwora
2012),
glikosidase
inhibitor (Lee
2000),
estrogenik
(Harris 1997)
3
0.906
0,0049%
C9H22N6
O2S
-
-
-
4
1.420
0,2361%
-
Metabolite Profiling of 96
7
5
1.786
0,0096%
C11H23N
O2
11-Aminoundecanoic
acid
11-Aminoundecanoic
acid
-
6
1.969
0,0041%
C10H23N
4O3P
Propanedioic acid, 2-
[[bis(1-
methylethyl)phosphi
nyl]methyl]-,
dihydrazide
2-
[(Diisopropylphosphory
l)methyl]malonohydrazi
de
-
7
2.084
0,0670%
C11H23N
O2
11-Aminoundecanoic
acid
11-Aminoundecanoic
acid
-
8
2.186
0,0306%
-
9
2.632
2,8001%
-
10
4.427
0,0282%
C15H27N
O5
Megalanthonine
[(1S,7R,7aR)-7-
Hydroxyhexahydro-1H-
pyrrolizin-1-yl]methyl
(2S,3S)-2,3-dihydroxy-
2-isopropylbutanoate
antifeedant and
antifungal
(Reina 1998)
11
4.930
0,0127%
C9H21N1
1O
-
-
-
12
5.342
0,2477%
-
13
5.479
0,0731%
-
14
5.662
0,0912%
-
15
5.925
0,0405%
C35H41N
3O
Cycloheptaneacetami
de, N-
(phenylmethyl)-α-[4-
[(5,6,7,8-tetrahydro-
4-methyl-9H-
pyrido[2,3-b]indol-9-
yl)methyl]phenyl]-
N-Benzyl-2-
cycloheptyl-2-{4-[(4-
methyl-5,6,7,8-
tetrahydro-9H-
pyrido[2,3-b]indol-9-
yl)methyl]phenyl}aceta
mide
-
16
6.211
0,0164%
-
17
6.474
0,0109%
-
18
6.840
0,0031%
-
-
-
-
19
7.206
0,2253%
C11H16O
3
1-Carboxy-3-
hydroxyadamantane
3-Hydroxy-1-
adamantanecarboxylic
acid
-
20
7.457
0,0010%
-
-
-
-
21
7.640
0,0242%
C12H25N
O2
Dodecanoic acid, 12-
amino-
12-Aminododecanoic
acid
-
22
8.006
0,1302%
C18H25N
O
Dextromethorphan
(9α,13α,14α)-3-
Methoxy-17-
methylmorphinan
Antitussive
(Manap 1999),
anticonvulsant
(Mohseni 2016),
neuroprotective
(Zhang 2004)
23
9.504
0,0908%
C20H31N
O
Trihexyphenidyl
1-Cyclohexyl-1-phenyl-
3-(1-piperidinyl)-1-
propanol
antiparkinson
antikolinergic
(Takahashi
1999), anti
oksidan (Ji
2008)
24
9.950
0,0080%
-
25
10.96
7
0,5387%
26
11.44
8
2,3323%
C16H35N
Hexadecylamine
1-Hexadecanamine
antibacteri,
adjuvant for
diphtheria,
tetanus toxoid,
and influenza
(Attwood 2012)
27
11.63
0
0,3879%
C17H37N
O2
2-Amino-2-
tetradecylpropane-
1,3-diol
2-Amino-2-tetradecyl-
1,3-propanediol
-
28
11.88
0,0775%
C19H18O
Benzylbutylphthalate
3-(1-Phenyl-2-
Estrogenik
8
2
4
pentanyl)phthalate
(Harris 1997)
29
12.11
1
0,0640%
C17H26O
5
Portentol
(1S,2S,3S,3'R,4R,4'R,5'
S,6'R,8R)-4'-Hydroxy-
1,3,3',5',6',8-
hexamethyltetrahydro-
6H,7H-spiro[5-
oxabicyclo[2.2.2]octane
-2,2'-pyran]-6,7-dione
Anticancer
(Schröckeneder
2012)
30
12.24
8
0,0123%
C15H33N
Pentadecylamine
1-Pentadecanamine
-
31
12.39
6
0,0027%
C19H41N
O2
1,2-Propanediol, 3-
(hexadecylamino)-
3-(Hexadecylamino)-
1,2-propanediol
-
32
12.69
4
0,6293%
C19H18O
4
Benzylbutylphthalate
3-(1-Phenyl-2-
pentanyl)phthalate
Estrogenik
(Harris 1997)
33
12.84
2
0,9778%
C21H37N
4-Pentadecylaniline
4-Pentadecylaniline
-
34
13.89
4
0,9962%
C23H41N
Benzylamine, N,N-
dioctyl-
N-Benzyl-N-octyl-1-
octanamine
-
35
15.07
2
16,7611%
C12H21N
25O5S
-
-
-
36
15.32
3
6,5543%
C12H21N
25O5S
-
-
-
37
15.98
7
26,3455%
C38H38N
5O11Cl
(1R,13S,16S,17R,28
R)-28-Amino-20-
chloro-17,25-
dihydroxy-5,8,10,24-
tetramethoxy-N-
methyl-15,29,31-
trioxo-22-oxa-
14,30,32-
triazahexacyclo[14.1
4.2.2
18,21
.1
2,6
.1
23,27
.0
7,
12
]hexatriaconta-
2(36),3,5
,7,9,11,18,20,23(33),
24,26,34-dodecaene-
13-carboxamide
(1R,13S,16S,17R,28R)-
28-Amino-20-chloro-
17,25-dihydroxy-
5,8,10,24-tetramethoxy-
N-methyl-15,29,31-
trioxo-22-oxa-14,30,32-
triazahexacyclo[14.14.2
.2
18,21
.1
2,6
.1
23,27
.0
7,12
]hex
atriaconta-2(36),3,5
,7,9,11,18,20,23(33),24,
26,34-dodecaene-13-
carboxamide
-
38
17.05
0
0,4132%
-
39
17.59
9
1,9907%
C35H36N
4O5
Pheophorbide A
3-[(3S,4S,21R)-14-
Ethyl-21-
(methoxycarbonyl)-
4,8,13,18-tetramethyl-
20-oxo-9-vinyl-3-
phorbinyl]propanoic
acid
Anticancer
(Cho, 2014)
40
18.43
3
37,6384%
C36H36N
5O6SCl
Benzamide, 4-[[3-
[[2-[(6-chloro-2-
methyl-4-
quinolinyl)amino]eth
yl][(4-
methoxyphenyl)sulfo
nyl]amino]-1-
oxopropyl]amino]-3-
methoxy-N-phenyl-
4-[(N-{2-[(6-Chloro-2-
methyl-4-
quinolinyl)amino]ethyl}
-N-[(4-
methoxyphenyl)sulfonyl
]-β-alanyl)amino]-3-
methoxy-N-
phenylbenzamide
-
41
19.64
5
0,0049%
-
42
20.96
0
0,0047%
-
43
21.10
9
0,0065%
C12N
-
-
-
44
21.32
6
0,0063%
-
45
21.50
9
0,0074%
-
Metabolite Profiling of 96
9
46
21.65
8
0,0150%
C12N
-
-
-
47
22.57
2
0,0466%
C7H10N2
2-Pyridylethylamine
2-(2-
Pyridinyl)ethanamine
-
10
