
LCA Histogram Distance for Rooted Labeled Caterpillars
Takuya Yoshino, Kohei Muraka and Kouichi Hirata
Kyushu Institute of Technology, Kawazu 680-4, Iizuka 820-8502, Japan
Keywords:
LCA Histogram Distance, Rooted Labeled Caterpillars, Path Histogram Distance, Complete Subtree
Histogram Distance.
Abstract:
An LCA histogram distance is an L
1
-distance between histograms consisting of triples of two nodes and their
least common ancestor (LCA) in two trees. In this paper, we show that the LCA histogram distance for cater-
pillars is always a metric, whereas that for trees is not. Then, we give experimental results for computing the
LCA histogram distance by comparing with the path histogram distance and the complete subtree histogram
distance for caterpillars.
1 INTRODUCTION
Comparing tree-structured data such as HTML and
XML data for web mining or RNA and glycan data
for bioinformatics is one of the important tasks for
data mining. Then, we deal with them as rooted la-
beled unordered trees, (trees, for short). In particular,
a caterpillar (cf. (Gallian, 2007)) is a tree transformed
to a path after removing all the leaves in it. Whereas
the caterpillars are very restricted and simple, there
are some cases containing many caterpillars in real
dataset, see Table 3 in Section 4.
The edit distance (Tai, 1979) is the most famous
distance measure between trees. It is formulated as
the minimum cost of edit operations, consisting of a
substitution, a deletion and an insertion, applied to
transform a tree to another tree and is always a metric.
Recently, Muraka et al. (Muraka et al., 2018) have de-
signed the algorithm to compute the edit distance be-
tween two caterpillars in O(λ
2
h
2
) time, where λ and h
are the maximum number of leaves and the maximum
height in two caterpillars, respectively. Then, this al-
gorithm runs in O(n
4
) time, where n is the maximum
number of vertices in two caterpillars.
A local frequency distance (Aratsu et al., 2009;
Kailing et al., 2004; Li et al., 2013) is formulated
as an L
1
-distance between histograms concerned with
local information. Whereas we can compute the lo-
cal frequency distance efficiently and they sometimes
provide the constant factor lower bound of the edit
distance, almost all of them is not a metric. In order
to compare caterpillars efficiently by using a metric,
a path histogram distance (Kawaguchi et al., 2018b)
and a complete subtree histogram distance (Akutsu
et al., 2013) are appropriate local frequency distances
for caterpillars.
A path histogram distance is an L
1
-distance be-
tween histograms consisting of paths from the root to
leaves in two trees (Kawaguchi et al., 2018b). It is
computable in linear time, always a metric for cater-
pillars, which is not a metric for trees in general,
and incomparable with the edit distance (Kawaguchi
et al., 2018b). On the other hand, as an extreme case,
for two paths with the same length such that every la-
bel in one path is a and that in another path is b, the
edit distance between them is the number of vertices
in a path but the path histogram distance is one.
A complete subtree histogram distance is an L
1
-
distance between histograms consisting of complete
subtrees in two trees (Akutsu et al., 2013). It is com-
putable in linear time, always a metric for trees and
greater than or equal to the edit distance (Akutsu et al.,
2013). On the other hand, as an extreme case, for
two paths with the same length such that the labels
of leaves are different, the edit distance between them
is one but the complete subtree histogram distance is
the number of vertices in two paths, which is the max-
imum value.
In this paper, we focus on an LCA his-
togram distance (Tatikonda and Parthasarathy, 2010),
which is an L
1
-distance between histograms con-
sisting of triples of two vertices and the LCA of
them with their depth. Whereas Tatikonda and
Parthasarathy (Tatikonda and Parthasarathy, 2010)
have claimed that the LCA histogram distance is a
metric for trees, in this paper, we give a counterex-
Yoshino, T., Muraka, K. and Hirata, K.
LCA Histogram Distance for Rooted Labeled Caterpillars.
DOI: 10.5220/0006951603070314
In Proceedings of the 10th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2018) - Volume 1: KDIR, pages 307-314
ISBN: 978-989-758-330-8
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
307

ample that their claim is false, even if the informa-
tion of depth is given, which is not well-known. On
the other hand, we show that the LCA histogram dis-
tance is a metric for caterpillars. By using the LCA
histogram distance, we can avoid not only the above
extreme cases but also the case that both the path his-
togram distance and the complete subtree histogram
distance are their maximum values but the edit dis-
tance is not. We can compute the LCA histogram dis-
tance in quadratic time.
Then, by using caterpillars in real data in Table 3
in Section 4, we give experimental results of comput-
ing the LCA histogram distance comparing with the
path histogram distance and the complete subtree his-
togram distance. Note that the maximum values of
the path histogram distance, the complete subtree his-
togram distance and the LCA histogram distance are
different. Then, by normalizing the distances to com-
pare them as experimental results, we compare the
running time, distributions and scatter charts of the
three distances.
2 PRELIMINARIES
A tree T is a connected graph (V, E) without cycles,
where V is the set of vertices and E is the set of edges.
We denote V and E by V(T) and E(T). The size of
T is |V| and denoted by |T|. We sometime denote
v ∈ V(T) by v ∈ T. We denote an empty tree (
/
0,
/
0)
by
/
0. A rooted tree is a tree with one vertex r chosen
as its root. We denote the root of a rooted tree T by
r(T).
Let T be a rooted tree such that r = r(T) and
u,v,w ∈ T. We denote the unique path from r to v, that
is, the tree (V
′
,E
′
) such that V
′
= {v
1
,.. .,v
k
}, v
1
= r,
v
k
= v and (v
i
,v
i+1
) ∈ E
′
for every i (1 ≤ i ≤ k − 1),
by UP
r
(v). The depth of v, denoted by d(v), is the
number of edges in UP
r
(v).
The parent of v(6= r), which we denote by par(v),
is its adjacent vertex on UP
r
(v) and the ancestors of
v(6= r) are the vertices on UP
r
(v) − {v}. We say that
u is a child of v if v is the parent of u and u is a de-
scendant of v if v is an ancestor of u. We call a vertex
with no children a leaf and denote the set of all the
leaves in T by lv(T).
We denote the set of all the children of v in T by
ch(v). The degreeof v, denoted by g(v), is the number
of children of v, that is, |ch(v)|, and the degree of T,
denoted by g(T), is max{g(v) | v ∈ T}. The height of
v, denoted by h(v), is max{|UP
v
(w)| | w ∈ lv(T[v])},
and the height of T, denoted by h(T), is max{h(v) |
v ∈ T}.
We use the ancestor orders < and ≤, that is, u < v
if v is an ancestor of u and u ≤ v if u < v or u = v.
We say that w is the least common ancestor (LCA, for
short) of u and v, denoted by u⊔v, if u ≤ w, v≤ w and
there exists no vertex w
′
∈ T such that w
′
≤ w, u ≤ w
′
and v ≤ w
′
.
Let T be a rooted tree (V, E) and v a vertex in T.
A complete subtree of T at v, denoted by T[v], is a
rooted tree T
′
= (V
′
,E
′
) such that r(T
′
) = v, V
′
=
{u ∈ V | u ≤ v} and E
′
= {(u, w) ∈ E | u,w ∈ V
′
}. For
a tree T
′
, we say that T
′
occurs in T at v if T
′
= T[v].
For a vertex v ∈ T, we call the occurrence number
of v in the preorder (resp., postorder) traversal on T
the preorder (resp., postorder) number of v and de-
note it by pre(v) (resp., post(v)). We say that u is to
the left of v in T if pre(u) ≤ pre(v) and post(u) ≤
post(v). We say that a rooted tree is ordered if a left-
to-right order among siblings is given; unordered oth-
erwise. We say that a rooted tree is labeled if each
vertex is assigned a symbol from a fixed finite alpha-
bet Σ. For a vertex v, we denote the label of v by l(v),
and sometimes identify v with l(v). In this paper, we
call a rooted labeled unordered tree a tree simply.
As the restricted form of trees, we introduce a
rooted labeled caterpillar (a caterpillar, for short) as
follows, which this paper mainly deals with.
Definition 1 (Caterpillar (cf., (Gallian, 2007))). We
say that a tree is a caterpillar if it is transformed to a
path after removing all the leaves in it. For a caterpil-
lar C, we call the remained path a backbone of C and
denote it by bb(C).
Next, we introduce an edit distance for trees.
Definition 2 (Edit operations (Tai, 1979)). The edit
operations of a tree T are defined as follows.
1. Substitution: Change the label of the vertex v in
T.
2. Deletion: Delete a non-root vertex v in T with par-
ent v
′
, making the children of v become the chil-
dren of v
′
. The children are inserted in the place
of v as a subset of the children of v
′
.
3. Insertion: The complement of deletion. Insert a
vertex v as a child of v
′
in T making v the parent
of a subset of the children of v
′
.
Let ε 6∈ Σ denote a special blank symbol and define
Σ
ε
= Σ ∪ {ε}. Then, we represent each edit operation
by (l
1
7→ l
2
), where (l
1
,l
2
) ∈ (Σ
ε
×Σ
ε
−{(ε,ε)}). The
operation is a substitution if l
1
6= ε and l
2
6= ε, a dele-
tion if l
2
= ε, and an insertion if l
1
= ε. For vertices v
and w, we also denote (l(v) 7→ l(w)) by (v 7→ w). We
define a cost function γ : (Σ
ε
× Σ
ε
\ {(ε,ε)}) 7→ R
+
on
pairs of labels. We often constrain a cost function γ to
be a metric, that is, γ(l
1
,l
2
) ≥ 0, γ(l
1
,l
2
) = 0 iff l
1
= l
2
,
γ(l
1
,l
2
) = γ(l
2
,l
1
) and γ(l
1
,l
3
) ≤ γ(l
1
,l
2
)+γ(l
2
,l
3
). In
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
308

particular, we call the cost function that γ(l
1
,l
2
) = 1
if l
1
6= l
2
a unit cost function.
Definition 3 (Edit distance (Tai, 1979)). For a cost
function γ, the cost of an edit operation e = l
1
7→ l
2
is given by γ(e) = γ(l
1
,l
2
). The cost of a sequence
E = e
1
,.. .,e
k
of edit operations is given by γ(E) =
∑
k
i=1
γ(e
i
). Then, an edit distance τ
TAI
(T
1
,T
2
) be-
tween trees T
1
and T
2
is defined as follows:
τ
TAI
(T
1
,T
2
) = min
γ(E)
E is a sequence
of edit operations
transforming T
1
to T
2
.
For n
i
= |T
i
| (i = 1, 2), it holds that 0 ≤
τ
TAI
(T
1
,T
2
) ≤ n
1
+ n
2
− 1.
Unfortunately, the problem of computing the edit
distance between trees is MAX SNP-hard (Zhang and
Jiang, 1994). On the other hand, Muraka et al. (Mu-
raka et al., 2018) have recently shown the following
theorem for caterpillars.
Theorem 1 ((Muraka et al., 2018)). For caterpil-
lars C
1
and C
2
, we can compute τ
TAI
(C
1
,C
2
) in
O(λ
2
h
2
) time, where λ = max{|lv(C
1
)|,|lv(C
2
)|} and
h = max{h(C
1
),h(C
2
)}.
As the previous local frequency distances to com-
pare caterpillars, we introduce the path histogram dis-
tance (Kawaguchiet al., 2018b) and the complete sub-
tree histogram distance (Akutsu et al., 2013).
Let T be a tree such that r = r(T). Then,
for v ∈ lv(T), we regard the path P = UP
r
(v)
such that V(P) = {v
1
,.. .,v
k
}, v
1
= r, v
k
= v and
(v
i
,v
i+1
) ∈ E(P) for every i (1 ≤ i ≤ k− 1) as a string
l(v
1
)··· l(v
k
) on Σ and denote it by s(r,v). Also we
say that a string s ∈ Σ
∗
occurs in T if there exists a leaf
v ∈ lv(T) such that s = s(r,v) and denote the number
of occurrences of s in T by f(s,T). Furthermore, we
define S (T) as {s(r,v) | r = r(T),v ∈ lv(T)}.
Definition 4 (Path histogram distance (Kawaguchi
et al., 2018b)). For a tree T, a path histogram H
P
(T)
of T consists of pairs hs, f(s,T)i for every s ∈ S (T).
For trees T
1
and T
2
, a path histogram distance
δ
P
(T
1
,T
2
) between T
1
and T
2
is defined as an L
1
-
distance between H
P
(T
1
) and H
P
(T
2
):
δ
P
(T
1
,T
2
) =
∑
s∈S(T
1
)∪S (T
2
)
| f(s,T
1
) − f(s,T
2
)|.
For λ = |lv(T)|, it is obvious that |H
P
(T)| ≤ λ and
∑
s∈S (T)
f(s,T) = λ.
We denote the set {T[v] | v ∈ T} of all the com-
plete subtrees in T by C (T). For c ∈ C (T), the num-
ber of occurrences of c in T by f(c,T).
Definition 5 (Complete subtree histogram dis-
tance (Akutsu et al., 2013)). For a tree T, a com-
plete subtree histogram H
CS
(T) consists of pairs
hs, f(s,T)i for every s ∈ C (T).
For trees T
1
and T
2
, a complete subtree histogram
distance δ
CS
(T
1
,T
2
) between trees T
1
and T
2
is defined
as an L
1
-distance between H
CS
(T
1
) and H
CS
(T
2
):
δ
CS
(T
1
,T
2
) =
∑
c∈C (T
1
)∪C (T
2
)
| f(c, T
1
) − f(c,T
2
)|.
For n = |T|, it is obvious that |H
CS
(T)| ≤ n and
∑
c∈C (T)
f(c,T) = n.
We summarize the properties of δ
P
and δ
CS
as fol-
lows (Akutsu et al., 2013; Kawaguchi et al., 2018a;
Kawaguchi et al., 2018b).
Theorem 2. Let C
1
and C
2
be caterpillars such that
n = max{|C
1
|,|C
2
|} and λ = max{|lv(C
1
)|,|lv(C
2
)|}.
1. δ
P
is a metric for caterpillars but not a metric for
trees in general.
2. δ
CS
is a metric for trees, so is for caterpillars.
3. We can compute δ
P
(C
1
,C
2
) and δ
CS
(C
1
,C
2
) in
O(n) time.
4. τ
TAI
(C
1
,C
2
) ≤ δ
CS
(C
1
,C
2
).
5. There exist C
1
and C
2
such that τ
TAI
(C
1
,C
2
) =
δ
CS
(C
1
,C
2
) = 1 but δ
P
(C
1
,C
2
) = O(λ).
6. There exist C
1
and C
2
such that δ
P
(C
1
,C
2
) = 2 but
τ
TAI
(C
1
,C
2
) = δ
CS
(C
1
,C
2
) = O(n).
3 LCA HISTOGRAM DISTANCE
Let T be a tree. Then, we say that p = ((l
1
,d
1
) :
{(l
2
,d
2
),(l
3
,d
3
)}) is an LCA pivot in T if there ex-
ist mutually distinct vertices v and w in T such that
l
1
= l(v ⊔ w), d
1
= d(v ⊔ w), l
2
= l(v), d
2
= d(v),
l
3
= l(w) and d
3
= d(w), respectively. We denote p by
a 6-tuple (l
1
d
1
: l
2
d
2
⊔ l
3
d
3
) simply. In this case, we
also say that p occurs in T and denote p by p(v, w).
We denote the number of the occurrences of p in T
by f(p,T). Furthermore, we denote the set of all the
LCA pivots in T by P (T), that is, P (T) = {p(v,w) |
(v,w) ∈ T × T,v 6= w}.
Definition 6 (LCA histogram distance). For a tree T,
an LCA histogram H
LCA
(T) of T consists of a pair
hp, f(p, T)i for every p ∈ P (T).
For two trees T
1
and T
2
, an LCA histogram dis-
tance δ
LCA
(T
1
,T
2
) between T
1
and T
2
is defined as an
L
1
-distance between H
LCA
(T
1
) and H
LCA
(T
2
):
δ
LCA
(T
1
,T
2
) =
∑
p∈P (T
1
)∪P (T
2
)
| f(p, T
1
) − f(p,T
2
)|.
For n = |T|, it is obvious that |H
LCA
(T)| ≤ n(n−
1)/2 and
∑
p∈P (T)
f(p,T) = n(n− 1)/2.
Example 1. Let C
1
and C
2
be caterpillars illustrated
in Figure 1.
Then, we obtain the histograms H
LCA
(C
1
)
and H
LCA
(C
2
) illustrated in Table 1. Note
LCA Histogram Distance for Rooted Labeled Caterpillars
309
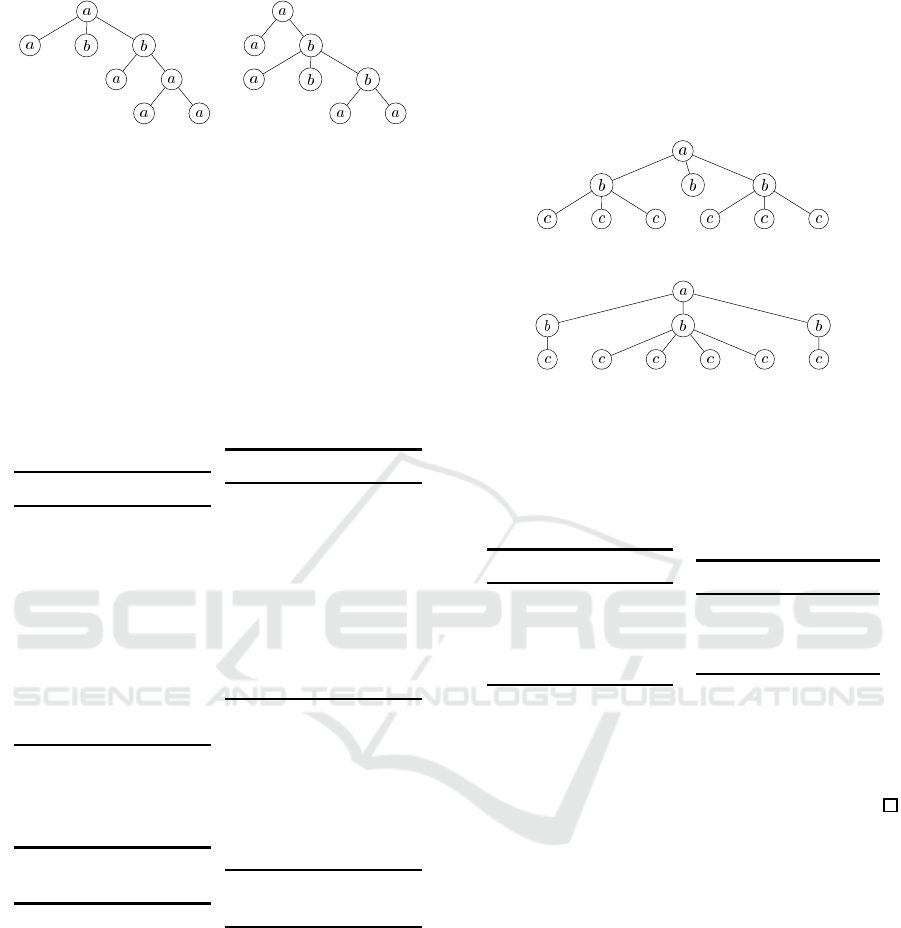
C
1
C
2
Figure 1: The caterpillars C
1
and C
2
in Example 1.
that, since |C
1
| = |C
2
| = 8, it holds that
∑
p∈P (C
i
)
f(p,C
i
) =
8
C
2
= 28 for i = 1, 2. Also,
the bold faces illustrate the LCA pivots occurring in
either H
LCA
(C
1
) or H
LCA
(C
2
) and its frequency, or
the frequencies of the LCA pivot if they are different
in H
LCA
(C
1
) and H
LCA
(C
2
).
Table 1: The histograms H
LCA
(C
1
) and H
LCA
(C
2
).
H
LCA
(C
1
) H
LCA
(C
2
)
LCA pivots freq.
(a0 : a1⊔ b1) 2
(a0 : a1⊔ a2) 2
(a0 : a1⊔ a3) 2
(a0 : b1⊔ b1) 1
(a0 : b1⊔ a2) 2
(a0 : b1⊔ a3) 2
(a0 : a0⊔ a1) 1
(a0 : a0⊔ b1) 2
(a0 : a0⊔ a2) 2
(a0 : a0⊔ a3) 2
(b1 : a2⊔ a2) 1
(b1 : a2⊔ a3) 2
(b1 : b1⊔ a2) 2
(b1 : b1⊔ a3) 2
(a2 : a3⊔ a3) 1
(a2 : a2⊔ a3) 2
LCA pivots freq.
(a0 : a1⊔ b1) 1
(a0 : a1⊔ a2) 1
(a0 : a1⊔ b2) 2
(a0 : a1⊔ a3) 2
(a0 : a0⊔ a1) 1
(a0 : a0⊔ b1) 1
(a0 : a0⊔ a2) 1
(a0 : a0⊔ b2) 2
(a0 : a0⊔ a3) 2
(b1 : a2⊔ b2) 2
(b1 : a2⊔ a3) 2
(b1 : b2⊔ b2) 1
(b1 : b2⊔ a3) 2
(b1 : b1⊔ a2) 1
(b1 : b1⊔ b2) 2
(b1 : b1⊔ a3) 2
(b2 : a3⊔ a3) 1
(b2 : b2⊔ a3) 2
Hence, it holds that:
δ
LCA
(C
1
,C
2
)
=
∑
p∈P (C
1
)∪P (C
2
)
| f(p,C
1
) − f(p,C
2
)|
=
∑
p∈P (C
1
)\P (C
2
)
f(p,C
1
) +
∑
p∈P(C
2
)\P (C
1
)
f(p,C
2
)
+
∑
p∈P (C
1
)∩P (C
2
)
| f(p,C
1
) − f(p,C
2
)|
= 9+ 14+ 5 = 28.
Whereas the LCA histogram distance seems to be
a metric, for example, Theorem 3.2 in (Tatikonda and
Parthasarathy, 2010), we show that it is not a metric
for trees as follows.
Theorem 3. There exist trees T
1
and T
2
such that
H
LCA
(T
1
) = H
LCA
(T
2
) but T
1
6= T
2
. Hence, the LCA
histogram distance is not a metric for trees in general.
Proof. Consider the trees T
1
and T
2
in Figure 2.
T
1
T
2
Figure 2: Trees T
1
and T
2
.
Then, we obtain the histogram H
LCA
(T
1
)(=
H
LCA
(T
2
)) illustrated in Table 2.
Table 2: The histogram H
LCA
(T
1
)(= H
LCA
(T
2
)).
LCA pivots freq.
(a0 : c2⊔ c2) 9
(a0 : b1⊔ c2) 12
(a0 : b1⊔ b1) 3
(a0 : a0⊔ b1) 3
LCA pivots freq.
(a0 : a0 ⊔ c2) 6
(b1 : c2⊔ c2) 6
(b1 : c2⊔ b1) 6
Here, since |T
1
| = |T
2
| = 10, it holds that
∑
p∈P (T
i
)
f(p,T
i
) =
10
C
2
= 45 for i = 1,2. Furthermore,
since the labels are not essential, this statement also
holds for unlabeled trees.
On the other hand, note that neither T
1
nor T
2
in
Figure 2 is a caterpillar. In the remainder of this sec-
tion, we discuss the LCA histogram distance between
caterpillars.
For caterpillars, the following lemma is obvious.
Lemma 1. Let p(v,w) = (l
1
d
1
: l
2
d
2
⊔ l
3
d
3
) ∈ P (C)
an LCA pivot in C. Then, the following statements
hold.
1. It holds that v⊔ w ∈ bb(C).
2. If v,w ∈ lv(C), then it holds that d
1
=
min{d
2
,d
3
} − 1. Also it holds that v⊔w = par(v)
if d
2
< d
3
, v⊔ w = par(w) if d
3
< d
2
and v ⊔ w =
par(v) = par(w) if d
2
= d
3
.
3. If v,w ∈ bb(C), then it holds that d
1
=
min{d
2
,d
3
}. Also it holds that v⊔w = v if d
2
< d
3
and v⊔ w = w if d
3
< d
2
.
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
310
4. Suppose that v ∈ lv(C) and w ∈ bb(C). If d
2
< d
3
,
then it holds that d
1
= d
3
and v ⊔ w = w. Other-
wise, that is, d
2
≥ d
3
, it holds that d
1
= d
2
−1 and
v⊔ w = par(v).
Then, the following theorem holds.
Theorem 4. For caterpillars, the LCA histogram dis-
tance is a metric.
Proof. By the definition, it is sufficient to show
that two caterpillars C
1
and C
2
are isomorphic iff
δ
LCA
(C
1
,C
2
) = 0. In other words, it is sufficient
to show that we can transform a caterpillar C from
H
LCA
(C) uniquely.
By Lemma 1.1, we can uniquely determine bb(C)
from P (C) because of l
1
and d
1
in p(v, w). Since
lv(C) = C \ bb(C), we can determine lv(C). Then, by
Lemma 1.2, we can determine the set of leaves with
depth i for every i (1 ≤ i ≤ d(C)).
For λ
i
= |lv(C
i
)| and n
i
= |C
i
| (i = 1,2), it holds
that 0 ≤ δ
P
(C
1
,C
2
) ≤ λ
1
+ λ
2
, 0 ≤ δ
CS
(C
1
,C
2
) ≤
n
1
+ n
2
and 0 ≤ δ
LCA
(C
1
,C
2
) ≤ (n
1
(n
1
− 1)+ n
2
(n
2
−
1))/2. Then, consider the extreme cases in Section 1.
Example 2. Let C
1
and C
2
be paths with length n.
Suppose that every vertex in C
1
is labeled by a and
that in C
2
by b. Then, it holds that τ
TAI
(C
1
,C
2
) = n,
δ
P
(C
1
,C
2
) = 1, δ
CS
(C
1
,C
2
) = 2n and δ
LCA
(C
1
,C
2
) =
2n(n − 1). Note that δ
P
(C
1
,C
2
), δ
CS
(C
1
,C
2
) and
δ
LCA
(C
1
,C
2
) are their maximum values.
Suppose that every vertex in C
1
and every non-
leaf vertex in C
2
is labeled by a and the leaf of C
2
is labeled by b. Then, it holds that τ
TAI
(C
1
,C
2
) = 1,
δ
P
(C
1
,C
2
) = 1, δ
CS
(C
1
,C
2
) = 2n and δ
LCA
(C
1
,C
2
) =
2(n − 1). Note that δ
P
(C
1
,C
2
) and δ
CS
(C
1
,C
2
) are
their maximum values but δ
LCA
(C
1
,C
2
) is not.
In particular, δ
P
(C
1
,C
2
) cannot distinguish the dif-
ference of labels between two paths C
1
and C
2
.
Furthermore, the following theorem holds.
Theorem 5. There exist caterpillars C
1
and C
2
satis-
fying the following statements.
1. τ
TAI
(C
1
,C
2
) = δ
CS
(C
1
,C
2
) = 1 but δ
P
(C
1
,C
2
) and
δ
LCA
(C
1
,C
2
) are their maximum values.
2. δ
P
(C
1
,C
2
) and δ
CS
(C
1
,C
2
) are their maximum
values but τ
TAI
(C
1
,C
2
) and δ
LCA
(C
1
,C
2
) are not.
Proof. 1. Let C
1
and C
2
be stars, that is, |bb(C
1
)| =
|bb(C
2
)| = 1, such that r(C
1
) = r
1
, r(C
2
) = r
2
, l(r
1
) 6=
l(r
2
), ch(r
1
) = ch(r
2
) and |ch(r
1
)| = |ch(r
2
)| = n−1.
Then, it is obvious that τ
TAI
(C
1
,C
2
) = δ
CS
(C
1
,C
2
) =
1 and δ
P
(C
1
,C
2
) = 2(n − 1). Also, since P (C
1
) ∩
P (C
2
) =
/
0, it holds that δ
LCA
(C
1
,C
2
) = 2n(n− 1).
2. Let C
1
and C
2
be caterpillars obtained by con-
necting λ leaves to the leaves of paths with length h,
where every vertex in C
1
and in a path in C
2
is la-
beled by a and every leaf in C
2
by b. Then, |C
1
| =
|C
2
| = h + λ = n. It is obvious that δ
P
(C
1
,C
2
) = 2λ
and δ
CS
(C
1
,C
2
) = 2(λ + h) = 2n, so they are the
maximum values. On the other hand, it holds that
τ
TAI
(C
1
,C
2
) = λ and δ
LCA
(C
1
,C
2
) = 2λ(n−1), where
their maximum values are 2n− 1 and 2n(n− 1).
By selecting every pair of vertices in two cater-
pillars, we can compute δ
LCA
(C
1
,C
2
) in O(n
2
) time,
because H
LCA
(C) = O(n
2
).
Note that the inequality that δ
P
< δ
CS
< δ
LCA
tends
to hold by the values of δ
P
, δ
CS
and δ
LCA
. Then, we
normalize δ
P
, δ
CS
and δ
LCA
by dividing their maxi-
mum values when comparing distances. We denote
the normalized distances of δ
P
, δ
CS
and δ
LCA
by δ
∗
P
,
δ
∗
CS
and δ
∗
LCA
, respectively. Then, the following ex-
ample shows that the inequality that δ
∗
P
< δ
∗
CS
< δ
∗
LCA
does not always hold.
Example 3. Consider caterpillars C
1
, C
2
and C
3
in
Figure 3.
C
1
C
2
C
3
Figure 3: Caterpillars C
1
, C
2
and C
3
in Example 3.
Then, we obtain δ
P
(C
i
,C
j
), δ
CS
(C
i
,C
j
),
δ
LCA
(C
i
,C
j
), δ
∗
P
(C
i
,C
j
), δ
∗
CS
(C
i
,C
j
) and δ
∗
LCA
(C
i
,C
j
)
for (i, j) = (1, 2),(1,3), (2,3) as follows.
(i, j) δ
P
δ
CS
δ
LCA
δ
∗
P
δ
∗
CS
δ
∗
LCA
(1,2) 4 3 10 2/3 1/3 5/8
(1,3) 2 4 6 1/3 2/5 3/10
(2,3) 2 3 4 1/3 1/3 1/4
Hence, the following statements hold:
δ
∗
CS
(C
1
,C
2
) < δ
∗
LCA
(C
1
,C
2
) < δ
∗
P
(C
1
,C
2
),
δ
∗
LCA
(C
1
,C
3
) < δ
∗
P
(C
1
,C
3
) < δ
∗
CS
(C
1
,C
3
),
δ
∗
LCA
(C
2
,C
3
) < δ
∗
P
(C
2
,C
3
) = δ
∗
CS
(C
2
,C
3
).
4 EXPERIMENTAL RESULTS
Table 3 illustrates the number (#cat) of caterpillars in
the datasets in N-glycans and all of the glycans from
KEGG
1
, CSLOGS
2
and dblp
3
datasets, whose num-
ber of data is denoted by #data.
1
Kyoto Encyclopedia of Genes and Genomes,
http://www.kegg.jp/
2
http://www.cs.rpi.edu/˜zaki/www-new/pmwiki.php
/Software/Software
3
http://dblp.uni-trier.de/
LCA Histogram Distance for Rooted Labeled Caterpillars
311
Table 3: The number of caterpillars in N-glycans and all-
glycans from KEGG, CSLOGS and dblp datasets.
dataset #cat #data %
N-glycans 514 2,142 23.996
all-glycans 8,005 10,704 74.785
CSLOGS 41,592 59,691 69.679
dblp 5,154,295 5,154,530 99.995
We deal with caterpillars for N-glycans, all-
glycans, CSLOGS and the selected 50,000 caterpil-
lars in dblp (we refer to dblp
−
). Table 4 illustrates
the information of such caterpillars. Here, ([a,b];c)
means that a, b and c are the minimum, the maximum
and the average number.
In the remainder of this section, we compare the
LCA histogram distance with the path histogram dis-
tance and the complete subtree histogram distance for
caterpillars.
Table 5 illustrates the running time of computing
δ
∗
P
, δ
∗
CS
and δ
∗
LCA
for N-glycans, all-glycans, CSLOGS
and dblp
−
.
Table 5 shows that, whereas we compute δ
∗
P
and
δ
∗
CS
in linear time and δ
∗
LCA
in quadratic time in theo-
retical, the running time of computing δ
∗
LCA
is within
twice for N-glycans and all-glycans, within thrice for
CSLOGS and about seven times for dblp
−
, respec-
tively, of computing δ
∗
CS
in experimental. The rea-
son why the running time of computing δ
∗
LCA
is not
so large is that the number of |H
LCA
| is not so large
except dblp
−
; For dblp
−
, |H
LCA
| is larger than others
because the number of leaves is large but the height is
small in Table 4.
Figure 4 illustrates the distributions of δ
∗
P
, δ
∗
CS
and
δ
∗
LCA
for N-glycans, all-glycans, CSLOGS and dblp
−
.
Figure 4 shows that almost of the distributions
concentrate near to 1, in particular, CSLOGS and
dblp
−
. On the other hand, for dblp
−
, the distributions
appear near to 0. For N-glycans and all-glycans, δ
∗
P
is
larger than δ
∗
CS
and δ
∗
CS
is larger than δ
∗
LCA
.
Figure 5 illustrates the detailed distributions of δ
∗
P
,
δ
∗
CS
and δ
∗
LCA
for N-glycans, all-glycans, CSLOGS
and dblp
−
, where the scopes of the distances of N-
glycans, all-glycans, CSLOGS and dblp
−
are [0.8,1],
[0.9,1], [0.995,1] and [0.99.1], respectively.
Note that, for dblp
−
, since the maximum value of
δ
∗
CS
is 0.992308 and the frequency is low, the distribu-
tion is just of δ
∗
P
and δ
∗
LCA
. Figure 5 shows that, near
to 1 and for N-glycans, all-glycans and CSLOGS, the
inequality of δ
∗
P
< δ
∗
CS
< δ
∗
LCA
holds.
Figure 6 illustrates the scatter charts of δ
∗
P
, δ
∗
CS
and
δ
∗
LCA
for N-glycans and all-glycans and Figure 7 illus-
trates those for CSLOGS and dblp
−
, and their cor-
relation coefficients (cc). Here, the representation of
0
10
20
30
40
50
60
70
80
90
0 0.2 0.4 0.6 0.8 1
percentage(%)
distance
LCA
CS
path
0
10
20
30
40
50
60
70
80
90
100
0 0.2 0.4 0.6 0.8 1
percentage(%)
distance
LCA
CS
path
N-glycans all-glycans
0
10
20
30
40
50
60
70
80
90
100
0 0.2 0.4 0.6 0.8 1
percentage(%)
distance
LCA
CS
path
0
10
20
30
40
50
60
0 0.2 0.4 0.6 0.8 1
percentage(%)
distance
LCA
CS
path
CSLOGS dblp
−
Figure 4: The distributions of δ
∗
P
, δ
∗
CS
and δ
∗
LCA
for N-
glycans, all-glycans, CSLOGS and dblp
−
.
0
10
20
30
40
50
60
70
80
90
0.8 0.82 0.84 0.86 0.88 0.9 0.92 0.94 0.96 0.98 1
percentage(%)
distance
LCA
CS
path
0
10
20
30
40
50
60
70
80
90
100
0.9 0.91 0.92 0.93 0.94 0.95 0.96 0.97 0.98 0.99 1
percentage(%)
distance
LCA
CS
path
N-glycans all-glycans
0
10
20
30
40
50
60
70
80
90
100
0.995 0.996 0.997 0.998 0.999 1
percentage(%)
distance
LCA
CS
path
0
10
20
30
40
50
60
0.99 0.992 0.994 0.996 0.998 1
percentage(%)
distance
LCA
CS
path
CSLOGS dblp
−
Figure 5: The detailed distributions of δ
∗
P
, δ
∗
CS
and δ
∗
LCA
for
N-glycans, all-glycans, CSLOGS and dblp
−
.
δ
∗
Y
/δ
∗
X
means that the number of pairs of caterpillars
with δ
∗
X
is pointed at the x-axis and that with δ
∗
Y
is
pointed at the y-axis.
Figures 6 and 7 show that, the scatter charts for N-
glycans and all-glycans in Figure 6 are more sparse
than those for CSLOGS and dblp
−
in Figure 7, be-
cause the number of caterpillars in N-glycans and
all-glycans is much smaller than that in CSLOGS
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
312

Table 4: The information of caterpillars in N-glycans, all-glycans, CSLOGS and dblp
−
.
dataset #vertices degree height #leaves #labels
N-glycans ([6,15];6.40) ([1,3];1.84) ([1,9];4.22) ([1,7];2.18) ([2,8];4.50)
all-glycans ([1,24];4.74) ([0,5];1.49) ([0,15];3.02) ([1,14];1.72) ([1,9];2.84)
CSLOGS ([2,404];5.84) ([1,403];3.05) ([1,70];2.20) ([1,403];3.64) ([2,168];5.18)
dblp
−
([7,244];11.96) ([6,243];10.94) ([1,3];1.02) ([6,243];10.94) ([7,13];9.86)
Table 5: The running time of computing δ
∗
P
, δ
∗
CS
and δ
∗
LCA
(msec.).
dataset δ
∗
P
δ
∗
CS
δ
∗
LCA
N-glycans 142 239 419
all-glycans 34,113 40,364 73,219
CSLOGS 1,017,730 1,361,343 3,439,560
dblp
−
1,980,062 3,534,120 24,633,812
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
path
CS
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
path
LCA
N-glycans, δ
∗
P
/δ
∗
CS
N-glycans, δ
∗
P
/δ
∗
LCA
cc = 0.402189 cc = 0.804891
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
CS
LCA
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
path
CS
N-glycans, δ
CS
/δ
LCA
all-glycans, δ
∗
P
/δ
∗
CS
cc = 0.356957 cc = 0.281586
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
path
LCA
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
CS
LCA
all-glycans, δ
∗
P
/δ
∗
LCA
all-glycans, δ
∗
CS
/δ
∗
LCA
cc = 0.571927 cc = 0.413714
Figure 6: The scatter charts of δ
∗
P
, δ
∗
CS
and δ
∗
LCA
for N-
glycans and all-glycans.
and dblp
−
. Also for all datasets, the scatter chart
for δ
∗
CS
/δ
∗
LCA
spreads more widely than those for
δ
∗
P
/δ
∗
LCA
and δ
∗
P
/δ
∗
CS
.
For Figure 6, the scatter charts for N-glycans have
the values on the line that y = 1 and, in particular, the
scatter charts of δ
∗
CS
/δ
∗
LCA
also have the values on the
line that x = 1. On the other hand, the scatter charts
for all-glycans have the values on the line that y = 1,
those of δ
∗
P
/δ
∗
CS
and δ
∗
CS
/δ
∗
LCA
the vales on the lines
that x = 1 and y = 0.
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
path
CS
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
path
LCA
CSLOGS, δ
∗
P
/δ
∗
CS
CSLOGS, δ
∗
P
/δ
∗
LCA
cc = 0.735274 cc = 0.841885
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
CS
LCA
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
path
CS
CSLOGS, δ
∗
CS
/δ
∗
LCA
dblp
−
, δ
∗
P
/δ
∗
CS
cc = 0.645293 cc = 0.568405
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
path
LCA
0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
CS
LCA
dblp
−
, δ
P
/δ
LCA
dblp
−
, δ
∗
CS
/δ
∗
LCA
cc = 0.980705 cc = 0.644874
Figure 7: The scatter charts δ
∗
P
, δ
∗
CS
and δ
∗
LCA
for CSLOGS
and dblp
−
and their correlation coefficients (cc).
LCA Histogram Distance for Rooted Labeled Caterpillars
313

For Figure 7, the scatter charts for CSLOGS have
the values on the line that y = 1 and those of δ
∗
CS
/δ
∗
LCA
have the values on the line that x = 1. On the other
hand, the scatter charts of δ
∗
P
/δ
∗
CS
for dblp
−
have the
values on the line that y = 1 and those of δ
∗
CS
/δ
∗
LCA
have the values on the line that x = 1. In particular,
the scatter charts for dblp
−
constitutes at most two
clusters, where one lies on the axis.
For correlation coefficients, which we denote by
cc(δ
∗
Y
/δ
∗
X
), the value of cc(δ
∗
P
/δ
∗
LCA
) is highest for all
the data. On the other hand, it holds that
cc(δ
∗
CS
/δ
∗
LCA
) < cc(δ
∗
P
/δ
∗
CS
) < cc(δ
∗
P
/δ
∗
LCA
)
for N-glycans and CSLOGS, whereas it holds that
cc(δ
∗
P
/δ
∗
CS
) < cc(δ
∗
CS
/δ
∗
LCA
) < cc(δ
∗
P
/δ
∗
LCA
)
for all-glycans and dblp
−
. For the values of corre-
lation coefficients, almost of the distances are related
for CSLOGS and dblp
−
, because cc(δ
X
/δ
Y
) is greater
than 0.6, just δ
∗
LCA
is related with δ
∗
P
for N-glycans,
and no distances are related for all-glycans. In par-
ticular, cc(δ
∗
P
δ
∗
LCA
) is greater than 0.8 for N-glycans,
CSLOGS and dblp
−
.
5 CONCLUSION
In this paper, we have introduced an LCA histogram
distance δ
LCA
between trees and shown that it is not
a metric for trees but is a metric for caterpillars. Fur-
thermore, we have given experimental results of com-
puting δ
LCA
for caterpillars, by comparing the path
histogram distance δ
P
and the complete subtree his-
togram distance δ
CS
(or their normalized distances
δ
∗
LCA
, δ
∗
P
and δ
∗
CS
).
It is a future work to design the algorithm to com-
pute δ
LCA
more efficiently, without constructing LCA
histograms explicitly, for example. It is also a future
work to analyze the relationship between δ
LCA
, δ
P
and
δ
CS
(or δ
∗
LCA
, δ
∗
P
and δ
∗
CS
) in more detail in experimen-
tal, in particular, as stated in Section 4, to analyze why
the correlation coefficients of δ
∗
P
and δ
∗
LCA
have been
high, and that in theoretical.
Furthermore, it is a future work to give experimen-
tal results for other data of caterpillars. Finally, it is
an important future work to analyze the relationship
between δ
LCA
and τ
TAI
(Muraka et al., 2018).
ACKNOWLEDGEMENTS
This work is partially supported by Grant-in-Aid
for Scientific Research 17H00762, 16H02870 and
16H01743 from the Ministry of Education, Culture,
Sports, Science and Technology, Japan.
REFERENCES
Akutsu, T., Fukagawa, D., Halld´orsson, M. M., Takasu, A.,
and Tanaka, K. (2013). Approximation and parame-
terized algorithms for common subtrees and edit dis-
tance between unordered trees. Theoret. Comput. Sci.,
470:10–22.
Aratsu, T., Hirata, K., and Kuboyama, T. (2009). Sibling
distance for rooted labeled trees. In JSAI PAKDD’08
Post-Workshop Proc. (LNAI 5433), pages 99–110.
Gallian, J. A. (2007). A dynamic survey of graph labeling.
Electorn. J. Combin., 14:DS6.
Kailing, K., Kriegel, H.-P., Sch¨onaur, S., and Seidl, T.
(2004). Efficient similarity search for hierarchical data
in large databases. In Proc. EDBT’04, pages 676–693.
Kawaguchi, T., Yoshino, T., and Hirata, K. (2018a). Path
histogram distance and complete subtree histogram
distance for rooted labeled caterpillars. (submitted).
Kawaguchi, T., Yoshino, T., and Hirata, K. (2018b). Path
histogram distance for rooted labeled caterpillars. In
Proc. ACIIDS’18 (LNAI 10751), pages 276–286.
Li, F., Wang, H., Li, J., and Gao, H. (2013). A survey on
tree edit distance lower bound estimation techniques
for similarity join on XML data. SIGMOD Record,
43:29–39.
Muraka, K., Yoshino, T., and Hirata, K. (2018). Computing
edit distance between rooted labeled caterpillars. In
Proc. FedCSIS’18 (to appear).
Tai, K.-C. (1979). The tree-to-tree correction problem. J.
ACM, 26:422–433.
Tatikonda, S. and Parthasarathy, S. (2010). Hashing tree-
structured data: Methods and applications. In Proc.
ICDM’10, pages 429–440.
Zhang, K. and Jiang, T. (1994). Some MAX SNP-hard re-
sults concerning unordered labeled trees. Inform. Pro-
cess. Lett., 49:249–254.
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
314
