
HEXTRATO: Using Ontology-based Constraints to Improve Accuracy
on Learning Domain-specific Entity and Relationship Embedding
Representation for Knowledge Resolution
Hegler Tissot
C3SL, Universidade Federal do Paran
´
a, Curitiba, Brazil
Keywords:
Knowledge Resolution, Knowledge Embedding, Link Prediction, Knowledge Completion, Electronic Health
Records.
Abstract:
This paper focuses the problem of learning the knowledge low-dimensional embedding representation for
entities and relations extracted from domain-specific datasets. Existing embedding methods aim to represent
entities and relations from a knowledge graph as vectors in a continuous low-dimensional space. Different
approaches have been proposed, being usually evaluated on standard benchmark knowledge graphs, such as
Wordnet and Freebase. However, the nature of such data sources prevents those methods of taking advantage
of more detailed and enriched metadata, lacking more accurate results on the evaluation tasks. In this paper, we
propose HEXTRATO, a novel embedding approach that extends a traditional baseline model TransE by adding
ontology-based constraints in order to better capture the relationships between categorised entities and their
symbolic representation in the vector space. Our method is evaluated on an adapted version of Freebase, on a
publicly available dataset used on machine learning benchmarks, and on two datasets in the clinical domain.
Our method outperforms the state-of-the-art accuracy on the link prediction task, evidencing the learnt entity
and relation embedding representation can be used to improve more complex embedding models.
1 INTRODUCTION
The problem of representing multi-relational data has
gained more attention in the last decade as long as
more knowledge bases become available and useful
as supporting resources for a variety of machine lear-
ning related applications. A knowledge graph (KG)
is a multi-relational dataset composed by entities
(nodes) and relations (edges). Freebase (Bollacker
et al., 2008), Google Knowledge Graph (Dong et al.,
2014), Wordnet (Fellbaum, 1998), and YAGO (Su-
chanek et al., 2007) are some well-known exam-
ples of multi-relational data. They provide reaso-
ning ability and can be used for inference, supporting
applications such as information retrieval, question-
answering systems (Gardner and Mitchell, 2015), link
prediction (Taskar et al., 2003), and knowledge reso-
lution (Lin et al., 2017).
In multi-relational data, each entity represents an
abstract concept or concrete entity of the world and
relationships are predicates that represent facts invol-
ving two entities. KGs are described in the form of
triples (h, r, t) – h and t are the head and tail enti-
ties (also known as subject and object) and r is the
predicate that represents the relation between h and
t. Knowledge embedding methods aim to represent
entities (h and t) and relations (r) as vectors in a con-
tinuous vector space, enforcing the embedding com-
patibility by using distinct scoring (loss) functions
to evaluate their representations, which implies some
transformations on the triple constituents (h, r, t), and
distinct algorithms to optimize the margin-based ob-
jective function.
KGs are usually created based on facts extracted
from unstructured or semi-structured data sources, so
they are typically inaccurate and incomplete. Lear-
ning the distributed representation of multi-relational
data provides an efficient tool to complete knowledge
bases without requiring extra knowledge. Thus, kno-
wledge base completion or link prediction became an
important task of automatically recovering missing
facts based on observed ones.
Embedding methods represent entities as a k-
dimensional vector in order to learn and operate on
the latent feature representation of the constituents
and on their semantic relatedness, by defining a sco-
ring function f (h,t) to measure the plausibility of the
triplet (h, r, t), where f (h,t) implies a transforma-
72
Tissot, H.
HEXTRATO: Using Ontology-based Constraints to Improve Accuracy on Learning Domain-specific Entity and Relationship Embedding Representation for Knowledge Resolution.
DOI: 10.5220/0006923700720081
In Proceedings of the 10th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2018) - Volume 1: KDIR, pages 72-81
ISBN: 978-989-758-330-8
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

tion on the pair of entities which characterises the
relation r. TransE (Bordes et al., 2013) is one of
the usual baseline methods that uses simple assump-
tions to achieve accurate and scalable results, pro-
ving to be effective and efficient even in complex
and heterogeneous multi-relational domains. After
providing the initial embedding representation in the
first learning steps, TransE is usually extended by
more complex models that use distinct techniques
and embedding representations to obtain better link
prediction performance on the benchmark datasets.
TransH (Wang et al., 2014), TransR (Lin et al., 2015),
and STransE (Nguyen et al., 2016) are some exam-
ples of other methods designed to learn and operate
on embedding representations based on TransE.
Although embedding methods have driven the at-
tention to the widely used standard benchmark data-
sets, we aim to apply knowledge representation met-
hods over more structured datasets, built with data
extracted from domain-specific information systems.
Such source systems are able to provide enriched me-
tadata and produce more dense KGs rather than the
sparse ones usually employed in the traditional eva-
luation protocols. We are particularly interested on
evaluating embedding learning methods over data ex-
tracted from patient electronic health records (EHR)
in order to create more accurate prediction models in
the clinical domain.
In this paper, we present HEXTRATO, a novel
embedding approach that extends the traditional ba-
seline model TransE by adding ontology-based con-
straints designed based on the source metadata in or-
der to better capture the relationships between enti-
ties and their symbolic representation in the vector
space. Experiments on the task of link prediction,
using an adapted version of Freebase, a publicly avai-
lable dataset used on machine learning benchmarks,
and two datasets from the clinical domain, show im-
provements of predictive accuracy over the traditional
baseline approach TransE and other similar approa-
ches. The results demonstrate our method improves
the accuracy on the evaluation task when dealing with
more structured data and metadata, evidencing the re-
sulting learnt entity and relation embedding represen-
tations can also be used to improve more complex em-
bedding models when dealing with domain-specific
categorised data.
2 RELATED WORK
Embedding models in general aim to represent enti-
ties in a k-dimensional vector space (or “embedding
space”), where k is a model hyper-parameter, so that
there is a specific similarity metric able to capture the
relationship between entities for any given relation
type, by learning how each entity interacts with other
entities with respect to all types of relations (Bordes
et al., 2011). Given a knowledge base set S of triplets
(h,r,t) composed of two entities h,t ∈ E and a rela-
tionship r ∈ R , where E denotes the set of entities
and R the set of relation types, the embedding mo-
del learns an embedding vector e ∈R
k
for each entity
and one or more embedding vectors (and/or matrices)
r ∈ R
k
(and/or r ∈ R
k×m
) for each relationship.
TransE (Bordes et al., 2013) is a baseline method
that uses simple assumptions to achieve accurate and
scalable results. TransE proved to be relatively ef-
fective and efficient by representing entities h, t and
a relation r by translation vectors h,t,r ∈ R
k
, chosen
so that the pair of embedded entities in a triple (h,r,t)
can be connected by r with low error (Equation 1).
h + r ≈t (1)
Although TransE is very efficient while achie-
ving predictive performance, it rifts on dealing with
certain kinds of relations, such as reflexive, one-
to-many, many-to-one, and many-to-many relations-
hips (Wang et al., 2014). Nevertheless, other met-
hods utilise TransE as a base model as part of the first
learning steps in order to provide the initial embed-
dings, aiming to learn better knowledge representa-
tions for complicated semantic correlations between
knowledge triples – e.g. by projecting the entity em-
bedding vector into a relation space using relation-
specific matrices. Some of these translation-based
embedding models are briefly described below.
TransH (Wang et al., 2014) models relations as
hyperplanes together with translation operations on
it. TransH overcomes the flaws regarding to those
kinds of relationships that TransE does not perform
well, by preserving the mapping properties of rela-
tions, and keeping the same model complexity and
running time of TransE. Each entity can have distinct
distributed representations when involved in different
relations, which allows entities to play different roles
in different relations. Each relation r is represented
by a vector r on a hyperplane with w
r
as the normal
vector. The entity embedding vectors h and t are first
projected to the hyperplane of w
r
(h
⊥
and t
⊥
). The
score function is similar to that used in TransE, but
using the projected embedding vectors instead (Equa-
tion 2).
f
r
(h,t) = kh
⊥
+ r −t
⊥
k
2
2
(2)
TransR (Lin et al., 2015) and ETransR (Lin et al.,
2017) model entity and relation embedding represen-
tation into separate distinct vector spaces, bridged by
a relation-specific matrix M
r
(a k-dimensional space
HEXTRATO: Using Ontology-based Constraints to Improve Accuracy on Learning Domain-specific Entity and Relationship Embedding
Representation for Knowledge Resolution
73

for entities and a m-dimensional space for relations).
These methods are mainly focused on modelling sin-
gle knowledge in continuous space instead of model-
ling the semantic relatedness between knowledges. In
these models, the entity and relation embedding di-
mensions are not necessarily identical. In ETransR,
however, all the results report k = m, which lead us to
conclude that: a) projecting entity embedding vectors
into lower dimensional spaces can lose some precious
information, and b) using higher dimensional spaces
do not necessarily add any further useful information
to the embedding model.
Structured Embedding or SE (Bordes et al., 2011)
and STransE (Nguyen et al., 2016) intend to account
for relationship asymmetry by using two relation-
specific projection matrices for entities h and t. SE
defines the score function by using two projected vec-
tors, so that:
f
r
(h,t) = kW
r,1
h −W
r,2
tk (3)
where f
r
(h,t) is large for corrupted triplets (and small
otherwise) in some subspace that depends on the re-
lationship r. STransE combines SE and TransE by
using relation-specific matrices W
r,1
and W
r,2
to iden-
tify the relation-dependent aspects, and a vector r to
capture the relationship between the entities h and
t. In STransE, a score function f
r
(h,t) (Equation
4) is used to minimize the margin-based objective
function, and performs better than the SE, TransE and
other state-of-the-art link prediction models.
f
r
(h,t) = kW
r,1
h + r −W
r,2
tk
l
1/2
(4)
TransT (Ma et al., 2017) is a recent attempt to in-
tegrate structured information and entity types in or-
der to describe the categories of entities. TransT con-
structs relation types from entity types and utilises
type-based semantic similarity to capture prior distri-
butions of entities and relations. However, it gerena-
tes multiple embedding representations of each entity
in different contexts.
Knowledge embedding methods are commonly
evaluated on standard benchmark datasets WN18 and
FB15K built with data extracted from Wordnet (Fel-
lbaum, 1998) and Freebase (Bollacker et al., 2008).
Reporting results (Lin et al., 2015; Nguyen et al.,
2016), however, evidence the lack of accuracy when
dealing with non-categorised data available in this tra-
ditional benchmark datasets. Type-based constraints
can support the statistical modelling with latent vari-
able models, by integrating prior knowledge on entity
and relation types, significantly improving these mo-
dels up to about 70% in link prediction tasks, especi-
ally when a low model complexity is enforced (Krom-
paß et al., 2015).
3 HEXTRATO
Our method couples the baseline embedding method
TransE with a set of ontology-based constraints in-
herited from the source metadata in order to improve
both the accuracy and validation performance when
dealing with more structured and well categorized
domain-specific data.
3.1 Ontology-based Constraints
3.1.1 Typed Entities
As long as the source database provides categorised
data and metadata, each resulting triple in the kno-
wledge base has both head and tail entities h and t
identified by a type. Each resulting triple is presen-
ted in the form (c
h
:h, r, c
t
:t), where c
h
and c
t
repre-
sent the types of h and t. Besides providing a cate-
gorised set of entities, the metadata also enriches the
definition of each relation r, by restricting the dom-
ain and range of r to set of entities h ∈ c
h
: E and
t ∈ c
t
: E , respectively. In the following example,
the relation hasGender is constrained by the dom-
ain patient and the range gender: (patient:P01,
hasGender, gender:male).
HEXTRATO uses independent vector spaces to
project each entity type, thus leading to a substan-
tial processing time improvement along the validation
process – related work models usually perform the
validation process every each 100 cycles along the
training step, whilst our method performs validation
after each 20 training cycles with similar processing
time comparing to previous works.
3.1.2 Isolating Values
Specific set of tail values can share the same entity
names and types when involved in different relations.
A very simple example to illustrate this condition is
the boolean type. When multiple relations r
1
and r
2
are defined with same range boolean, they end up by
sharing the possible entities boolean:true and bool-
ean:false in the boolean vector space. However, this
correlation between r
1
and r
2
does not necessarily ex-
ist. We set the relations sharing tail types that should
be taken as independent relations, by isolating the as-
sociated values in relation-specific types. Effectively,
given two relations r
1
and r
2
both defined with the
same range type
t
of tail entities, we set each relation
to isolate the tail values by creating an independent
set of tail entities for each relation, i.e. independent
vector spaces for each relation.
For example, the relations isPregnant and
isSmoker are both defined with the same range
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
74

boolean: (patient:?,isPregnant,boolean:?)
and (patient:?,isSmoker,boolean:?). Howe-
ver, isPregnant and isSmoker should be taken as
independent properties for a given patient, and it
should not be expected to have any correlations
between those entities by sharing the tail enti-
ties boolean:true and boolean:false. By isolating
their values, each relation creates its own set of
boolean values, boolean:isPregnant true and bool-
ean:isPregnant false for the relation isPregnant, and
boolean:isSmoker true and boolean:isSmoker false
for the relation isSmoker.
3.1.3 Disjoint Sets
By learning the distributed representation of multi-
relational data, knowledge embedding models can ef-
ficiently deal with the semantic relatedness of their
constituents. Similar entities are expected to be found
near to each other in the vector space, while dissi-
milar entities should be placed apart. However, this
expected result can be harmed when learning the em-
bedding representation for dense graphs, especially
when combining independent types of relations to
describe the subject entities. By imposing a minimum
disjoint dissimilarity (distance margin) among the en-
tities belonging to specific types on the tail side, we
avoid the model converging to undesirable solutions.
In very dense graphs, we observed multiple tail values
associated with uncorrelated types of relations found
very close to each other, leading to a model that mi-
mics a random probability of choices.
For instance, by setting the type gender as a
disjoint set, a minimum disjoint margin distance be-
tween the entities gender:male and gender:female is
enforced in the beginning of each training step. The
disjoint margin is an additional hyper-parameter in
our approach, but for the experimental results it was
fixed as
√
k
8
for each k-dimensional space evaluated.
3.1.4 Functional Relations
In a functional relation r, for each head entity h, there
can be at most one distinct tail entity t such that (h,r,t)
is true, which is equivalent of saying the cardinality of
the relation r is ≤ 1. Combining typed and functional
relations with disjoint tail sets proved to be very ef-
fective on learning the embedding representation of
multi-relational data, by narrowing the process of se-
lecting corrupted triples along the training process.
By way of illustration, considering the following
true positive triple (patient:P01, hasGender,
gender:male), in which the relation hasGender is
set as functional and the type gender is a disjoint set,
the process of electing a corrupted tail along the trai-
ning process is straightly redirect to pick up all the re-
maining tail entities from the gender set, in this case
gender:female would be the only alternative.
3.2 Embedding Representation
Among previous embedding methods, TransE is a
promising baseline, as it is simple and efficient
while achieving predictive performance. However,
we find that there are flaws in TransE when dealing
with relations mapping properties of reflexive/one-
to-many/many-to-one/many-to-many. Few previous
works discuss the role of these mapping properties in
embedding. Some advanced models with more free
parameters are capable of preserving these mapping
properties, e.g. TransH (Wang et al., 2014). Howe-
ver, the model complexity and running time is signi-
ficantly increased accordingly. Our method follows
the idea presented by TransE, coupling this baseline
model with the ontology-based constraints previously
described in order to improve accuracy in domain-
specific knowledge bases.
Despite the great expressiveness of the previously
proposed embedding models, they can be complex
to model, hard to interpret, and expensive in terms
of training computational costs. Besides, we ob-
served in empirical experiments they are susceptible
to either overfitting in higher embedding spaces, or
under-fitting due to multiple local minima along the
optimization process. Indeed, according to (Bengio
et al., 2005), lower k-dimensional spaces are appro-
priate for achieving good results because a density
estimator can misbehave in high dimensions when
there is no smooth low-dimensional manifold captu-
ring the distribution. In our approach, we target lower
k-dimensional models (e.g. k < 100) favouring a dis-
tributed representation that is rather cheap in memory
and potentially keep the generalization ability.
Given a training set S of triplets (c
h
:h, r, c
t
:t) our
model learns embedding vectors for the entities and
the relations. Each categorised entity c:e is represen-
ted by a embedding vector e
c
∈ R
k
, and each rela-
tion r is represented by a embedding vector r ∈ R
k
.
Similarly as it was defined in TransE, for each rela-
tion r there is a score function f
r
(Equation 5) that
represents a dissimilarity using a p-norm metric (in
our experiments we used p = 2), such that the score
f
r
(h
c
h
,t
c
t
) of a plausible triple (c
h
:h, r, c
t
:t) is smaller
than the score f
r
(h
0
c
h
,t
0
c
t
) of a implausible triple (c
h
:h’,
r, c
t
:t’).
f
r
(h
c
h
,t
c
t
) = kh
c
h
+ r −t
c
t
k
2
(5)
HEXTRATO: Using Ontology-based Constraints to Improve Accuracy on Learning Domain-specific Entity and Relationship Embedding
Representation for Knowledge Resolution
75

In order to learn knowledge embedding represen-
tation, our method uses Stochastic Gradient Descent
(SGD) (Robbins and Monro, 1951) to minimize a
margin-based loss functions L:
L =
∑
(c
h
:h,r,c
t
:t)∈S
(c
h
:h
0
,r,c
t
:t
0
)∈S
0
[γ + f
r
(h
c
h
,t
c
t
) − f
r
(h
0
c
h
,t
0
c
t
)]
+
(6)
where, γ is the margin parameter, S is the set
of correct triples, S
0
is the set of incorrect triples
(c
h
:h’,r,c
t
:t) ∪ (c
h
:h,r,c
t
:t’), and [x]
+
= max(0,x).
In TransE, incorrect triples ((h
0
,r,t)∪(h, r,t
0
)) are
generated by randomly corrupting either h or t in a
correct triple (h,r,t) ∈ S using different probabilities
for entity replacement (Wang et al., 2014). We follow
the same idea as presented in TransE, but the entity
replacement is randomly chosen from the set of enti-
ties belong to the corresponding type of each relation
domain and range instead.
Entity and relation embedding vectors are initia-
lised with the random uniform normalized initializa-
tion (Glorot and Bengio, 2010). The set of golden
triples is then randomly traversed multiple times al-
ong the training process up to the maximum of 1,000
iterations, such that each training step produces a cor-
rupted triple for each correct triple. HEXTRATO in-
troduces a disjoint verification step, performed once
before each training cycle, in which the disjoint mar-
gin is enforced among each set of disjoint entity types.
Finally, at the end of each training iteration, we
impose a L2-norm constraint for the embedding vec-
tors of each entity (khk
2
≤ q and ktk
2
≤ q) in order
to prevent the training process to minimize the loss
function L by artificially increasing the entity embed-
ding norms (no regularization constraint is given to
the relation embedding vectors). The constant q = 1
is commonly used in previous work, but it tends to
produce small embedding vector values for higher va-
lues of k in a k-dimensional space. In order to better
exploit the range of possible embedding values in the
interval [-1,+1], we define the max magnitude con-
straint for each entity as:
q = max
1,
√
k
2
(7)
3.3 Evaluation Datasets
In order to evaluate the effectiveness of our method
and the ability of improving the baseline accuracy
obtained from TransE in domain-specific data, we
conducted experiments on two real datasets extracted
from InfoSaude (Tissot and Dobson, 2018), a Electro-
nic Health Record (EHR) system.
1
The system mana-
ges and tracks patient records, being used to meet
the needs of several integrated public health centres
in the city of Florianopolis/Brazil by integrating dif-
ferent information structures to provide required out-
puts, such as the Outpatient Information and Ambu-
latory Care Individual reports, and summarizing data
on the type of care, pregnancies, procedures perfor-
med on the patient, applied vaccines and drug pres-
criptions. Statistics about the evaluation datasets are
given in Table 1:
Table 1: Statistics of domain-specific benchmark datasets,
given by the number of entities, relations, and triples in
each dataset split – training (LRN), validation (LVD), tu-
ning (TUN) and test (TST) sets.
EHR Datasets
# Demographics Pregnancy
Entities 2237 3088
Relations 6 5
LRN 13875 14588
VLD 463 1997
TUN 475 2093
TST 532 2090
Both EHR datasets are totally de-identified. Ages
are converted to a range of values to avoid determi-
ning the actual year of birth. New independent se-
quential IDs are assigned to each patient – patients
with more than one admission have distinct IDs in
each EHR dataset. No additional sensitive patient
data is included in any of the datasets. Table 2 pre-
sents the types of entities involved in each kind of re-
lation in the evaluations datasets, as well as the num-
ber of triples available for each kind of relation.
In addition to the EHR datasets, we used an adap-
ted version of FB15K dataset (FB15K-Typed), in
which each entity was categorised based on the des-
cripton of their corresponding relations, so that ma-
king it possible to compare our results with previous
work. We also report results on the Mushroom
2
da-
taset in order to motivate further experiments and im-
provements in the link prediction task. Both datasets
are available for download.
3
EHR-Demographics
This dataset comprises a set of 2,185 randomly se-
lected patients who had at least one admission bet-
ween 2014 and 2016. Each patient is described by a
1
Not publicly available – a synthetic sample is available
at https://github.com/HeglerTissot/hextrato
2
https://archive.ics.uci.edu/ml/datasets/mushroom
3
https://github.com/HeglerTissot/hextrato/
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
76
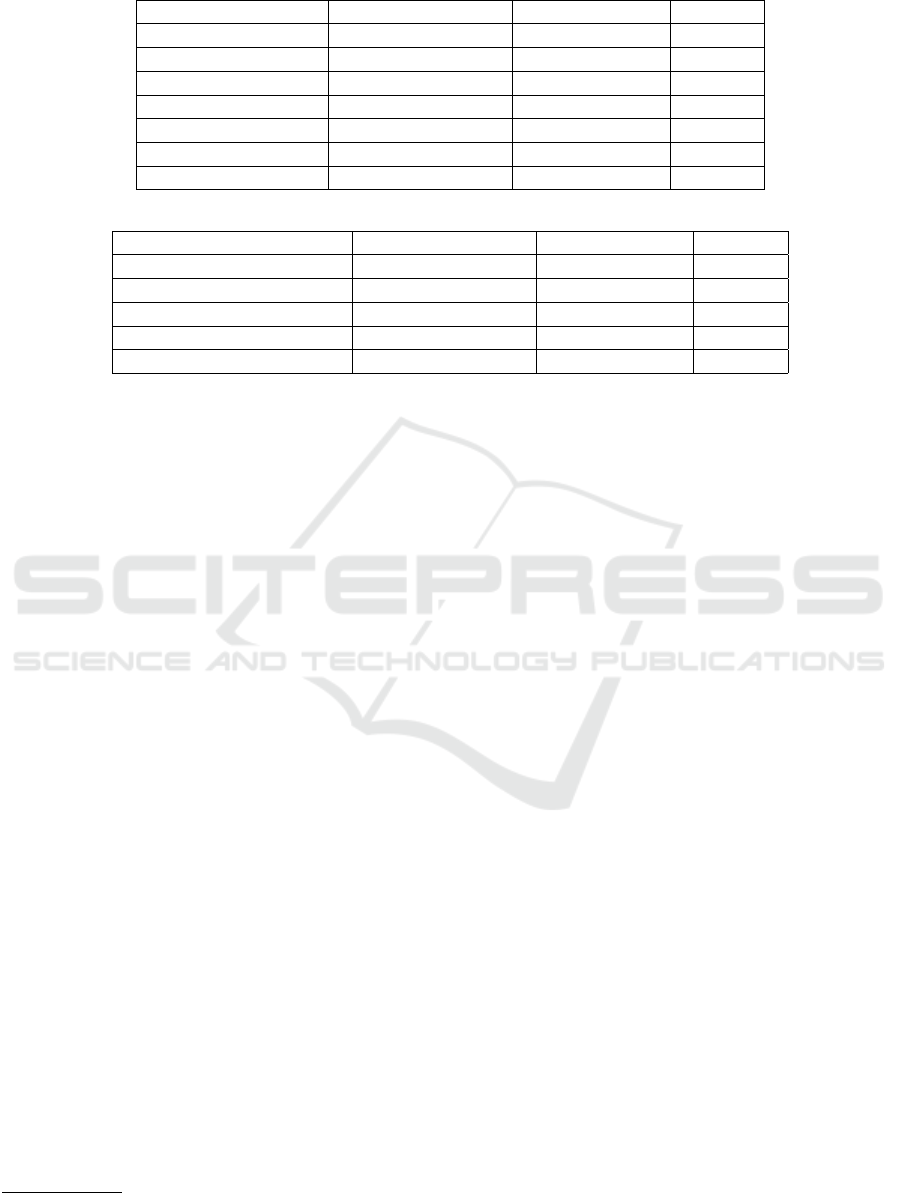
Table 2: Relations and corresponding entity types (domain and range) found in each domain-specific benchmark datasets.
Relation Domain (head type) Range (tail type) # Triples
hasGender patient gender 2,185
ageRange patient interval 2,185
hasMaritalStatus patient maritalStatus 1,844
hasMaxEducation patient education 1,815
isSmoker patient boolean 2,185
isPregnant patient boolean 506
inSocialGroup (N:N) patient socialGroup 4,625
(a) EHR-Demographics dataset
Relation Domain (head type) Range (tail type) # Triples
ageYearsWhenLMP patient interval 2,879
hadAbortion patient boolean 2,879
ageWeeksWhenInterrupted patient interval 2,879
ICDBeforeLMP (N:N) patient ICD 5,776
ICDAfterLMP (N:N) patient ICD 6,355
(b) EHR-Pregnancy dataset
set of basic demographic information, including gen-
der, age (range in years) in the admission, marital sta-
tus (unknown for about 15% of the patients), educa-
tion level, and two flags indicating whether the patient
is known to be either a smoker or pregnant, and the
social group. Social groups are assigned to each pa-
tient according to a diverse set of rules mainly based
on demographic and historical data. Social groups are
further used to determine which social programs each
patient can be offered to join.
EHR-Pregnancy
This dataset includes a set of 2,879 randomly selected
pregnant female patients from which pregnancy was
inadvertently and abnormally interrupted before the
expected date of birth. Each patient is described by
age (range in years) by the known date of last men-
strual period (LMP), whether the patient had an abor-
tion (regardless of reason), and a list of ICD-10 (Inter-
national Classification of Diseases) codes
4
registered
either before or after the LMP date. This dataset has
been used in order to identify correlations between
pre and post clinical conditions on pregnant patients
with abnormal pregnancy termination.
3.4 Evaluation Protocol
A commonly used evaluation protocol for knowledge
embedding methods includes a Link Prediction (LP)
task on the test set. LP is a typical question answering
task which aims at completing a triple (h, r,t) with
4
http://www.who.int/classifications/icd/en/
h or t missing, by predicting t given (h, r, ?) or pre-
dicting h given (?,r,t), where ? denotes the missing
element. Rather than giving one best answer, this task
is focused on ranking the plausibility of a set of can-
didate entities in descending order of similarity sco-
res, calculated by inducing the score function f
r
(h,t)
and recording the rank of the correct missing entity.
HEXTRATO is evaluated by predicting t given (c
h
:h,
r, c
t
:?) or predicting h given (c
h
:?, r, c
t
:t).
Overall results in the related work are usually pre-
sented by reporting the following commonly used
scores as evaluation metrics: a) Mean Rank (MR); b)
Mean Reciprocal Rank (MRR) of correct entities; and
c) the proportion of correct entities in top-N ranked
entities (Hits@N, with N usually equals 10). MRR is
an improved measure of Mean Rank [8] which calcu-
lates the average rank of all the entities (relations) and
calculates the average reciprocal rank of all the enti-
ties (relations). Compared with Mean Rank, MRR is
less sensitive to outliers. A good link predictor should
achieve lower MR or higher MRR and Hits@N. As
long as we aim to deal with more consistent and ca-
tegorised data, we also focus on achieving better per-
formance on the prediction task by comparing ranking
metrics with lower values of N, such as Hits@1 and
Hits@3.
Corrupted triples may also exist in a KG, which
should be also considered as correct from the training
set for instance, flawing the evaluation metrics. Thus,
the LP task may under-estimate those approaches that
rank corrupted but correct triples high. Hence, in or-
der to avoid such a misleading behaviour, all the tri-
ples that appear either in the training, validation, tu-
ning or test set are usually removed from the list of
corrupted triples, ensuring that all corrupted triples do
HEXTRATO: Using Ontology-based Constraints to Improve Accuracy on Learning Domain-specific Entity and Relationship Embedding
Representation for Knowledge Resolution
77
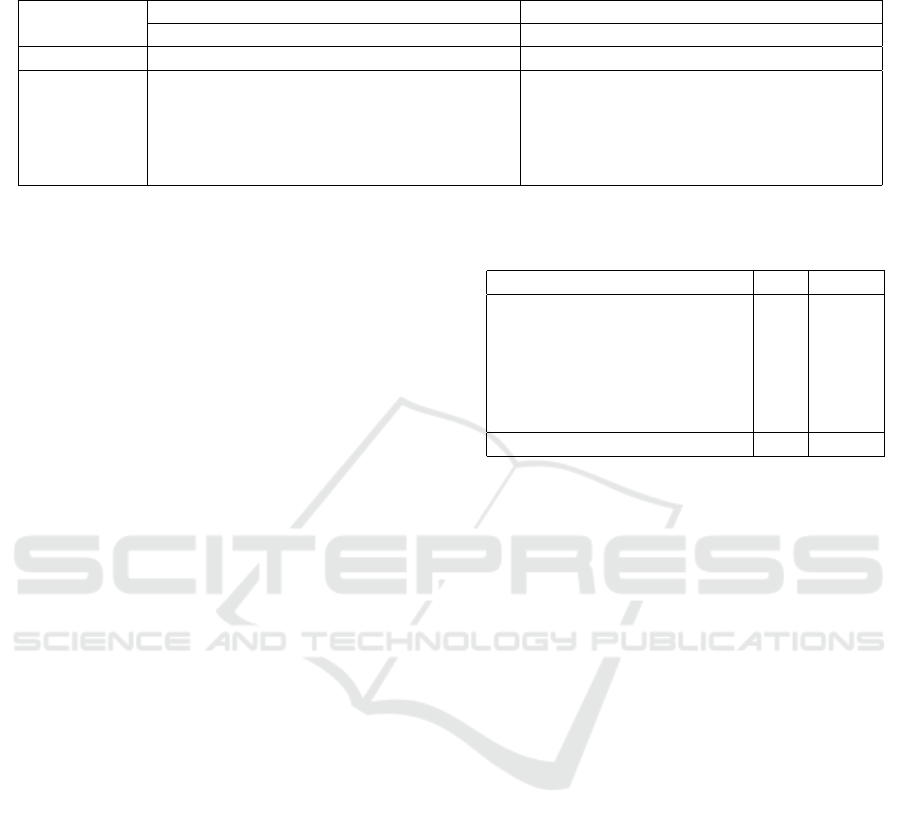
Table 3: Evaluation results for the Link Prediction task – Mean Reciprocal Rank (MRR), Mean Rank (MR), Hits@1 (H@1),
Hits@3 (H@3), Hits@10 (H@10) on two EHR datasets.
EHR-Demographics EHR-Pregnancy
MRR MR H@1 H@3 H@10 MRR MR H@1 H@3 H@10
TransE 0.3787 6.01 0.1523 0.4812 0.9173 0.227 103.87 0.103 0.264 0.457
HEXTRATO
(H1) 0.492 3.44 0.266 0.635 0.9530 0.260 27.29 0.146 0.273 0.498
(H2) 0.469 3.65 0.261 0.560 0.9530 0.236 28.17 0.119 0.244 0.502
(H3) 0.505 3.43 0.281 0.634 0.9530 0.270 27.80 0.150 0.288 0.537
(H4) 0.506 3.42 0.282 0.635 0.9531 0.279 26.30 0.153 0.303 0.555
not belong to the data set. In previous works, results
on the evaluation datasets are usually reported as both
“Raw” (possibly flawed) and “Filtered”. In this work,
we report the results referring to the former (“Raw”),
which we believe it provides a clearer view on the
ranking performance for categorised datasets.
We used a grid search on validation set
in order to select the learning rate λ among
{0.001,0.01,0.1}, the margin hyper-parameter γ
among {0.5,1.0,2.0,4.0}, and selected the best mo-
del by early stopping using the average of MMR score
calculated on predicting t on the validation sets, the
embedding dimension k among {8,16,32, 64}. The
dissimilarity measure was set to the L2-norm dis-
tance, and the optimal parameters are determined ac-
cording to performance accuracy on the validation set.
Ten distinct instances of each model were indepen-
dently trained for each set of hyper-parameters. After
traversing all the training triplets at most 1,000 epo-
chs, the best model is chosen by comparing the scores
against a tuning set. Final results are then calculated
over the test set.
4 RESULTS
In order to compare HEXTRATO against previous
works, we performed an initial experiment using an
adapted version of FB15K dataset (FB15K-Typed).
Table 4 compares the link prediction results of
HEXTRATO with results reported in previous work.
The lowest mean rank on the validation set was obtai-
ned when using the L2-norm, k = 32, λ = 0.01, γ =
2.0. Although HEXTRATO does not use projection
matrices for each relation as usually reported by other
methods that extend TransE, it outperforms previous
state-of-the-art methods in “Raw” scores.
Overall results for the EHR datasets in Table 3 re-
port the “raw” Mean Reciprocal Rank, Mean Rank,
and Hists@N scores calculated as the score for pre-
dicting t subtask. The lowest mean rank on the tuning
set was obtained when using the L2-norm, k = 32,
Table 4: Link prediction results – “Raw” Mean Rank (MR)
and Hits@10 (H@10) on FB15K.
Method MR H@10
TransE (Bordes et al., 2013) 243 0.349
SE (Bordes et al., 2011) 273 0.288
TransH (Wang et al., 2014) 212 0.457
TransR (Lin et al., 2015) 198 0.482
STransE (Nguyen et al., 2016) 219 0.516
TransT (Ma et al., 2017) 199 0.533
HEXTRATO (H1) 116 0.535
λ = 0.01, γ = 1.0. We started by running the origi-
nal TransE model on the two evaluation datasets. We
then applied our approach, cumulatively adding each
constraint described in Section 3:
(H1) We added types to each entity, which im-
plicitly set range and domain for each relation, and
restrict the set of ranked entities being evaluated al-
ong the link prediction task. This constraint added
substantial improvement comparatively to the origi-
nal TransE model in both EHR datasets.
(H2) We then coupled the previous attempt (H1)
with disjoint sets of tail entities. All tail types were
set as disjoint groups in both datasets – the patient
type was kept as a non-disjoint set, so that the model
would not enforce minimum disjoint distance among
the patients, allowing them to converge into semantic
similar clusters.
(H3) The disjoint set model (H2) was extended,
so that some of the relations were defined as functio-
nal. Although no significant improvement in the sco-
res could be observed, this constraint proved to faci-
litate the step of choosing corrupted tails in order to
produce incorrect triples along the training process.
(H4) We reached the best scores by isolating va-
lues from those relations that originally share types,
such as boolean and interval.
At the current stage we are only evaluating our
model against the baseline TransE model for the EHR
datasets. Further experiments are required in order to
test more complex models that usually extend or use
TransE as a baseline and check whether our proposal
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
78

Table 5: Resulting scores for each relation in the EHR-Demographics dataset – Mean Reciprocal Rank (MRR), Mean Rank
(MR), Hits@1 (H@1), and Hits@3 (H@3) for the best model (H4) in the Link Prediction task.
EHR-Demographics
Relations MRR MR H@1 H@3
hasGender 0.8194 1.36 0.6389 N/A
ageRange 0.2339 9.42 0.0930 0.2326
hasMaritalStatus 0.5747 2.10 0.2414 0.8621
hasMaxEducation 0.3966 3.63 0.1852 0.3704
isSmoker 0.9286 1.14 0.8571 N/A
inSocialGroup 0.4557 3.28 0.1994 0.6168
Table 6: Resulting scores for each relation in the EHR-Pregnancy dataset – Mean Reciprocal Rank (MRR), Mean Rank (MR),
Hits@1 (H@1), and Hits@3 (H@3) for the best model (H4) in the Link Prediction task.
EHR-Pregnancy
Relations MRR MR H@1 H@3
ageYearsWhenLMP 0.3869 4.08 0.1623 0.4755
hadAbortion 0.8401 1.32 0.6801 N/A
ageWeeksWhenInterrupted 0.3603 5.66 0.1628 0.4286
ICDBeforeLMP 0.0784 52.30 0.0117 0.0417
ICDAfterLMP 0.1218 32.56 0.0319 0.0909
fits into them.
Finally, Tables 5 and 6 detail the resulting scores
for each relation for the best model (H4) highlighted
boldface in Table 3. For the relations where tail en-
tities belong to the type boolean (isSmoker in EHR-
Demographics and hadAbortion in EHR-Pregnancy)
or gender (hasGender in EHR-Demographics) we do
not present the score Hits@3 – it is not applicable as
these relations have only two possible values to be
ranked, so that the resulting score is obviously equals
1. Within the EHR-Demographics dataset, there were
no examples of triples with relation hadAbortion in
the test set, so that the resulting scores for this speci-
fic relation is not being presented in Table 5.
By analysing the results from Table 6, it
becomes evident those many-to-many relations
(ICDBeforeLMP and ICDBeforeLMP) impose most
of the challenge on the LP task. However, the re-
sults presented in Table 5 contrast that assumption
(inSocialGroup). Within the EHR-Demographics da-
taset, both ageRange and inSocialGroup relations
have approximately 20 possible tail values each. Alt-
hough the relation inSocialGroup has cardinality N:N,
it presents better scores than the results referring to
the relation ageRange, which has cardinality (N:1).
Firstly, the social groups assigned to each patient take
into consideration both demographic and clinical his-
torical data, so that, as long as some of this demo-
graphic data is available within the dataset, the re-
sulting model can more easily reason on predicting
what groups should be assigned to each patient. Fi-
nally, the relation ageRange went through a discreti-
sation of a continuous variable age with original va-
lues ranging from 0 to 99. Embedding models are not
designed to deal with continuous values and some in-
formation is supposedly lost along the discretisation
process.
In order to motivate further experiments on publi-
cly available datasets we finally report the preliminary
results on the Mushroom dataset. Table 7 compares
HEXTRATO and TransE based on “raw” Mean Re-
ciprocal Rank, Mean Rank, and Hists@N scores cal-
culated as the score for predicting t subtask. In ad-
dition we also present the accuracy of each model,
based on the Hists@1 score for the relation has class.
The highest accuracy on the tuning set was obtained
when using the L2-norm, k = 64, λ = 0.1, γ = 1.0
for TransE, and using the L2-norm, k = 64, λ = 0.01,
γ = 2.0 for HEXTRATO (H4). Results from the at-
tempts using distinct values ok k along this described
set of hyperarameters are also presented to demon-
strate how changes increasing the dimensionality of
the low embedding space positively affect our model.
5 CONCLUSIONS
In this paper, we present HEXTRATO, a novel know-
ledge embedding approach that couples previous ba-
seline TransE model with ontology-based constraints
in order to better capture the relationships between
entities and their symbolic representation in the vec-
tor space.
Experimental benchmark results on an adapted
HEXTRATO: Using Ontology-based Constraints to Improve Accuracy on Learning Domain-specific Entity and Relationship Embedding
Representation for Knowledge Resolution
79

Table 7: Evaluation results for the Link Prediction task on the Mushroom dataset (Entities=8487, Relations=23, Tri-
ples={153057, 9525, 9564, 18942} for training, validation, tuning and test sets) – Mean Reciprocal Rank (MRR), Mean
Rank (MR), Hits@1, Hits@3, Hits@5, Hits@10, and Accuracy (equivalent to Hits@1 on predicting the relation has class).
MRR MR Hits@1 Hits@3 Hits@5 Hits@10 Accuracy
TransE 0.565 472.32 0.466 0.643 0.682 0.718 53.1%
HEXTRATO (H4)
k = 8 0.717 2.054 0.553 0.856 0.955 0.993 88.6%
k = 16 0.763 1.856 0.619 0.892 0.961 0.994 89.3%
k = 32 0.804 1.712 0.683 0.914 0.964 0.994 90.7%
k = 64 0.814 1.688 0.703 0.915 0.965 0.996 95.3%
version of Freebase, on a publicly available da-
taset, and on two domain-specific datasets show
HEXTRATO outperforms previous state-or-the-art
methods in the link prediction task when using cate-
gorised entities. Some of the directions in which this
work can be extended include:
TransE-like extended models. Learning embed-
ding representation from more structured knowledge
sources can benefit from the inherit enriched meta-
data. HEXTRATO is a constraint-based method that
extends TransE in order to obtain an initial baseline
for the evaluation task when dealing with domain-
specific categorised datasets. We plan to evaluate our
method coupled with more complex embedding mo-
dels originated from TransE.
Many-to-many relationships. Normalising N:N
relations can make an embedding model more flexi-
ble. However, it adds additional level of complexity
in terms of learning semantically related entities. Alt-
hough preliminary experiments did not show effective
improvement over previously applied constraints, we
believe further investigation can demonstrate whether
more specific conditions can lead our model to reach
better results.
Activation functions. More complex embedding
models deal with projection matrices and rely on sim-
ple linear neural networks. We plan to investigate
whether alternatively coupling ontology-based con-
straints with non-linear activation functions, such as
RELUs, Sigmoid, or Tanh, can improve the embed-
ding model performance on domain-specific datasets.
Hybrid approaches. Distinct sets of relation em-
bedding representations can be more effectively learnt
from distinct approaches. Tightening state-of-the-art
bounds by combining different methods into a hybrid
approach in which each relation can be represented by
a distinct embedding model can produce models that
are more flexible on learning distinct types of relati-
onships between entities within a dataset.
Unseen entities. The primordial assumption when
dealing with any kind of machine learning model is
the ability of such resulting model on generalising.
Embedding models are weak regarding to this aspect.
Validation and test sets are required to be designed
with entities and relations that appear at least once in
the training set. We plan to investigate how embed-
ding models coupled with ontology-based constraints
can be used to learn low-embedding representation
for unseen entities along the validation, tuning and
test steps.
REFERENCES
Bengio, Y., Larochelle, H., and Vincent, P. (2005). Non-
local manifold parzen windows. In Weiss, Y.,
Sch
¨
olkopf, B., and Platt, J., editors, Advances in
Neural Information Processing Systems 18 (NIPS’05),
Cambridge, MA. MIT Press.
Bollacker, K., Evans, C., Paritosh, P., Sturge, T., and Taylor,
J. (2008). Freebase: A collaboratively created graph
database for structuring human knowledge. In Procee-
dings of the 2008 ACM SIGMOD International Con-
ference on Management of Data, SIGMOD ’08, pages
1247–1250, New York, NY, USA. ACM.
Bordes, A., Usunier, N., Garcia-Duran, A., Weston, J., and
Yakhnenko, O. (2013). Translating embeddings for
modeling multi-relational data. In Burges, C. J. C.,
Bottou, L., Welling, M., Ghahramani, Z., and Wein-
berger, K. Q., editors, Advances in Neural Information
Processing Systems 26, pages 2787–2795. Curran As-
sociates, Inc.
Bordes, A., Weston, J., Collobert, R., and Bengio, Y.
(2011). Learning structured embeddings of know-
ledge bases. In Conference on Artificial Intelligence.
Dong, X., Gabrilovich, E., Heitz, G., Horn, W., Lao, N.,
Murphy, K., Strohmann, T., Sun, S., and Zhang, W.
(2014). Knowledge vault: A web-scale approach to
probabilistic knowledge fusion. In Proceedings of
the 20th ACM SIGKDD International Conference on
Knowledge Discovery and Data Mining, KDD ’14,
pages 601–610, New York, NY, USA. ACM.
Fellbaum, C., editor (1998). WordNet: an electronic lexical
database. MIT Press.
Gardner, M. and Mitchell, T. (2015). Efficient and expres-
sive knowledge base completion using subgraph fe-
ature extraction. In Proceedings of the 2015 Con-
ference on Empirical Methods in Natural Language
Processing, pages 1488–1498. Association for Com-
putational Linguistics.
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
80

Glorot, X. and Bengio, Y. (2010). Understanding the dif-
ficulty of training deep feedforward neural networks.
In Teh, Y. W. and Titterington, D. M., editors, Pro-
ceedings of the Thirteenth International Conference
on Artificial Intelligence and Statistics (AISTATS-10),
volume 9, pages 249–256.
Krompaß, D., Baier, S., and Tresp, V. (2015). Type-
constrained representation learning in knowledge
graphs. In Proceedings of the 13th International Se-
mantic Web Conference (ISWC).
Lin, H., Liu, Y., Wang, W., Yue, Y., and Lin, Z. (2017). Le-
arning entity and relation embeddings for knowledge
resolution. Procedia Computer Science, 108(Supple-
ment C):345 – 354. International Conference on Com-
putational Science, ICCS 2017, 12-14 June 2017, Zu-
rich, Switzerland.
Lin, Y., Liu, Z., Sun, M., Liu, Y., and Zhu, X. (2015). Le-
arning entity and relation embeddings for knowledge
graph completion. In Proceedings of the Twenty-Ninth
AAAI Conference on Artificial Intelligence, AAAI’15,
pages 2181–2187. AAAI Press.
Ma, S., Ding, J., Jia, W., Wang, K., and Guo, M. (2017).
Transt: Type-based multiple embedding representati-
ons for knowledge graph completion. In The Euro-
pean Conference on Machine Learning and Principles
and Practice of Knowledge Discovery in Databases.
Nguyen, D. Q., Sirts, K., Qu, L., and Johnson, M. (2016).
Stranse: a novel embedding model of entities and rela-
tionships in knowledge bases. CoRR, abs/1606.08140.
Robbins, H. and Monro, S. (1951). A stochastic approxi-
mation method. Ann. Math. Statist., 22(3):400–407.
Suchanek, F. M., Kasneci, G., and Weikum, G. (2007).
Yago: A core of semantic knowledge. In Procee-
dings of the 16th International Conference on World
Wide Web, WWW ’07, pages 697–706, New York,
NY, USA. ACM.
Taskar, B., fai Wong, M., Abbeel, P., and Koller, D. (2003).
Link prediction in relational data. In in Neural Infor-
mation Processing Systems.
Tissot, H. and Dobson, R. (2018). Identifying misspelt
names of drugs in medical records written in portu-
guese. HealTAC-2018: Unlocking Evidence Contai-
ned in Healthcare Free-text.
Wang, Z., Zhang, J., Feng, J., and Chen, Z. (2014). Know-
ledge graph embedding by translating on hyperplanes.
In Brodley, C. E. and Stone, P., editors, AAAI, pages
1112–1119. AAAI Press.
HEXTRATO: Using Ontology-based Constraints to Improve Accuracy on Learning Domain-specific Entity and Relationship Embedding
Representation for Knowledge Resolution
81
