
Computer-aided Formal Proofs about Dendritic Integration
within a Neuron
Oph
´
elie Guinaudeau
1
, Gilles Bernot
1
, Alexandre Muzy
1
, Daniel Gaff
´
e
2
and Franck Grammont
3
1
Universit
´
e C
ˆ
ote d’azur, CNRS, I3S, France
2
Universit
´
e C
ˆ
ote d’azur, CNRS, LEAT, France
3
Universit
´
e C
ˆ
ote d’azur, CNRS, LJAD, France
Keywords:
Single Neuron Modelling, Dendrites, Signal Integration, Formal Methods, Model Checking.
Abstract:
This article is threefold: (i) we define the first formal framework able to model dendritic integration within
biological neurons, (ii) we show how we can turn continuous time into discrete time consistently and (iii) we
show how a Lustre model checker can automatically perform proofs about neuron input/output behaviours
owing to our framework.
Our innovative formal framework is a carefully defined trade-off between abstraction and biological relevance
in order to facilitate proofs. This framework is hybrid: inputs entering the synapses as well as the soma output
are discrete signals made of spikes but, inside the dendrites, we combine signals quantitatively using real
numbers. The soma potential is inevitably specified as a differential equation to keep a biologically accurate
modelling of signal accumulation. This prevents from performing simple formal proofs. This has been our
motivation to discretize time. Owing to this discretization, we are able to encode our neuron models in Lustre.
Lustre is a particularly well suited flow-based language for our purpose. We also encode in Lustre a property of
input/output equivalence between neurons in such a way that the model checker Kind2 is able to automatically
handle the proof.
1 INTRODUCTION
Many studies suggest that there is a strong interdepen-
dence between morphology and information process-
ing capabilities of a neuron (Bianchi et al., 2012; Mo-
han et al., 2015; Hu and Vervaeke, 2017). In this pi-
oneering work, we make use of formal methods from
computer science to investigate how single neurons
process information, with a particular emphasis on
their dendritic morphology.
Neurons are the building blocks of the brain.
They are highly connected and communicate through
electrical impulses. Typically, neurons receive those
signals on branched extensions called dendrites, at
specific locations named synapses (Figure 1). All
the stimuli are finally integrated at the soma. If the
resulting signal accumulation is strong enough, it is
transmitted to neighbouring neurons through the axon
which is another kind of extension. There are many
types of neurons (Mel, 1994; Stuart et al., 2016)
and some of them are structurally and functionally
very different from the standard one described above,
depending on the respective sizes on the constituting
Figure 1: Schematic representation of a standard biological
neuron.
sub-parts.
There can be between a few to several tens thou-
sands of synapses as a result of the connectivity
level of the neuron. The incoming signals interact
continually in time and space and while propagat-
ing through the dendrites towards the soma, signals
are significantly modified. Indeed, many parame-
ters of information-processing such as signal filtering,
response speed and connectivity, are strongly influ-
enced by the dendrites properties (Williams and Stu-
Guinaudeau, O., Bernot, G., Muzy, A., Gaffé, D. and Grammont, F.
Computer-aided Formal Proofs about Dendritic Integration within a Neuron.
DOI: 10.5220/0006680500490060
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 3: BIOINFORMATICS, pages 49-60
ISBN: 978-989-758-280-6
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
49

art, 2002; Stuart et al., 2016). Therefore, it makes this
sophisticated structure a key determinant of neuronal
computation.
A combination of experimental and theoretical
work is required to make the link between structure
and function of neurons. Nowadays and despite re-
cent advances, dendrites are still difficult to study
experimentally mainly because of their small size.
Theoretical modelling frameworks are thus helpful to
overcome these limitations as they provide a rigor-
ous support for understanding how such complex be-
haviours emerge. Since Rall’s pioneering work, some
theoretical models have been dedicated to dendrites
structure and physiology (see Section 2). However,
these models mainly focus on microscopic biophysi-
cal aspects of dendrites physiology, making difficult
the scaling from a computational point of view. On
the other side, most models focusing on this compu-
tational efficiency neglect dendritic morphology and
integration properties.
Our goal is to use formal methods from computer
science to prove properties on complete neurons and
allow automated identification of parameters, at the
cost of a minimal set of simplifying assumptions.
We define here a model of neuron sufficiently sim-
ple to use formal methods while taking into account
the dendritic structure. Basically, we propose a rele-
vant trade-off between abstraction and biological rel-
evance. This model is hybrid as its inputs and output
are discrete while we consider linear equations on real
numbers within the neuron.
The next section presents some relevant existing
single neuron models. In Section 3, we introduce
our model with mathematical definitions of its com-
ponents and its dynamics. We then explain in Sec-
tion 4, how to discretize time in order to prove some
properties on neurons using model checking.
2 STATE OF THE ART
Numerous single neuron models are found in the liter-
ature. Depending on the research objectives, they can
be classified on a variety of criteria: computational or
biophysical, discrete or continuous, punctual or struc-
tural, with rate coding or temporal coding, etc.
The purpose of this section is not to give the huge
exhaustive list of existing models, it rather aims at po-
sitioning our work with respect to some of them.
Computational Versus Biophysical Models. Op-
posing computational to biophysical models is a clas-
sic way to characterize single neuron models (Brette,
2003). In the case of computational models, neurons
are highly abstract in order to facilitate the study of
global behaviours under certain hypotheses. A good
example to illustrate this type of models, is the formal
neuron of McCulloch and Pitts (M&P) (McCulloch
and Pitts, 1943). It has multiple inputs (by analogy
with synapses in biological neuron) and a unique out-
put (which can be compared to the axon). The output
is a binary variable which is calculated as a function
of the weighted sum of the inputs. If the sum ex-
ceeds a given threshold, the neuron becomes active
(its state is equal to 1), otherwise, it becomes inactive
(its state is equal to 0). Despite its apparent simplic-
ity, this model is remarkably powerful as such neuron
networks allow to implement any calculable function.
However, this kind of very simple models is of low
interest for a biological understanding of the neuronal
functioning.
On the contrary, biophysical models aim at repre-
senting in details the physico-chemical mechanisms
driving biological functions. The most famous and
widely used biophysical model is perhaps Hodgkin
and Huxley’s (H&H) (Hodgkin and Huxley, 1952). It
describes the action potential generation at the axon
hillock, based on ionic channels dynamics.
Our work, is a trade-off between those two types
of models. We are more interested in how the neuron
“computes” to relate input signals to an output, rather
than in the precise biochemical mechanisms involved
in this input/output function. Nevertheless, most of
our definitions are sensible abstractions of biophysi-
cal processes (Section 3).
Discrete Versus Continuous Models. M&P’s neu-
ron is a typical discrete model as the values are dis-
crete (either 0 or 1) and the time is discrete too (the
state of the neuron is calculated at each step).
On the contrary, H&H’s model is continuous
as it is governed by a set of differential equa-
tions. Most of the biophysical models are, by
nature, continuous. It is worth mentioning an-
other well-known continuous model: the Integrate-
and-Fire (I&F) model (Lapicque, 1907; Brunel and
Van Rossum, 2007). It represents the membrane as an
electrical circuit and describes the electrical potential
of the neuron with a differential equation. There are
many extensions of this model including the Leaky
I&F, quadratic and exponential versions (Brette and
Gerstner, 2005; B
¨
orgers, 2017).
Our work is a hybrid formalism. Unlike the I&F
model which does not explicitly represent the elec-
trical impulses, we focus on them: the sequence of
impulses actually constitutes the input/output of our
neuron. This input/output is thus discrete while the
electrical potential of our neuron is continuous (see
Section 3). We share similarities with the work of
Maass (Maass, 1999; Maass, 1996), as signals ex-
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
50

changed by neurons are discrete. Moreover, mod-
elling of synapses uses piecewise linear functions.
However, Maass does not focus on dendritic integra-
tion and is rather interested in networks.
Punctual Versus Structural Models. As already
mentioned, punctual models represent a neuron as a
“point,” totally ignoring its morphology. Most of sin-
gle neuron models are punctual including all the pre-
viously discussed ones. In some studies, artificial de-
lays are added to neurons in order to compensate the
lack of morphology (Izhikevich, 2006).
Some models focus on the neuron structure. This
is the purpose of Cable Theory (CT) which was first
used to describe the propagation of electrical signals
through submarine cables. It was then applied to neu-
rons in the 1960’s by Rall, a pioneer of dendrites mod-
elling. Dendrites are seen as cylinders in which signal
propagates following a second order partial differen-
tial equation with dendrites physico-chemical proper-
ties as parameters (Rall, 1959; Rall, 1962). This equa-
tion has been solved analytically for some particular
conditions and it is widely used as part of simulators
such as GENESIS (Bower, 2003).
All in all, punctual models facilitate the use of for-
mal methods. However, they do not allow the study
of dendritic integration.
Rate Coding Versus Temporal Coding Models. A
classic point of view is that activity of neurons is en-
coded by their firing frequency. This rate coding has
been long at the basis of many neuron models. As a
simple example, there are second generation neurons
which are similar to M&P’s neuron but with a contin-
uous activation function instead of a threshold gate.
Their output is a sequence of real numbers which
can be seen as an instantaneous frequency. It im-
plies that the exact times of firing are not taken into
account, thus ignoring synchronization phenomena
experimentally observed in the brain (Brette, 2003).
However, temporal coding is of growing interest.
With this approach, the most important variable is the
electrical impulse per se and not the frequency. The
I&F is one of the most widely used model of this type.
In this paper, we are interested in the role of the den-
drites in the neuronal function. Therefore, we need
a non punctual model. Moreover, our goal is to use
formal methods to prove properties on neurons. As
a matter of fact, the larger the number of parameters,
the harder the model validation (Popper, 1963), espe-
cially with computer-aided proofs. Purely biophysi-
cal models are thus not suitable for our purpose. We
decided to define a new framework dedicated to our
formal study, and we decided to adopt the temporal
coding paradigm. This framework is described in the
following section along with the biological founda-
tions and the hypotheses we made.
3 HYBRID FORMAL NEURON
MODEL
A tree being defined as a root node to which are re-
cursively attached children trees, our single neuron
model can be defined as dendritic trees connected to
a root soma (denoted ∇) (Figure 2). In our model, we
ignore the axon as, in first approximation, it transmits
the signal faithfully. Dendrites are divided into com-
partments, each one being delimited by either branch-
ing points or synapses. A formal neuron is thus a
forest of dendritic trees composed of formal synapses
(leafs) and compartments (branches) linked together
at branching points. Each tree of the forest is rooted
on ∇. The following subsections give the formal def-
initions and the dynamics of these basic components.
We note N the set of all neurons and, given a
neuron N, we note Sy(N) the set of its synapses and
Co(N) the set of its compartments.
Figure 2: Schematic representation of our single neuron
model.
We can see the neuron as a system which receives
discrete inputs at synapses, converts them into a con-
tinuous signal and re-discretizes it in the soma. Elec-
trical signals processed by neurons are basically ionic
charge flows along the tree, making the potential lo-
cally changing. From our formal point of view, we
consider an abstract notion of charges/potential using
real numbers.
3.1 Synapses and Input Signal
Synapses are the input ports of neurons, the locations
where they receive electrical signals coming from
other neurons. In the brain, signals are sequences of
electrical impulses called action potentials or spikes.
When a spike (sent by another neuron) reaches a
synapse, it triggers a local variation of the neuron
Computer-aided Formal Proofs about Dendritic Integration within a Neuron
51
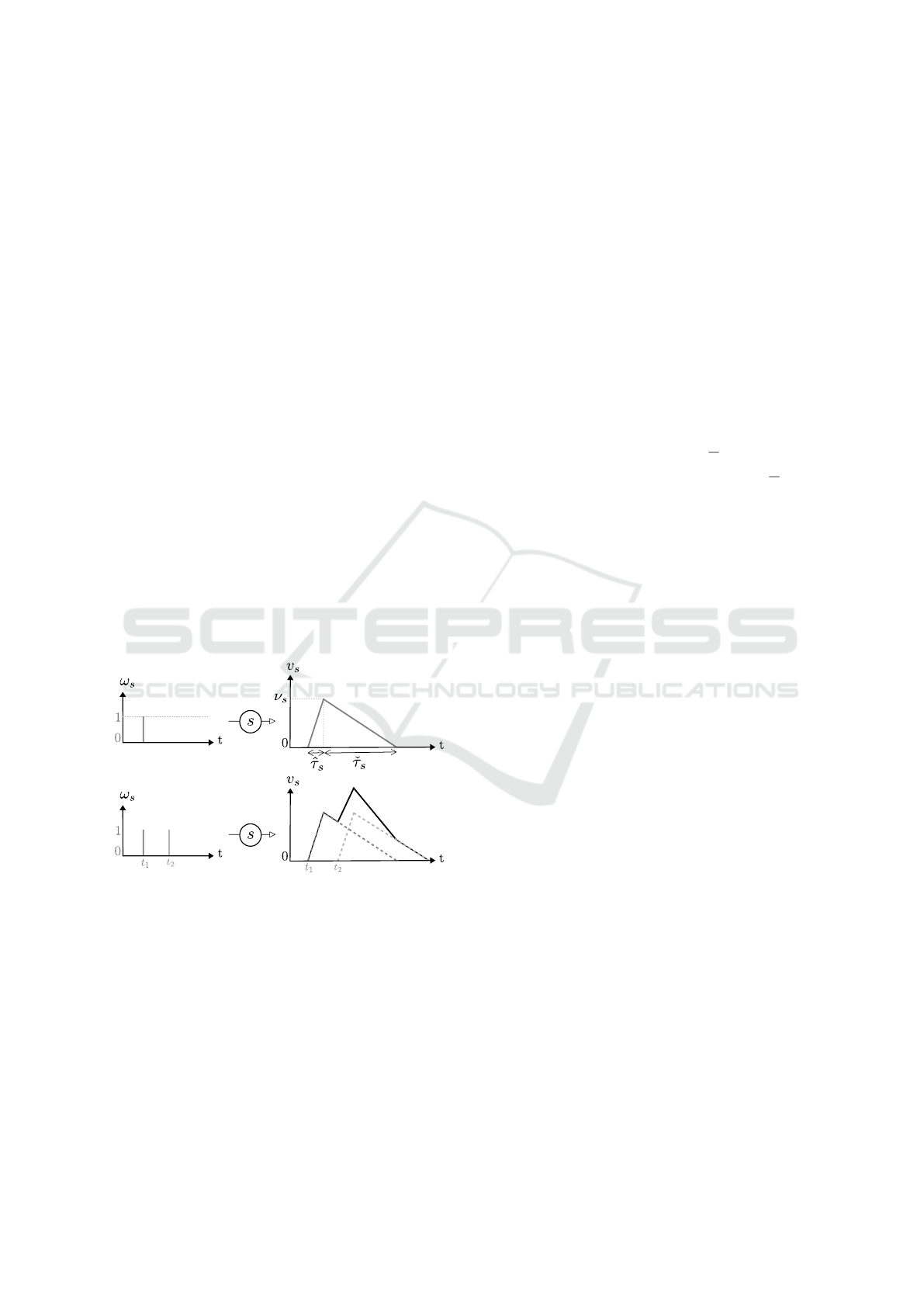
electrical potential. This effect, called post-synaptic
potential (PSP), is due to ionic movements through
the membrane. Depending on the synapse, the PSP
can be either excitatory or inhibitory, respectively in-
creasing or decreasing the potential.
In our modelling, we focus on spike times. An in-
put signal (ω) is a function of time which is equal to
1 at spike times and 0 otherwise. Each spike is con-
verted by the synapse into a continuous trace compa-
rable to a PSP. For each synapse s receiving a spike,
the trace reaches a maximum absolute value (ν
s
) after
a certain delay (
ˆ
τ
s
) and take a certain time (
ˇ
τ
s
) be-
fore being back to the resting value (zero here, see
Figure 3). Even though it is described as exponen-
tial by biophysical models (Rall, 1959; Rall, 1962;
Rall, 1967), we assume that this potential perturba-
tion is linear. This approximation is justified by ex-
perimental observations showing that the kinetics do
not strictly follow theory (Koch, 2004). As men-
tioned before, piecewise linear functions were also
used in (Maass, 1999) to represent PSPs.
The potential increase triggered by one spike is
most of the time not sufficient for the neuron to reach
its threshold and transmit the signal. The PSPs are
added if the spikes received at a synapse are separated
by a sufficiently small time interval: it is called tem-
poral summation. We reproduce this phenomenon by
summing the traces (Figure 3).,
Figure 3: Trace of signals on a synapse s.
Formal definitions are given below.
Definition 1. [Synapse] A synapse s is a tuple (ν
s
,
ˆ
τ
s
,
ˇ
τ
s
) where ν
s
is a non-zero real number called maxi-
mal potential of a spike for s. If ν
s
> 0 then s is said
excitatory, otherwise it is said inhibitory.
ˆ
τ
s
and
ˇ
τ
s
are strictly positive real numbers respectively called
rise time and descent delay of the potential.
Definition 2. [Signal] A signal is a function ω :
IR
+
→ {0,1} such that: ∃r ∈ IR
∗+
,∀h ∈ IR
+
,(ω(h) =
1 ⇒ (∀h
0
∈]h,h + r[,ω(h
0
) = 0)) The carrier of ω is
defined by: Car(ω) = {h ∈IR
+
|ω(h) = 1}. Moreover:
• A signal such that Car(ω) is a singleton {u} is
called a spike at the time u, noted ω
u
.
• Given a neuron N, an input signal for N is a family
of signals S = {ω
s
}
Sy(N)
indexed by the synapse
identifiers of Sy(N).
Remark: Obviously, a signal ω can be split into a
sum of spikes at times separated by at least r: ω =
∑
u∈Car(ω)
ω
u
.
The ultimate goal of the following mathematical con-
struction is to build a signal ω
∇
called output signal,
given an initial state and an input signal S .
Definition 3. [Trace of a signal] The trace of a spike
ω
u
on a synapse s is the function v
s,ω
u
defined by:
• If h 6 u then v
s,ω
u
(h) = 0;
• If u 6 h 6 u +
ˆ
τ
s
then v
s,ω
u
(h) =
ν
s
ˆ
τ
s
(h −u);
• If u +
ˆ
τ
s
6 h 6 u +
ˆ
τ
s
+
ˇ
τ
s
then v
s,ω
u
(h) =
ν
s
ˇ
τ
s
(u +
ˆ
τ
s
+
ˇ
τ
s
−h);
• If u +
ˆ
τ
s
+
ˇ
τ
s
6 h then v
s,ω
u
(h) = 0
Given an input signal ω
s
on a synapse s, the
trace of ω
s
is defined by the real function v
s,ω
s
=
∑
u∈Car(ω
s
)
v
s,ω
u
.
This definition simply encodes Figure 3.
3.2 Compartments
So far, synapses convert discrete inputs into contin-
uous signals. Those signals then spread through the
dendrites. In this work, we only consider propaga-
tions towards the soma even though it is known that
there exist back-propagation mechanisms (from soma
to dendritic tips) (H
¨
ausser et al., 2000; Remme et al.,
2010). It is also important to note that we do not take
into account active properties of the dendrites even
though it is clear that dendritic voltage-gated chan-
nels play a considerable role in their function (Cook
and Johnston, 1997; Johnston and Narayanan, 2008;
Stuart et al., 2016). In particular, we do not consider
dendritic spikes (Sun et al., 2014; Stuart and Sprus-
ton, 2015; Manita et al., 2017). Therefore, this work
is under the hypothesis of fully passive dendrites: It is
currently the price to pay in order to be able to apply
formal methods at this stage.
In passive dendrites, signals are attenuated while
spreading. Basically, the flow of ionic charges under-
goes a leak because of channels embedded in the neu-
ronal membrane (Figure 4). For biophysical reasons,
the more the potential is different between the extra-
cellular media and the inside of the neuron, the more
the attenuation is: biological systems usually tend to
return to equilibrium. It was well described by CT
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
52

model but with the main disadvantage of involving
many parameters (Section 2). In our model, we di-
vide the dendrites into small homogeneous compart-
ments. Inspired by the biophysical theory, we charac-
terise compartments using only two parameters: the
time for crossing it (δ) and the attenuation applied in
a non linear manner to the signal (α < 1). At the end
of a compartment, the signal is thus multiplied by the
α factor after a time δ (Figure 4). We consider a con-
stant throughput whatever the value of the signal. The
two parameters α and δ are specific to a given com-
partment. Concretely they are related to its length,
diameter and membrane properties such as resistance
and capacitance. Intuitively, the parameters α and δ
are a convenient way to hide possibly very sophisti-
cated processes. It constitutes a valuable compromise
between biophysical precision and preservation of the
ability to perform formal proofs. Also, biophysics
suggests that
ˆ
τ and
ˇ
τ are elongated within a compart-
ment (Rall, 2011). Here, for the sake of simplicity,
we include the total distortion directly in the synapse
parameters as we know the synapse locations.
Figure 4: Schematic representation of a compartment from
biophysical (left) and computational (right) points of view.
Definition 4. [Compartment] A compartment is a
couple c = (δ
c
, α
c
) where δ
c
is a real number greater
or equal to zero and α
c
is a real number such that
α
c
∈]0,1]. If δ
c
= 0, then α
c
= 1.
Additional definitions given below introduce
terms and concepts used further.
Terminology 1. Given a neuron N and a compart-
ment y
c
−→ z, y and z are respectively called the input
node and output node of c. Moreover, c is called the
input compartment of z and we note In(z) the set of
input compartments of z.
A predecessor of y
c
−→ z is a compartment in N of the
form x
c
0
−→ y and we note Pred(c) the set of the prede-
cessors of c.
Definition 5. [Contributors] Let N be a neuron. The
family CCo(c) indexed by c ∈Co(N) is defined induc-
tively as follows:
• ∀c ∈ Co(N), ∀c
0
∈ Pred(c), if δ
c
0
6= 0 then c
0
∈
CCo(c);
• ∀c ∈ Co(N), ∀c
0
∈ Pred(c), if δ
c
0
= 0 then
CCo(c
0
) ⊂CCo(c).
CCo(c) is called the set of contributor compartments
of c. Moreover, the family of sets CSy(c) indexed by
c ∈Co(N) is defined inductively as follows:
• ∀c ∈Co(N), if the input node of c is a synapse s,
then s belongs to CSy(c);
• ∀c ∈ Co(N) and ∀c
0
Pred(c), such that δ
c
0
= 0
then CSy(c
0
) ⊂CSy(c).
CSy(c) is called the set of synaptic contributors of c,
3.3 Soma
The soma (or cell body) of the neuron integrates all
the charges coming form the dendrites. When the re-
sulting potential reaches a given threshold, a spike is
generated at the axon hillock and transmitted to other
neurons. From that time, the neuron enters an ab-
solute refractory period (ARP) during which it can-
not emit any spike even if its potential is over the
threshold. This is due to ionic channels which are in-
activated and cannot generate any electrical impulse.
Then, the neuron is subject to a relative refractory pe-
riod (RRP) during which a greater signal than usual
is needed to trigger a spike.
We model the ARP with a infinite threshold mak-
ing it unreachable, as it was done in (Maass, 1999).
The RRP is represented by an abnormally high thresh-
old (θ+
ˆ
θ), progressively returning to its normal value
(θ). Moreover, as in dendrites, there is a leak (γ) at the
soma:
Definition 6. [Soma] A soma is a tuple ∇ = (θ,
ˆ
θ, ρ,
ˆ
ρ, γ) where θ is the normal threshold,
ˆ
θ is the thresh-
old augmentation, ρ is the duration of the absolute
refractory period,
ˆ
ρ is the duration of the relative re-
fractory period and γ is the leak. They are all strictly
positive real numbers.
To be able to compute when the neuron fires, one
needs to know the value of the soma potential (p)
at each time. Moreover, because of refractory pe-
riods, one needs to know the time elapsed since the
last emitted spike (denoted e). The couple (e, p) can-
not reach all values because for some values, it would
have been necessary to produce a spike before reach-
ing them (thus reinitializing e and subtracting θ from
p). We call nominal values the possible values for the
couple (e, p) (Figure 5).
Definition 7. [Nominal] Given a soma ∇ = (θ,
ˆ
θ, ρ,
ˆ
ρ, γ), a couple (e, p) where e ∈ IR
+
and p ∈IR, is said
nominal if (e < ρ) or (p < θ) or (p < θ +
ˆ
θ(ρ−e)
ˆ
ρ
). We
note Nominal(∇) the set of all the nominal couples.
The state of the soma is the value of the couple
(e, p) at a given time (see Definition 8). Thanks to
the nominal concept, we were able to formalize soma
Computer-aided Formal Proofs about Dendritic Integration within a Neuron
53
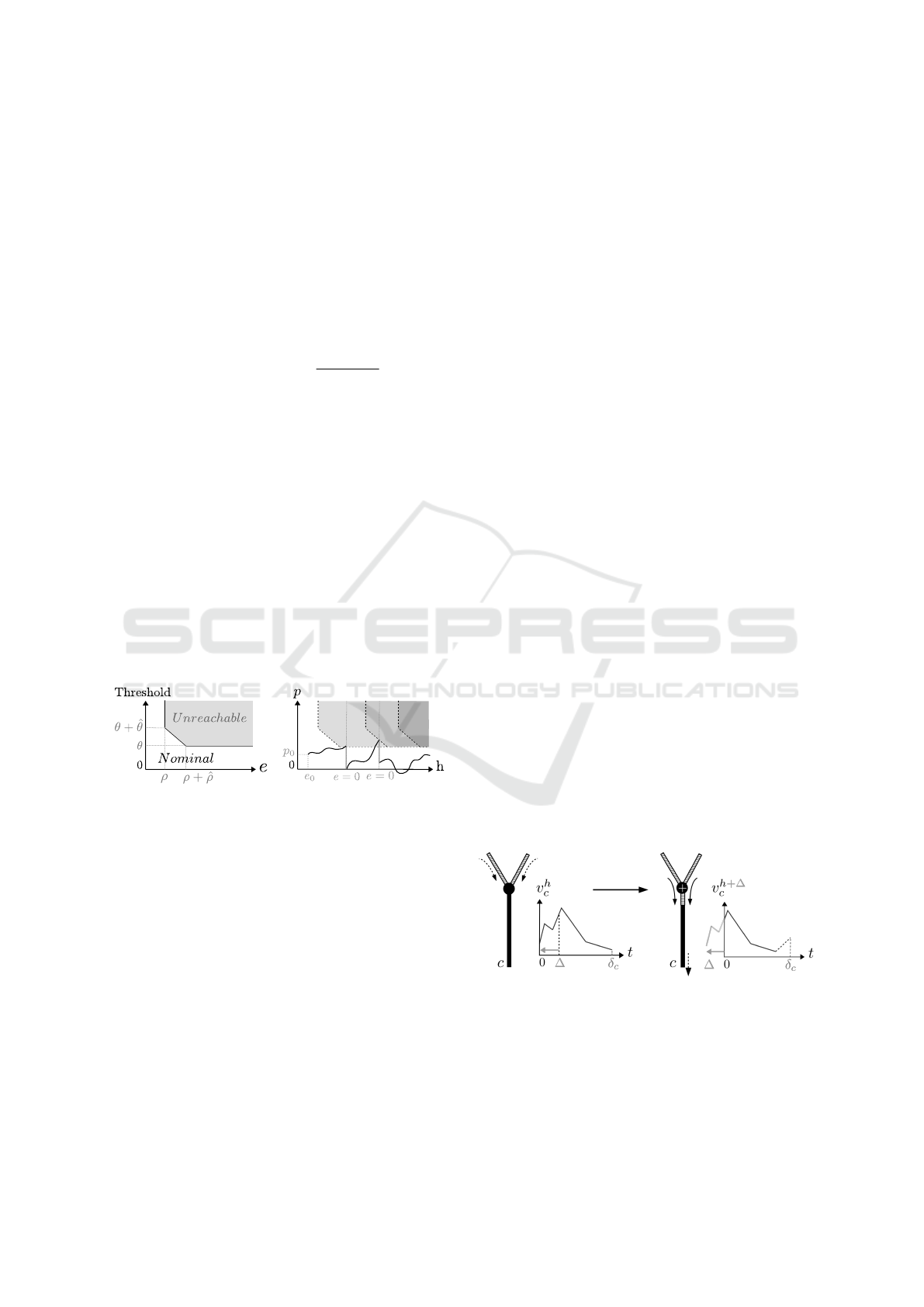
dynamics, meaning how p changes in time. This is
well defined in Definition 10 with Lemma 1 as a basis.
The expression of the derivative is compatible with
the Leaky I&F model.
Lemma 1. [Technical lemma]
Given a soma ∇ = (θ,
ˆ
θ, ρ,
ˆ
ρ, γ), there exists a unique
family of functions P
F
: Nominal(∇) ×IR
+
→ IR in-
dexed by the set of continuous and lipschitzian func-
tions F : IR
+
→IR, such that for any couple (e
0
, p
0
) ∈
Nominal(∇), P
F
satisfies:
• P
F
(e
0
, p
0
,0) = p
0
• ∀h ∈IR
+
the right derivative
dP
F
(e
0
,p
0
,h)
dh
exists and
is equal to F(h) −γ.P
F
(e
0
, p
0
,h)
• ∀h ∈IR
+∗
, ` = lim
t→h
−
(P
F
(e
0
, p
0
,t)) exists and:
– if (h+e
0
,`) ∈Nominal(∇) then P
F
(e
0
, p
0
,h) is
differentiable, therefore P
F
(e
0
, p
0
,h) = `,
– otherwise, for any t > h, P
F
(e
0
, p
0
,t) =
P
G
(0,` −θ,t −h) where G is defined by:
∀u ∈ IR
+
, G(u) = F(u + h).
In other words (Figure 5), from an initial condition
(e
0
, p
0
), the potential changes following its derivative.
It depends on signals coming from the dendrites (F),
while taking into account the leak (γ). Every time the
potential reaches the threshold, its value is updated
(by subtracting θ) and e is reset to zero (keeping the
couple (e, p) nominal). Then, the potential follows
again its derivative with the new couple (e, p) as ini-
tial condition.
Figure 5: Nominal (e,p) couples and soma dynamics.
3.4 State of a Neuron and Dendrites
Dynamics
Intuitively, the state of a neuron is the potential value
at each point. So that it is not only the state of the
soma (e, p) but also the state of the dendrites (denoted
V in Definition 8). According to our compartment
definition, we only observe the value of the potential
at the beginning and at the end of a compartment (by
applying the attenuation factor α). Accordingly, we
define the state of a compartment c at a time t as the
value of the potential at the end of c between time t
and time t + δ
c
. Given α
c
, δ
c
being the time needed
for crossing c, it is thus possible to find the potential
value at any point of c by “looking into the future”
(Figure 6).
Definition 8. [State of a neuron] The state of a neu-
ron N is a triplet η = (V,e, p) where:
• V is called the dendritic state of the neuron. V is
a family of functions, indexed by Co(N), the set
of the compartments of N; each function is of the
form v
c
: [0,δ
c
] →IR where δ
c
is the crossing delay
of the compartment c.
• e ∈ IR
+
represents the elapsed time since the last
spike and p ∈ IR is called the soma potential.
and such that the two following conditions are satis-
fied:
• for each compartment c:
v
c
(δ
c
) ∼
∑
c
0
∈CCo(c)
v
c
0
(0)
!
α
c
where by convention the comparator ”∼” is ”=”
if the input node of c is a branching point, ”>” if
its input node is an excitatory synapse, ”6” if its
input node is an inhibitory synapse.
• the couple (e, p) is nominal for the soma of N.
We note ζ
N
the set of all possible states of the neuron
N.
From the state of a neuron, it is possible to define
its dynamics (see Definition 10). Soma dynamics
is mainly governed by its derivative as seen before.
For dendrites, because signals spread unidirectionally
through compartments and then from one compart-
ment to another, it is possible to calculate the suc-
cession of states by shifting as shown in Figure 6 (see
Definition 9 and Theorem 1). As a simple example,
the potential at the end of a compartment c at a time
h + δ
c
, is equal to the potential at the beginning of c
at time h attenuated by α
c
. The potential at the be-
ginning of c at time h is itself equal to the sum of the
potentials at the end of its contributors compartments
at this given time.
Figure 6: State of a compartment and ∆-shift (Definition 9).
Definition 9. [∆-shift] Given a neuron N and an in-
put signal S = {ω
s
}
s∈Sy(N)
, let V
h
= {v
h
}
c∈Co(N)
be a dendritic state of N at time h and let ∆ ∈ IR
such that 0 6 ∆ 6 in f ({δ
c
| c ∈ CCo(c)}). For any
compartment c ∈ Co(N), the ∆-shift of v
h
c
(noted ∆-
shift(v
h
c
,S )), is the function v : [0,δ
c
] → IR such that:
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
54
• ∀t ∈ [0, δ
c
−∆], v(t) = v
h
c
(t + ∆);
• ∀t ∈ [δ
c
−∆, δ
c
], v(t) = (
∑
c
0
∈CCo(c)
v
h
c
0
(t −δ
c
+∆) +
∑
s∈CSy(c)
v
s
(h +t −δ
c
+ ∆)) ×α
c
.
By extension, the family composed of the ∆-shifts of
all the v
c
∈V is called the ∆-shift of V according to
S and it is noted ∆-shift(V, S ).
Theorem 1. [Compartments dynamics] Given an
initial dendritic state V
I
of a neuron N and an input
signal S = {ω
s
}
s∈Sy(N)
. There exists a unique den-
dritic state family {V
h
}
h∈IR
+
such that:
• V
0
= V
I
;
• for any ∆ such that 0 6 ∆ 6 in f ({δ
c
|c ∈Co(N) ∧
δ
c
> 0}) and for any h, V
h+∆
=∆-shift(V
h
,S ).
Definition 10. [Dynamics of a neuron] Given a neu-
ron N, a state η
I
= (V
I
,e
I
, p
I
) and an input signal
S = {ω
s
}
s∈Sy(N)
, the dynamics of N, according to S
with η
I
as initial state, is the function d
N
: IR
+
→ ζ
N
defined by:
• d
N
(0) = η
I
• ∀h ∈IR
+
, d
N
(h) = η
h
= (V
h
,e
h
, p
h
) where:
– V
h
is defined as in Theorem 1;
– Consider beforehand the lipschitzian function
F(u) =
∑
c∈In(∇)
v
u
c
(0). According to Technical
lemma 1, there exists a unique function P
F
such
that P
F
(e
I
, p
I
,0) = p
I
and ∀u,
dP
F
(e
I
,p
I
,u)
dh
=
F(u) −γ.P
F
(e
I
, p
I
,u). If P
F
(e
I
, p
I
,u) is con-
tinuous on the interval ]0,h], then e = e
I
+ h.
Otherwise, let u
0
be the greatest u such that
P
F
(e
I
, p
I
,u) is discontinuous, then e = h −u
0
.
– Considering the previous P
F
function, p =
P
F
(e
I
, p
I
,h).
4 SIMULATION OF THE HYBRID
MODEL AND MODEL
CHECKING
Our goal is to use our framework to prove particu-
lar properties of neurons. Our framework defines a
hybrid process of signal integration via the dendritic
tree, preserving linear equations. However, the inte-
gration at the soma inevitably induces a differential
equation in order to preserve the essence of the bio-
logical functioning. This is unfortunately detrimental
to computer-aided formal proving approaches. There-
fore, the following step was to discretize time. Obvi-
ously, the smaller the time step is and the more ac-
curate the result will be. But, one must also consider
the computational cost. The proper trade-off largely
depends on the system we are interested in. When
considering neuron spiking, events are usually as-
sumed to be synchronous when happening in a given
1 to 5 milliseconds time window (Izhikevich, 2006;
Brette, 2012; Gr
¨
un et al., 1999). Therefore, our time
step should be less than 1 millisecond. According
to (Bower, 2003), 0.01 millisecond is an appropriate
choice when looking at the shape of the action poten-
tial. Here, as we consider discrete spikes, we have
chosen 0.1 millisecond as a reasonable time step (∆t).
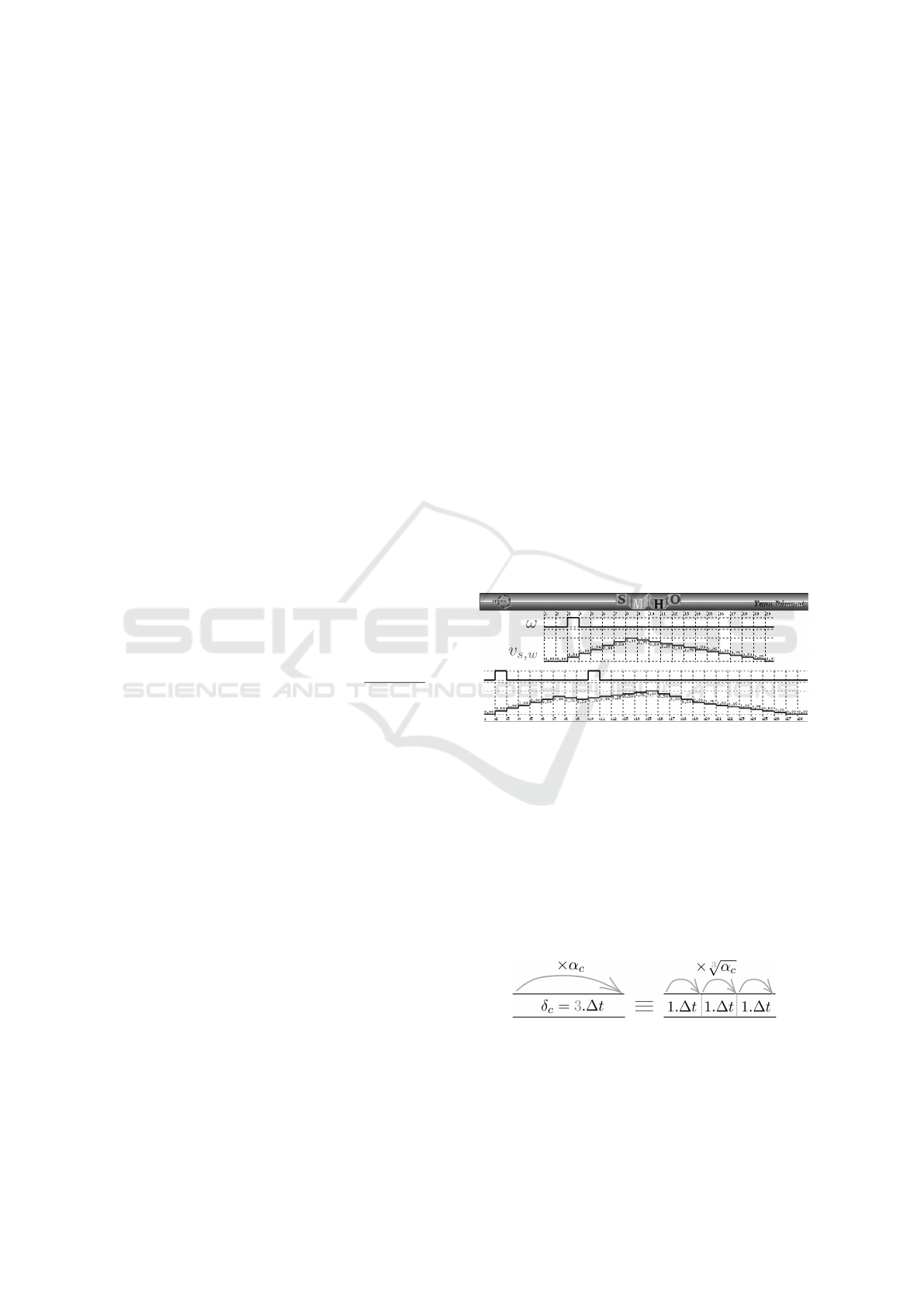
4.1 Timing Discretization
For simulation and model checking purposes, all the
temporal parameters have to be expressed in multiples
of ∆t.
Synapses. Usually,
ˆ
τ (sometimes called time-to-
peak) goes from one to tens of milliseconds and
ˇ
τ is
most of the time greater (Williams and Stuart, 2002;
Magee and Cook, 2000). This makes our time step ap-
propriate, as a sufficiently large number of points will
be considered in the trace of the signal (Figure 7).
Figure 7: Simulation of a trace (v
s,ω
) of an input signal (ω)
on a synapse (s) with arbitrary parameters (ν = 3,
ˆ
τ = 6
and
ˇ
τ = 12). It is encoded in Lustre and simulated with
luciole. On the top is represented a trace in response to
a single spike. On the bottom, the trace exhibits temporal
summation.
Compartments. The delay for crossing a compart-
ment (δ) being expressed in multiples of ∆t, com-
partments smaller than 1∆t will be considered as
null. This assumption may lead to kind of clusters of
synapses, which are observed experimentally (Stuart
et al., 2016; G
¨
okc¸e et al., 2016).
Figure 8: Division of a compartment into sub-
compartments for proper discretization.
For each compartment, we define n = δ/∆t sub-
compartments (noted subC, see Figure 8). Sub-
compartments parameters are defined as follows:
Computer-aided Formal Proofs about Dendritic Integration within a Neuron
55

δ
subC
= 1∆t and α
subC
=
n
√
α (where n is the num-
ber of sub-compartments in the original compartment
and α is the attenuation of the original compartment).
Given this representation, the dynamics is done by
shifting all the sub-compartments at a time.
Soma. The major motivation for discretization is the
time discretization at the soma. Basically, we chose
to replace a differential equation with a difference
to be solved at discrete time intervals (∆t). From
Lemma 1, we have:
dP
dt
= F(t) −γ.P(t). Once time is
discretized, since P is piecewise linear, this equation
is equivalent to
P(t+∆t)−P(t)
∆t
= F(t) −γ.P(t). We thus
have P(t +∆t)−P(t) = (F(t)−γ.P(t)).∆t. Therefore,
we should use the following equation:
P(t + ∆t) = F(t).∆t + P(t).(1 −γ.∆t).
Linearity properties (resulting from Definition 3)
make this discretization exact. However, spikes times
are approximated and this could lead to some error
accumulation. Nevertheless, there is a limited risk
of missing any spikes as the ARP is in the order
of a millisecond, largely limiting the maximal fre-
quency (Johnson, 2003).
4.2 Formal Proofs using Model
Checking: a Simple Example
Based on our discretized model, we can use model
checking in order to prove properties on neurons with
dendrites in a computer-aided manner. Model check-
ing allows one to automatically verify if a model sat-
isfies a given property. We have chosen to encode our
model in Lustre and using Kind2 as model checker
(with z3 solver).
Lustre. Lustre is a synchronous programming lan-
guage, working with flows. It benefits from a formal
definition and it is particularly suited to reactive sys-
tems. A program in Lustre consists of nodes. A node
computes outputs based on algebraic equations, ac-
counting for input variables. All variables are typed.
Local variables can be declared using the keyword
var. The body of the node is composed of set of
equations always true, surrounded by the keywords
let and tel. For this reason, Lustre is a declarative
language and not imperative. A node is declared as
follows:
node nameOftheNode(input1: typeInput1; ...)
returns(output1: typeOutput1; ...);
var localVar1: typeVar1; ...;
let
Equations;
tel
In Lustre, a variable does not carry a given value
but a function of time, that is an infinite sequence of
values. As an example, to define a variable x, we
should use the following syntax:
x = initial value -> induction principle;
The values carried by a variable are computed by
a temporal recurrence expression which can depend
on other variables. The initial value is the first
value of the infinite sequence. It splits up the other
values by the operator -->. Lustre has some elemen-
tary basic types (bool, int, real) and usual operators
can be used on them ( +,-,*, etc.; and,or,not;
if then else). As an example, if a variable x is the
infinite flow (x
1
, x
2
,...) and another variable y is the
infinite flow (y
1
, y
2
,...) then, the variable z = x + y is
the infinite flow (z
1
= x
1
+ y
1
, z
2
= x
2
+ y
2
,...).
Moreover there exists in Lustre an additional op-
erator to deal with logical time: pre(). It acts as a
memory by providing access to the previous value of
a variable at a given time. For instance if the variable
x is defined by x = 0 -> pre(x)+1, the flow will be
equal to 0 at time 0, equal to 0+1 = 1 at time 1, equal
to 1 + 1 = 2 at time 2, and so on.
Any node can be simulated by a dedicated Lustre sim-
ulator called luciole (see Figure 7).
Kind2. There are several model checkers for Lus-
tre. Kind2 relies on SMT (Satisfiabitily Modulo The-
ories) based k-induction and it proved to be efficient
contrary to others model checkers such as lesar, nbac,
luke and rantanplan (De Maria et al., 2016).
Kind2 is able to prove mathematical properties en-
coded as Lustre node: We write, in Lustre, the prop-
erty as a boolean variable and Kind2 verifies if it is
always true. As an example, to check if a variable x
is equal to a variable y, meaning that at each time step
the values of the two variables are the same, one can
write the following node:
node property(x, y : int) returns (proof: bool);
let
proof = x = y;
tel
Here, the output of the node property returns the
value of proof which is true when x = y and false
otherwise. The property is thus satisfied if proof is
always true whatever the values of x and y, at any time
step.
A Simple Example.
As a first property to check, we have decided to
compare the outputs of simple neurons for a given in-
put and given initial states.
Let us consider the two neurons illustrated in Fig-
ure 9. Following Definition 10 and since both neu-
rons have the same soma parameters, it is sufficient
to compare the output of their dendritic forests. We
have thus ignored the soma at first. We have en-
coded synapses and compartments in Lustre by defin-
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
56
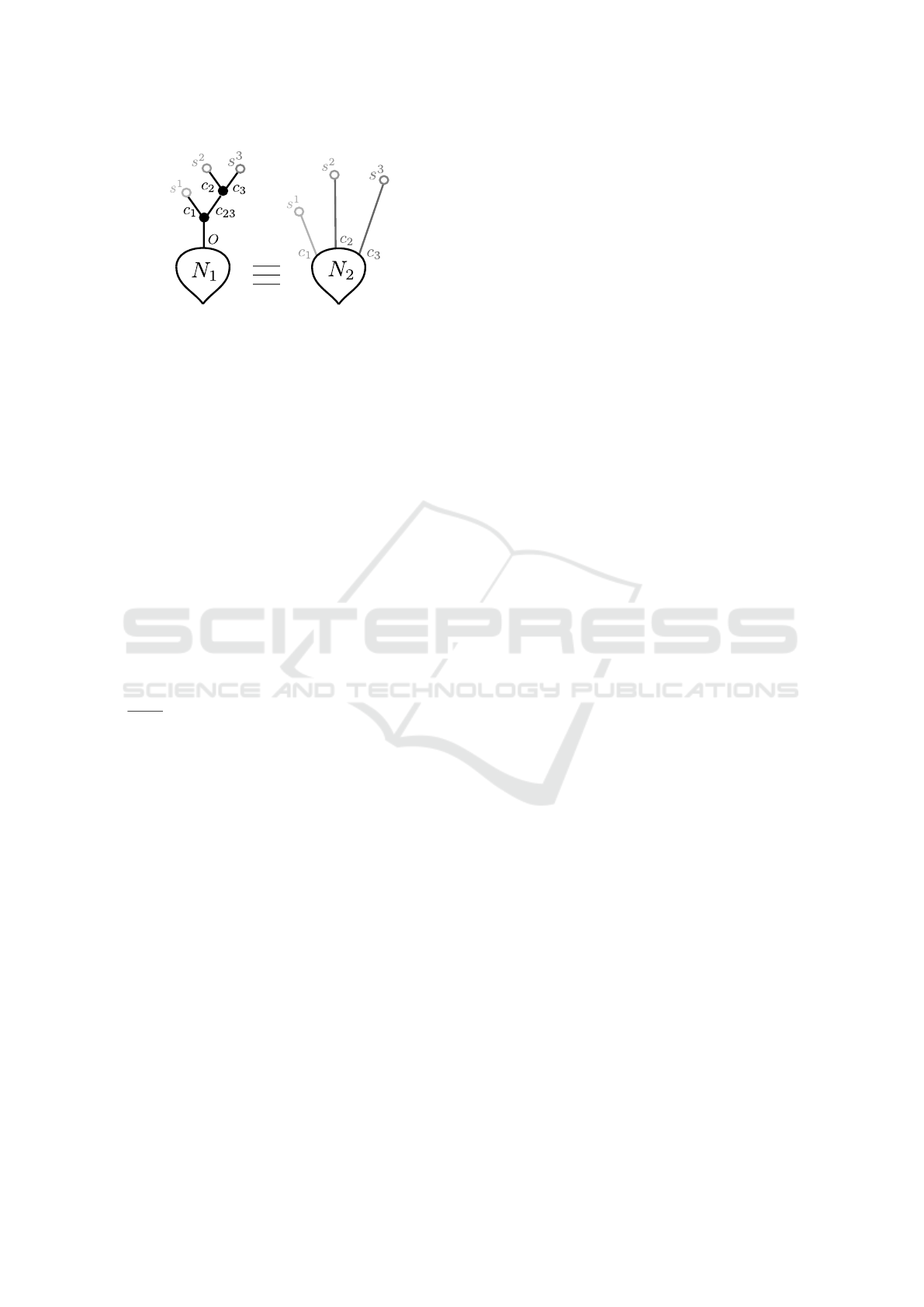
Figure 9: Example of two simple neurons. Their parameters
are δ = 1 and α = 0.5 for all black compartments (in neuron
N
1
), δ = 2 and α = 0.25 for the light grey compartment (c
1
in neuron N
2
), δ = 3 and α = 0.125 for dark gray compart-
ments (c
2
and c
3
in neuron N
2
). Synapses s
1
are defined
by ν = 5,
ˆ
τ = 5,
ˇ
τ = 15, synapses s
2
are defined by ν = 3,
ˆ
τ = 6,
ˇ
τ = 12 and synapses s
3
are defined by ν = −5,
ˆ
τ = 5,
ˇ
τ = 10. N
1
and N
2
both have the same soma parameters.
ing nodes for each of them. Below, we show as
an example, how a compartment can be defined for
δ = 3.∆t (with α = 0.125). Compartments with dif-
ferent parameters are encoded in the same way.
node comp3dt (I, init1, init2, initO : real)
returns (O : real);
var c1, c2, alphaSubC : real;
let
alphaSubC = 0.5;
c1 = init1 -> pre(I) * alphaSubC;
c2 = init2 -> pre(c1) * alphaSubC;
O = initO -> pre(c2) * alphaSubC;
tel
As shown in Figure 8, the compartment is divided into
3 sub-compartments with δ
subC
= 1.∆t and α
subC
=
3
√
0.125 = 0.5. The node comp3dt takes as inputs
real variables: the input (I) of the compartment
(representing the signals coming from the contribu-
tor compartments or synapses) and the initial states
(init1, init2, initO) of each sub-compartment.
It then computes the output (O) of the whole com-
partment (with initO as initial value). The variables
(c1) and (c2) are the outputs of respectively the first
and the second sub-compartments. The output of the
first sub-compartment is computed from the value of
the input I, one ∆t before (using the operator pre()).
Similarly, the output of the second sub-compartment
is computed from the output of the first one, and so
on.
To encode synapses, one node is required for each
couple of parameters (
ˆ
τ,
ˇ
τ). The node presented below
encodes a simple excitatory synapse with ν = 1,
ˆ
τ = 2
and
ˇ
τ = 4.
node synapseE2_4 (omega : bool)
returns (vs : real);
var nu, hatTau, checkTau : real;
o1, o2, o3, o4, o5 : bool;
let
nu = 1.0;
hatTau = 2.0;
checkTau = 4.0;
o1 = false -> pre(omega);
o2 = false -> pre(o1);
o3 = false -> pre(o2);
o4 = false -> pre(o3);
o5 = false -> pre(o4);
vs = 0.0 -> (if o1
then nu/hatTau
else 0.0)
+ (if o2
then 2.0*nu/hatTau
else 0.0)
+ (if o3
then nu-nu/checkTau
else 0.0)
+ (if o4
then nu-2.0*nu/checkTau
else 0.0)
+ (if o5
then nu-3.0*nu/checkTau
else 0.0);
tel
The node synapseE2_4 takes as input the boolean
variable omega that represents the spikes sequence
and computes the output vs that is the trace of the
input signal omega on the synapse. The local vari-
ables nu, hatTau and checkTau are respectively the
maximal potential variation triggered by a spike, the
rise time and the descent delay. As shown in Fig-
ure 3, the contribution of a spike to the trace at a
given time greatly depends on the time elapsed since
the spike occurrence. The local boolean variable o1
is true if omega was true one time step before the cur-
rent time, meaning that there was a spike at the pre-
vious time. Similarly, o2 is true if omega was true
two time steps before the current time, meaning that
there was a spike two times before, and so on. In
this case, we have to look up to
ˆ
τ +
ˇ
τ −1 = 5 time
steps back because beyond this time window a spike
no longer influences the trace. In order to produce a
temporal summation, the trace is computed from the
contributions of all the preceding spikes. In the same
way, we define nodes for inhibitory synapses called
synapseIx_y where nu is a negative real number.
Once synapses and compartments are encoded as
Lustre nodes, it is possible to define the entire den-
dritic forest for both neurons:
node neuron1(I1, I2, I3 : bool)
returns (O : real);
var S1, S2, S3, C1, C2, C3, C23,
initC1, initC2, initC3, initC23,
initO : real;
let
initC1 = 0.0; initC2 = 0.0;
initC3 = 0.0; initC23 = 0.0;
initO = 0.0;
S1 = synapseE5_15(I1);
S2 = synapseE6_12(I2);
Computer-aided Formal Proofs about Dendritic Integration within a Neuron
57

S3 = synapseI5_10(I3);
C1 = comp1dt(S1, initC1);
C2 = comp1dt(S2, initC2);
C3 = comp1dt(S3, initC3);
C23 = comp1dt(C2 + C3, initC23);
O = comp1dt(C1 + C23, initO);
tel
node neuron2(I1, I2, I3 : bool)
returns (O : real);
var S1, S2, S3, C1, C2, C3,
initC11, initC12, initC21, initC22,
initC23, initC31, initC32, initC33,
initO : real;
let
initC11 = 0.0; initC12 = 0.0;
initC21 = 0.0; initC22 = 0.0;
initC23 = 0.0; initC31 = 0.0;
initC32 = 0.0; initC33 = 0.0;
initO = 0.0;
S1 = synapseE5_15(I1);
S2 = synapseE6_12(I2);
S3 = synapseI5_10(I3);
C1 = comp2dt(S1,initC11,initC12);
C2 = comp3dt(S2,initC21,initC22,initC23);
C3 = comp3dt(S3,initC31,initC32,initC33);
O = initO -> C1 + C2 + C3;
tel
Both nodes (neuron1 and neuron2) compute a
boolean output (O) from boolean inputs I1, I2, I3
(representing sequences of spikes at synapses). As
mentioned before, the two neurons have the same
set of synapses (Figure 9). The real variables
S1, S2, S3 are the outputs of the nodes encoding
respectively synapses s
1
, s
2
, s
3
with I1, I2, I3 as
inputs. They are thus used as inputs for compart-
ments comp1dt, comp2dt, comp3dt whose outputs
C1, C2, C3, C23 are, in turn, used as inputs for
other compartments. It allows ultimately to compute
the output of the whole dendritic forest. For the sake
of simplicity, compartment initial states were fixed to
zero in both neurons. Under this condition and by
considering constant inputs (always equal to true or
false), we have proven that these two particular neu-
rons (Figure 9) always produce the same outputs. All
the possible inputs combinations (2
3
= 8) were veri-
fied. Below is the Lustre node encoding the property
with I1 and I2 being always true and I3 being always
false:
node equivalence(gost : bool, epsilon: real)
returns (proof: bool);
var I1, I2, I3 : bool;
let
I1 = true;
I2 = true;
I3 = false;
proof = abs(neuron1(I1, I2, I3)
- neuron2(I1, I2, I3)) < epsilon;
tel
Because neurons compute with real numbers,
rounding errors in computation prevent to define
proof as the simpler equality neuron1(I1, I2,
I3) = neuron2(I1, I2, I3);. In fact, with our
example, this equality version the proof succeeds too:
neuron1 and neuron2 make the same approxima-
tions.
We have also proven that the property is satis-
fied for other arbitrary inputs. As an example, we
used I1 being true on 1 time step in 50, I2 be-
ing true on 1 time step in 10 and I3 being true on
1 time step in 20.
For constant inputs, the model checker is remark-
ably efficient. For inputs with frequencies, we reach
the limits of kind2 on a standard laptop, as the proof
takes several hours.
Here, proofs have been entirely automatically han-
dled by the model checker. Even though this example
is simple, the ability of Kind2 to fully perform proofs
without human guidance is encouraging.
5 CONCLUSION
We defined here the first hybrid formal model of neu-
ron taking into account the morphology and its key
role in neuronal computation. Our aim was to use for-
mal methods from computer science to prove proper-
ties based on this framework. In order to make au-
tomatic proofs, we have used model checking. The
main interest of this proof-based approach is the ex-
ploration of the whole space of possibilities inside a
single proof. However, it requires models with dis-
crete time. We thus proposed relevant abstractions of
the neuron structure and dynamics with a subsequent
time discretization.
Nevertheless, the number of parameters is still
huge and it increases as we get closer to the biolog-
ical reality. A “standard” neuron has thousands of
synapses located on its dendrites making the system
very complex.
Here, we successfully used model checking on a
simple example. We proved that two particular neu-
rons with different dendritic structures can have the
same input/output function. This first result empha-
sises the crucial role of delays and attenuations rather
than the importance of the precise dendrites morphol-
ogy. It is partly a consequence of the hypotheses and
choices we made. If we would like to incorporate
some other biological properties (such as shunting)
to our model, it would probably change this basic re-
sult (H
¨
ausser and Mel, 2003; Paulus and Rothwell,
2016; Gorski et al., 2017). But, even though our
modelling relies on several simplifying assumptions,
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
58

it constitutes a first track to investigate the impact of
the morphology on the neuron function, the first for-
mal one.
As a direct continuation of this work, it would be
interesting to make more general proofs and possi-
bly with more complex and biologically relevant ex-
amples. The ultimate aim would be to automatically
find constraints on parameters, such as observed de-
lays provided by dendrites, for a model to satisfy a
given behaviour. In another research direction, we
think about building neuronal circuits. This would
probably bring insights on how the neuronal structure
could enable the emergence of complex behaviour at
a larger scale.
REFERENCES
Bianchi, S., Stimpson, C. D., Bauernfeind, A. L., Schapiro,
S. J., Baze, W. B., McArthur, M. J., Bronson, E.,
Hopkins, W. D., Semendeferi, K., Jacobs, B., et al.
(2012). Dendritic morphology of pyramidal neu-
rons in the chimpanzee neocortex: regional special-
izations and comparison to humans. Cerebral cortex,
23(10):2429–2436.
B
¨
orgers, C. (2017). Quadratic integrate-and-fire (qif) and
theta neurons. In An Introduction to Modeling Neu-
ronal Dynamics, pages 51–55. Springer.
Bower, J.M., B. D. (2003). The book of GENESIS: explor-
ing realistic neural models with the General Neural
Simulation System. Internet Edition.
Brette, R. (2003). Mod
`
eles impulsionnels de r
´
eseaux de
neurones biologiques. PhD thesis, Universit
´
e Pierre et
Marie Curie-Paris VI.
Brette, R. (2012). Computing with neural synchrony. PLoS
computational biology, 8(6):e1002561.
Brette, R. and Gerstner, W. (2005). Adaptive exponen-
tial integrate-and-fire model as an effective descrip-
tion of neuronal activity. Journal of neurophysiology,
94(5):3637–3642.
Brunel, N. and Van Rossum, M. C. (2007). Lapicques 1907
paper: from frogs to integrate-and-fire. Biological cy-
bernetics, 97(5-6):337–339.
Cook, E. P. and Johnston, D. (1997). Active dendrites re-
duce location-dependent variability of synaptic input
trains. Journal of neurophysiology, 78(4):2116–2128.
De Maria, E., Muzy, A., Gaff
´
e, D., Ressouche, A., and
Grammont, F. (2016). Verification of temporal prop-
erties of neuronal archetypes modeled as synchronous
reactive systems. In International Workshop on Hy-
brid Systems Biology, pages 97–112. Springer.
G
¨
okc¸e, O., Bonhoeffer, T., and Scheuss, V. (2016). Clusters
of synaptic inputs on dendrites of layer 5 pyramidal
cells in mouse visual cortex. eLife, 5:e09222.
Gorski, T., Veltz, R., Galtier, M., Fragnaud, H., Telenczuk,
B., and Destexhe, A. (2017). Inverse correlation pro-
cessing by neurons with active dendrites. bioRxiv.
Gr
¨
un, S., Diesmann, M., Grammont, F., Riehle, A., and
Aertsen, A. (1999). Detecting unitary events without
discretization of time. Journal of neuroscience meth-
ods, 94(1):67–79.
H
¨
ausser, M. and Mel, B. (2003). Dendrites: bug or feature?
Current opinion in neurobiology, 13(3):372–383.
H
¨
ausser, M., Spruston, N., and Stuart, G. J. (2000). Di-
versity and dynamics of dendritic signaling. Science,
290(5492):739–744.
Hodgkin, A. L. and Huxley, A. F. (1952). A quantitative
description of membrane current and its application to
conduction and excitation in nerve. The Journal of
physiology, 117(4):500–544.
Hu, H. and Vervaeke, K. (2017). Synaptic integration in
cortical inhibitory neuron dendrites. Neuroscience.
Izhikevich, E. M. (2006). Polychronization: computation
with spikes. Neural computation, 18(2):245–282.
Johnson, L. R. (2003). Essential medical physiology. Aca-
demic Press.
Johnston, D. and Narayanan, R. (2008). Active dendrites:
colorful wings of the mysterious butterflies. Trends in
neurosciences, 31(6):309–316.
Koch, C. (2004). Biophysics of computation: information
processing in single neurons. Oxford university press.
Lapicque, L. (1907). Recherches quatitatives sur
l’excitation electrique des nerfs traitee comme polari-
sation. J. Physiol. Pathol. Gen., 9:620–635.
Maass, W. (1996). Lower bounds for the computational
power of networks of spiking neurons. Neural com-
putation, 8(1):1–40.
Maass, W. (1999). Computing with spiking neurons. Pulsed
neural networks, 85.
Magee, J. C. and Cook, E. P. (2000). Somatic epsp am-
plitude is independent of synapse location in hip-
pocampal pyramidal neurons. Nature neuroscience,
3(9):895.
Manita, S., Miyakawa, H., Kitamura, K., and Murayama,
M. (2017). Dendritic spikes in sensory perception.
Frontiers in cellular neuroscience, 11.
McCulloch, W. S. and Pitts, W. (1943). A logical calculus
of the ideas immanent in nervous activity. The bulletin
of mathematical biophysics, 5(4):115–133.
Mel, B. W. (1994). Information processing in dendritic
trees. Neural computation, 6(6):1031–1085.
Mohan, H., Verhoog, M. B., Doreswamy, K. K., Eyal,
G., Aardse, R., Lodder, B. N., Goriounova, N. A.,
Asamoah, B., B. Brakspear, A. C., Groot, C., et al.
(2015). Dendritic and axonal architecture of individ-
ual pyramidal neurons across layers of adult human
neocortex. Cerebral Cortex, 25(12):4839–4853.
Paulus, W. and Rothwell, J. C. (2016). Membrane resis-
tance and shunting inhibition: where biophysics meets
state-dependent human neurophysiology. The Journal
of physiology, 594(10):2719–2728.
Popper, K. R. (1963). Conjectures and refutations: the
growth of scientific knowledge.
Rall, W. (1959). Branching dendritic trees and motoneu-
ron membrane resistivity. Experimental neurology,
1(5):491–527.
Computer-aided Formal Proofs about Dendritic Integration within a Neuron
59

Rall, W. (1962). Theory of physiological properties of den-
drites. Annals of the New York Academy of Sciences,
96(1):1071–1092.
Rall, W. (1967). Distinguishing theoretical synaptic poten-
tials computed for different soma-dendritic distribu-
tions of synaptic input. Journal of neurophysiology,
30(5):1138–1168.
Rall, W. (2011). Core conductor theory and cable properties
of neurons. Comprehensive physiology.
Remme, M. W., Lengyel, M., and Gutkin, B. S. (2010).
Democracy-independence trade-off in oscillating den-
drites and its implications for grid cells. Neuron,
66(3):429–437.
Stuart, G., Spruston, N., and H
¨
ausser, M. (2016). Dendrites.
Oxford University Press.
Stuart, G. J. and Spruston, N. (2015). Dendritic integra-
tion: 60 years of progress. Nature neuroscience,
18(12):1713–1721.
Sun, Q., Srinivas, K. V., Sotayo, A., and Siegelbaum, S. A.
(2014). Dendritic na+ spikes enable cortical input to
drive action potential output from hippocampal ca2
pyramidal neurons. Elife, 3:e04551.
Williams, S. R. and Stuart, G. J. (2002). Dependence of
epsp efficacy on synapse location in neocortical pyra-
midal neurons. Science, 295(5561):1907–1910.
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
60
