
Comparison of Decision Tree, Neural Network, Statistic Learning,
and k-NN Algorithms in Data Mining of Thyroid Disease Datasets
Wafaa Al Somali and Riyad Al Shammari
Department of Health Informatics, King Saud Bin Abdul-Aziz University for Health Sciences, Riyadh, Saudi Arabia
Keywords: Data Mining, Knowledge Discovery, Thyroid Dataset.
Abstract: Massive information contained in medical datasets presents challenge to the practitioners in diagnosing
diseases or determining health status of patients. Data mining is therefore required to help users obtaining
valuable information from a very complex data collection. In this study, we explored several methods of data
mining in order to improve the quality of a dataset which is related to diagnosis of thyroid disease. Several
classifiers were trained on the dataset and compared to previous study by Akbas
̧
et al (2013). The performance
improvement was examined in order to determine the best classifier that can be executed. Findings revealed
that decision tree (J48) algorithm outperformed all other algorithms in terms of accuracy, Kappa, Matthew’s
correlation coefficient (MCC), and receiver operating characteristics (ROC) with respective values of 0.994,
0.951, 0.953, and 0.987. Classification using J48 was found to be better than those conducted by Akbas
̧
et al.
In contrast, IBK algorithm showed the poorest performance, particularly Kappa and MCC. The size of tree
generated from J48 and Logistic Model Tree (LMT) varied greatly. Integration of single classifier with
AdaBoost classifier mostly resulted in higher accuracy. However, AdaBoost did not improve the performance
of NaïveBayes, IBK and RandomForest algorithms. These results were consistent with the previous study
using AdaBoost-based ensemble classifier.
1 INTRODUCTION
Medical datasets are growing rapidly in size.
Therefore, data mining (DM) is inseparable from the
process of establishing knowledge-based decision
(Yeh et al., 2008).
Over the last decades, studies have
been exploring the DM technique with optimum
results. The main task in DM of medical dataset is
classification (Olafsson et al., 2008).
The labelled
subjects are partitioned into predefined groups or
classes by using a suitable DM model, resulting in
new instances. Knowledge discovery in DM might be
achieved through an ‘if – then’ rules, knowledge
representation, and many more (Mohamadi et al.,
2008).
2 BACKGROUND
Health care related datasets are usually huge and
complex. Thyroid disease dataset has taken from UCI
Machine Learning repository). Thyroid disease
dataset has 7200 instances, with both continuous and
discrete inputs. The purpose of this dataset is to
determine whether the patient has hypothyroid,
hyperthyroid, or normal thyroid function.
Hypothyroid is a condition where insufficient level of
thyroid hormones are produced. Hyperthyroid, on the
contrary, is an abnormality where thyroid hormones
are produced in exceeding level. The attributes in the
datasets are based on clinical observation which
measures the percentage of T3-resin uptake, serum
thyroxin level, serum triiodothyronine level, basal
thyroid-stimulating hormone, and maximal absolute
difference of thyroid stimulating hormone level
(Chang et al.. 2010, Ozyilmaz and Yildirim. 2002).
It is an exhaustive task to diagnose thyroid disease
by only interpreting clinical observations and
examining medical history of the patient. Moreover,
thyroid disease is not only related to the production of
thyroid hormones. Other abnormalities such as
thyroid cancer might be confused with
hyperthyroidism (Ozyilmaz and Yildirim. 2002).
An
appropriate DM model will solve this problem and
help users to diagnose a disease more accurately.
Somali, W. and Shammari, R.
Comparison of Decision Tree, Neural Network, Statistic Learning, and k-NN Algorithms in Data Mining of Thyroid Disease Datasets.
DOI: 10.5220/0006479602410246
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 5: HEALTHINF, pages 241-246
ISBN: 978-989-758-281-3
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
241

2.1 Data Mining
Data mining (DM) is a process of obtaining valuable
data from a huge-sized data collection
(Giannopoulou, 2008).
Therefore, DM is an
important task in generating knowledge-based
decisions. A high number of DM programs has been
developed using various algorithms and approaches,
commonly known as classifiers. A classifier
determines a model for a collection of instances or
target values by predicting general rules.
3
In clinical
DM process, classification is often performed on
previous data stored in medical records and databases
in order to improve the quality of the data (Ressom et
al.. 2008, Iavindrasana et al.. 2010, Fernandez et al..
2010).
2.2 Thyroid Disease Data Mining
Various algorithm has been used in classification of
thyroid disease dataset, including decision tree, ANN,
RBF, MLP with BP and CSFNN (Ozyilmaz and
Yildirim. 2002, Fernandez et al., 2010).
In another
study, a high number of algorithms was applied,
including C4.5, Random Tree, Logistic Regression,
k-NN, Naïve Bayes, and SVM. Performance
evaluation was conducted by determining accuracy,
rate of error, and tree size (Jacob and Ramani, 2012).
We compared our data mining to a study by Akbas
̧
et al. (2013). The study describes the performance
improvement in diagnosis of thyroid cancer by
training and testing datasets using several classifiers
(Akbas
̧
et al., 2013).
Five individual classifiers were
used and compared for their accuracy, kappa,MCC
and ROC values: BayesNet, NaÏveBayes, SMO, IBk
and Random Forest. Ensemble classifiers were also
used in addition to individual classifiers, by
combining them with AdaboostMI. Algorithm
process comprised of three stages: dataset training,
dataset testing, and reclassification. All data mining
was performed on WEKA developer version 3.7.13.
Improvement in the diagnosis of thyroid cancer was
indicated by ROC graphs. Random Forest was found
to be the most accurate classifier (Akbas
̧
et al., 2013).
The value for accuracy, kappa, MCC, and ROC in
Random Forest were 0.99, 0.937, 0.941, and 0.998,
respectively (Akbas
̧
et al., 2013). In combination with
AdaboostMI, the value for accuracy, kappa, MCC,
and ROC in Random Forest remained the highest,
being 0.991, 0.939, 0.940, and 0.998, respectively
(Akbas
̧
et al., 2013).
In this study, we proposed several methods of data
mining of Thyroid dataset. The objectives of this
study were to obtain a classification algorithm that
can improve the quality of Thyroid dataset, to apply
several classifiers by training and testing on the
dataset and compare the results with previous study,
and to measure the performance improvement by
each algorithm in order to determine the best
classifier that can be executed.
3 METHODOLOGY
3.1 Individual Classifier
Classification of Thyroid disease dataset (retrieved
https://archive.ics.uci.edu/ml/datasets/Thyroid+Dise
ase), consisted of 21 attributes and 7200 instances,
were performed. The thyroid dataset was divided into
two groups: training group (3772 instances) and
testing group (3428 instances). In the pre-processing
the obtained dataset converted it to CSV (comma
separated values) format, the class column values
have been changed it from numeric to nominal.
Classifier algorithms used in this study were neural
network (Multiple Layer Perceptron/MLP) and
decision tree classifiers (J48 and Logistic Model
Tree/LMT). These classifiers were chosen because
thyroid disease dataset comprises of continuous and
discrete values. Thyroid dataset was also classified
using several algorithms used by Akbas
̧
et al (2013),
including BayesNet, NaiveBayes, sequential minimal
optimization (SMO), IBk, and RandomForest.
3.2 Ensemble Classifier
Individual classifiers were integrated into an
ensemble classifier, together with AdaBoost, to
evaluate whether ensemble classifier can improve the
performance of individual classifier. All DM
procedures was performed on WEKA developer
version 3.7.13.
3.3 Performance Evaluation
Performance improvement evaluation was conducted
by determining classification accuracy, Kappa
statistics, Matthews correlation coefficient (MCC),
receiver operating characteristics (ROC) value,
decision tree size (only for decision tree classifier.
3.4 Experiment
In this study, training and testing datasets were
separated for each classification method. This
technique was chosen to remove unnecessary data.
Thyroid dataset is a very large dataset and removal of
HEALTHINF 2018 - 11th International Conference on Health Informatics
242

unnecessary data was required. Accuracy was
determined by using equation 1, where 1 was
considered as the highest accuracy. Kappa statistics
was used to assess the reliability of cohesion between
two data, as shown in equation 2. Higher Kappa
means higher reliability and zero is considered the
worst result. Matthew’s correlation coefficient was
calculated by using equation 3, while ROC was
calculated by using equation 4.
4 RESULTS AND DISCUSSION
4.1 Individual Classifiers
In our proposed study, Thyroid dataset was classified
by using J48 tree, LMT tree, and multiple layer
perceptron (MLP) algorithms. The results of
performance evaluation are shown in Table 1. It was
found that J48 resulted in highest accuracy, Kappa,
and MCC. The highest ROC values was obtained
using LMT tree. The tree size in J48 was 23,
significantly higher than the tree size in LMT tree
(only 5). There were 12 leaves in J48 and only 3
leaves in LMT. In comparison with decision tree
models (J48 and LMT), MLP as a neural network
resulted in lower accuracy, Kappa, MCC, and ROC.
This finding is contrary to our expectation, where
decision tree algorithm was more likely to have lower
classification accuracy because the high variance in
decision tree that potentially lead to overfitting. We
also expected that neural network would perform
better on a large dataset. Kotsiantis (2007) has
reported that neural network would take longer time
to execute in comparison with decision trees
(Kotsiantis, 2007).
However, neural network are
usually able to provide learning more easily in
comparison to decision trees (Bostanci and Bostanci,
2012).
In this study, classification using other algorithms
were also performed in order to compare our findings
to the study conducted by Akbas
̧
et al (2013). The
results, summarized in Table 2, were consistent.
RandomForest was the most accurate classifier for the
Thyroid dataset. It also produced the highest Kappa,
MCC, and ROC. IBK algorithm was found to be the
poorest model in terms of all performance indicators.
Among all classification models, J48 showed the
best performance. Decision tree model is expected to
have a great performance on discrete or categorical
datasets, including Thyroid disease (Kotsiantis,
2007).
However, it might be not suitable for a very
large datasets or when the splitting of nodes are based
on more than one feature (Ozyilmaz and Yildirim.
2002),(Kotsiantis, 2007).
IBK showed poor accuracy,
reliability of cohesion, and ROC because IBK is a K-
Nearest Neighbor (KNN) classifier that is more
suitable for nominal and numerical data (Bostanci and
Bostanci, 2012).
4.2 Ensemble Classifiers
In this study, combination of classifier with Adaptive
Boosting algorithm (AdaBoost) resulted in higher
accuracy in all classifiers except NaïveBayes, IBK
and RandomForest models, as shown in Table 3 and
4. Moreover, Kappa, MCC and ROC values were
successfully improved in LMT model. Increased
accuracy in BayesNet and SMO was in accordance
with previous study by Akbas
̧
et al (2013).
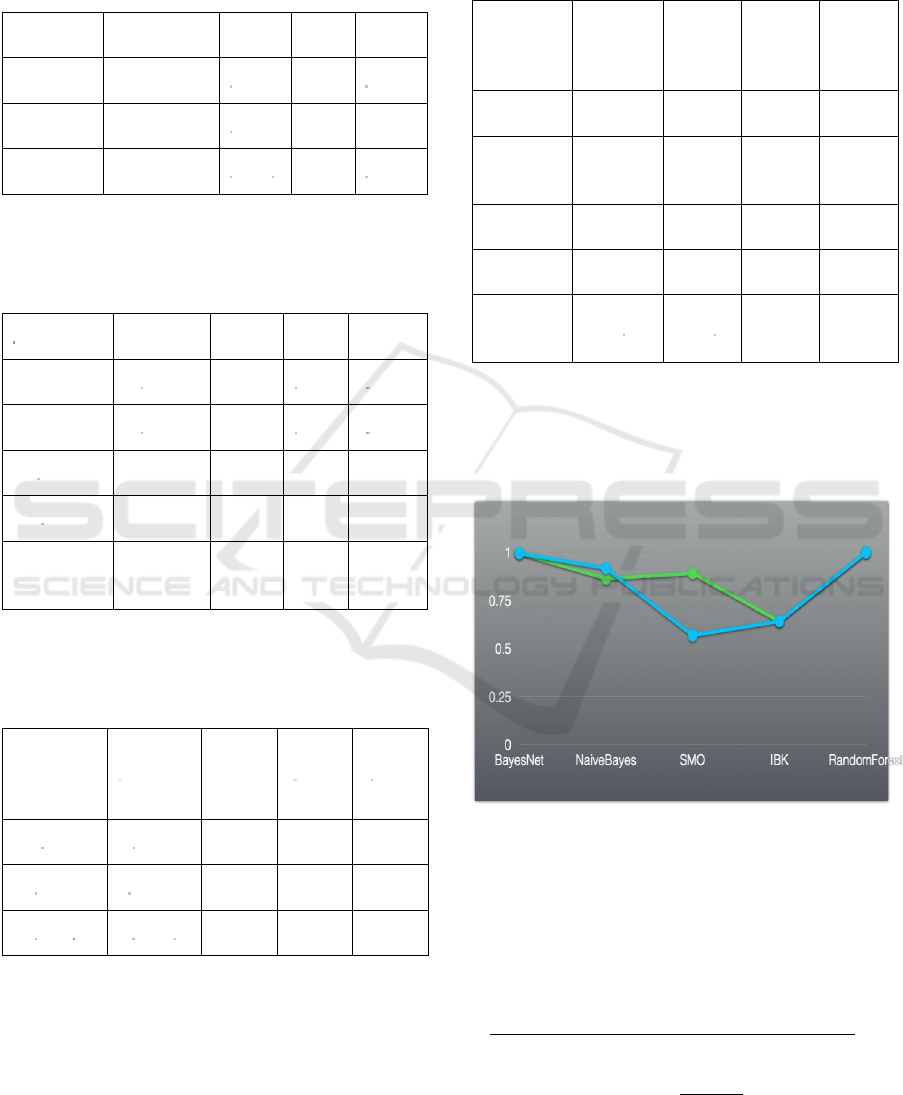
Improvement of ROC value was obtained only for
SMO model, as shown in Figure 1. None of
performance indicators were improved in
NaïveBayes, IBK and RandomForest models, which
has been similarly found by Akbas
̧
et al (2013).
Merging two algorithms into a combinatorial
classifier can overcome the shortages of individual
classifier. For instance, ensemble classifier generally
has higher accuracy. However, combinatorial
classifier is expected to take a long time in dataset
training (Zaïane and Zilles, 2013).
This is because the
increased data to be stored and the storage must be
conducted after dataset training (Kotsiantis, 2007).
Moreover, two classifiers or more should be run upon
input query. Lastly, although combinatorial classifier
can provide higher accuracy, the relationships
become less comprehensible (the rule learning is not
clear) (Kotsiantis, 2007).
Therefore, ensemble
classifier is recommended only for a purpose of
seeking the highest possible accuracy (Kotsiantis,
2007).
AdaBoost classifies dataset by using a number of
weak classifiers that concentrate on misclassification
of instances obtained from training using various
weight distribution (Elhenawy et al., 2015).
Misclassified instances from a classifier were passed
in the next iteration/training using another classifier,
generating a ‘cascading’ hypothesis (Zaïane and
Zilles, 2013).
These multiple training steps causes
long execution time in AdaBoost classification.
Ensemble classification with AdaBoost would be
even longer. This is the main challenge in using
combination of classifier with AdaBoost (Zaïane and
Zilles, 2013).
Comparison of Decision Tree, Neural Network, Statistic Learning, and k-NN Algorithms in Data Mining of Thyroid Disease Datasets
243
5 TABLES
Table 1: Performance Indicators for Decision Trees (J48
and LMT) in comparison with Neural Network Model
(MLP).
Classifier
Accuracy
Kappa
MCC
ROC
J48 trees
0.994
0.951
0.953
0.987
LMT trees
0.992
0.924
0.927
0.996
MLP
0.973
0.715
0.718
0.971
Table 2: Performance Indicators for Statistic Learning
Method (BayesNet and NaiveBayes), Instance-Based
Learning Method (IBK), Sequential Minimal Optimization
(SMO) and Decision Tree (RandomForest).
Classifier
Accuracy
Kappa
MCC
ROC
BayesNet
0.975
0.761
0.765
0.994
NaiveBayes
0.952
0.401
0.421
0.916
SMO
0.935
0.210
0.318
0.566
IBK
0.922
0.138
0.144
0.640
Random
Forest
0.992
0.932
0.936
0.999
Table 3: Performance Indicators for Ensemble Classifiers
using AdaBoost in combination with Decision Trees (J48
and LMT) in comparison with Neural Network Model
(MLP).
Ada Boost
+
Classifier
Accuracy
Kappa
MCC
ROC
J48
0.994
0.948*
0.951
0.999
LMT
0.992*
0.932*
0.934*
0.993*
MLP
0.973
0.715
0.718
0.971
* higher values compared to single classifier
Table 4: Performance Indicators for Ensemble Classifiers
using AdaBoost in combination with Statistic Learning
Method (BayesNet and NaiveBayes), Instance-Based
Learning Method (IBK), Sequential Minimal Optimization
(SMO) and Decision Tree (RandomForest).
Ada Boost
+
Classifier
Accuracy
Kappa
MCC
ROC
BayesNet
0.988*
0.884*
0.886*
0.995*
Naïve
Bayes
0.952
0.401
0.421
0.862
SMO
0.940*
0.375*
0.425*
0.890*
IBK
0.922
0.138
0.144
0.640
Random
Forest
0.993
0.930
0.934
0.999
* higher values compared to single classifier
6 FIGURES
Figure 1: ROC graphics obtained from individual classifier
(blue line) and combined classifier (green line).
Improvement of ROC value was obtained only for SMO
model.
7 EQUATIONS
(1)
(2)
HEALTHINF 2018 - 11th International Conference on Health Informatics
244

where p
a
is ratio of cohesions and p
e
is probability of
cohesion by coincidence
(3)
where TP is the number of true positives, TN is the
number of true negatives, FP is the number of false
positives, and FN is the number of false negatives
(4)
8 LIMITATION
The limitation of this paper is the pre-processing of
dataset that was not mentioned in details. However,
during running some algorithms compatibility issue
occurred with train and test dataset. WEKA developer
version 3.7.13 addresses this by using class input
mapped classifier. Secondly, this study did not
include statistical comparison of classification
performance and thus, how significant the difference
in performance among all classifiers is unknown.
9 CONCLUSIONS
In this study, Thyroid dataset has been classified
using various decision trees, neural network, Statistic
Learning, and k-NN algorithms. Decision tree J48
model was found to be the best classifier based on
accuracy, Kappa, MCC, and ROC. This model also
outperformed other classifiers used in previous study,
either as single classifier or in combination with
Adaptive Boosting algorithm. Deciding the best
algorithm that can be used in data mining of thyroid
dataset can simply be based on the classification
accuracy, which is the closeness of measured value to
the actual value. The vast majority of studies claimed
that the most common predictor of the optimum
classification algorithm is classification accuracy.
Improved accuracy can be achieved by combining
two classifiers. In this study, integration of different
algorithms into a combinatorial classifier has
successfully overcome the shortages of some
classifiers (SMO and BayesNet). However, there are
considerations to weigh in, particularly the type of
dataset. Moreover, classification using combinatorial
algorithm for large dataset would be a time-
consuming procedure. Therefore, it is suggested that
further study on optimization of combinatorial
classification for numerical, nominal, and discrete
datasets would be very beneficial.
REFERENCES
Akbas
̧
, A., Turhal, U., Babur, S., & Avci, C. (2013).
Performance Improvement with Combining Multiple
Approaches to Diagnosis of Thyroid Cancer.
Engineering, 05(10), 264-267.
Yeh, W.C., Chang, W.W., & Chung, Y.-Y. (2008). A new
hybrid approach for mining breast cancer pattern using
discrete particle swarm optimization and statistical
method. Expert Systems with Applications. Available
online 21 October 2008.
Olafsson, S., Li, X., & Wu, S. (2008). Operations research
and data mining. European Journal of Operational
Research, 187(3), 1429–1448.
Mohamadi, H., Habibi, J., Abadeh, M. S., & Saadi, H.
(2008). Data mining with a simulated annealing based
fuzzy classification system. Pattern Recognition, 41(5),
1824–1833.
Chang, W., Yeh, W., & Huang, P. (2010). A hybrid
immune-estimation distribution of algorithm for mining
thyroid gland data. Expert Systems With Applications,
37(3), 2066-2071.
Ozyilmaz, L., Yildirim. (2002) T. Diagnosis of Thyroid
Disease Using Artificial Neural Network Methods.
Proceedings of the 9
th
International Conference on
Neural Information Processing (ICONIP’02), 4, 2033-
2037.
Giannopoulou, E.G. (2008). Data Mining in Medical and
Biological Research, InTech, November, ISBN 978-
953-7619-30-5.
Ressom, W., Varghese, R.S., Zhang, Z., Xuan, J., and
Clarke, R. (2008) Classification Algorithms for
phenotype prediction in genomic and Proteomics Front
BioScience.
Iavindrasana, J., Hidki, A., Cohen, G., Geissbuhler, A.,
Platon, A., Poletti, P.A., Müller, H.J. (2010). Journal of
Digit Imaging, Comparative performance analysis of
state-of-the-art classification algorithms applied to lung
tissue categorization. Depeursinge. 23(1), 18-30.
Fernandez, A., Duarte, A., Hernandez, R., Sanchez, A.
(2010). GRASP for Instance Selection in Medical Data
Sets. AISC, 74, 53-60.
Jacob, S.B., Ramani, R.G. (2012). Mining of Classification
Patterns in Clinical Data through Data Mining
Algorithms. International Conference on Advances in
Computing Communiations and Informatics, 997-1003.
Kotsiantis, S.B. (2007). Supervised Machine Learning: A
Review of Classification Techniques. Informatica, 31,
249-268.
Bostanci, B. and Bostanci, E. (2012). An Evaluation of
Classification Algorithms Using Mc Nemar’s Test.
Advances in Intelligent Systems and Computing,
pp.15-26.
Comparison of Decision Tree, Neural Network, Statistic Learning, and k-NN Algorithms in Data Mining of Thyroid Disease Datasets
245

Zaïane, O. and Zilles, S. (2013). Advances in artificial
intelligence. Berlin: Springer.
Elhenawy, M., Jahangiri, A., Rakha, H. and El-Shawarby,
I. (2015). Classification of Driver Stop/Run Behavior at
the Onset of a Yellow Indication for Different Vehicles
and Roadway Surface Conditions Using Historical
Behavior. Procedia Manufacturing, 3, pp.858-865.
HEALTHINF 2018 - 11th International Conference on Health Informatics
246
