
Framework for Collection of Electrophysiology Data
Petr Jeˇzek
1
, Roman Mouˇcek
1
, Jakub Krauz
1
, Jaroslav Hoˇsek
1
, Yann Le Franc
2,3,4
, Thomas Wachtler
4
and Jan Grewe
4
1
Department of Computer Science and Engineering, New Technologies for the Information Society,
Faculty of Applied Sciences, University of West Bohemia, Plzen, Czech Republic
2
e-Science Data Factory S.A.S.U., Paris, France
3
Theoretical Neurobiology and Neuroengineering lab University of Antwerp, Wilrijk, Belgium
4
Department Biologie II, Ludwig-Maximilians-Universit¨at M¨unchen, Planegg-Martinsried, Germany
Keywords:
Electrophysiology, Mobile App, odML, EEGBase, Experimental Metadata.
Abstract:
Experiments in electrophysiology produce a lot of unstructured metadata collected in electrophysiology
databases. The data are usually accessed through a web interface implemented on the top of data model
respecting given data format. A lot of experiments are conducted outside the laboratory where access to these
databases is not always available. The usage of mobile devices such as tablets or smart phones seems to be
a practical solution, but users would welcome the same structured user interface such as they know from a
common computer. When user interfaces of electrophysiology databases are tailored to a unique data struc-
ture, they cannot be easily reused on a mobile device. As a solution, a mapping of a general data structure
to a graphical template is proposed. This mapping is implemented in a framework that generates a template
representing the database structure. The parsing process is driven by supplemented annotations added to the
code. Next, an Android tool visualizing a graphical layout generated from the template is developed. A use
case study is presented on a database of EEG/ERP experiments.
1 INTRODUCTION
Large collections of data sets are obtained during
electrophysiological experiments. With increasing
number of experiments a crucial task is to provide
metadata descriptions. These metadata usually in-
clude experimental constraints and scenarios, and in-
formation about tested subject. Raw data without de-
scriptive metadata are meaningless. At present, due
to the penetration of computers into medicine col-
lections of electrophysiology, metadata are moved
from paper forms to specialized electrophysiological
databases. These databases are accessed via desktop
or web based user interfaces using common comput-
ers. However, there are common situations when a
classic computer is not available. These situations
occur in environments such as hospitals or prisons
where a wireless connection is usually unavailable. In
addition, a lot of experiments are conducted outside
laboratories using portable measuring devices. In this
case data are obtained offline and then must be back-
ward synchronized with data stored in an electrophys-
iological database. Electrophysiological databases
are based on specific data models. User interfaces im-
plemented on top of these modelsare non-transferable
to other data models. Development of various user
interfaces is time consuming, and they are difficult to
maintain, consequently it reduces usability. As a so-
lution we present an annotation based framework that
aim is to generate a template for client applications.
This template contains description of metadata struc-
ture of an electrophysiological database. A graphical
layout for a specific device is then generated accord-
ing to the obtained template. As a first pilot system we
have implemented an Android based system that gen-
erates a template for mobile devices such as tablets
or cell-phones. These devices are connected to the
electrophysiological database through the framework
API.
The structure of the paper is following. First,
we briefly introduce existing electrophysiological
databases and mobile-based approaches. Then, we
describe a proposed mapping that maps the data
model of a database to a template expressed in a trans-
port format. Next, we introduce an implementation
the mapping in the framework. The rest of the pa-
558
Ježek P., Moucek R., Krauz J., Hošek J., Le Franc Y., Wachtler T. and Grewe J..
Framework for Collection of Electrophysiology Data.
DOI: 10.5220/0005275605580565
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2015), pages 558-565
ISBN: 978-989-758-068-0
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

per describes validation of the presented approach as
a part of a use-case study in which integration of the
framework in EEGBase (Jezek and Moucek, 2012) is
demonstrated.
2 STATE OF THE ART
Research in the electrophysiologydomain faces a lack
of tools and infrastructure for collecting and process-
ing experimental data. There are several initiatives
implementing and providing tools for data process-
ing, electrophysiological databases or hardware de-
vices. We provide a brief overview of these mem-
bers of the international neuroinformaticscommunity,
most of which are involved in the activities coordi-
nated by the International Neuroinformatics Coordi-
nating Facility (INCF)
1
:
• CRCNS (Teeters et al., 2008) provides market-
place and discussion forum for sharing tools and
data in neuroscience.
• Helmholtz (Davison et al., 2013) is a framework
for creating neuroscience databases that are cus-
tomized to the needs of individual neurophysiol-
ogy labs.
• Carmen (Watson et al., 2007) is a virtual labora-
tory that enables a storage of experimental data,
experimental protocols, or analysis code. It de-
signs workflows from the stored code and run
them remotely.
• INCF Dataspace (INC Working Group, 2013) en-
ables interested research groups to connect to a
distributed data file system based on iRods
2
.
• G-Node
3
provides tools for data access, data man-
agement and data sharing, including a data shar-
ing platform (Sobolev et al., 2014) based on com-
mon data models for electrophysiological data
and metadata (Garcia et al., 2014; Grewe et al.,
2011). To structure metadata, G-Node has devel-
oped odML, an XML schema for the creation of
complex metadata structure in computer-readable
format(Greweet al., 2011). The odML data model
is used as standard representation within the G-
Node infrastructure. An odML mobile editor
(Le Franc et al., 2014) is currently being devel-
oped as a standalone app for Android and iOS
platform. This mobile application enables users to
create odML structure natively on the app using
a Graphical User Interface and to acquire on the
1
http://www.incf.org/
2
http://irods.org/
3
http://www.g-node.org/
fly experimental metadata. These metadata can
be processed or shared. In addition, reusable tem-
plates can be designed and stored inside the device
and filled in later.
• EEGBase (Jezek and Moucek, 2012) is a database
of well-described EEG/ERP experimental data
sets that enables users to upload, download and
manage EEG/ERP experiments. A mobile EEG-
Base client (Jezek and Moucek, 2013) enables
users to collect data from EEG/ERP experi-
ments and provides an online synchronization
with EEGBase.
The first version of the EEGBase database was
accessible via a web based user interfaces that al-
lows acquiring, managing and sharing the experimen-
tal metadata from a desktop computer. However, it
is not always possible to access the web interface in
the course of an experiment. We therefore extended
the framework to support the use of mobile devices
for acquiring experimental metadata (Moucek et al.,
2014). Our first version was limited to the metadata
model predefined in the database. Any modification
of the metadata model would require a complete up-
date of the User Interface for the mobile client. As
creating and maintaining new user-interfaces is time-
consuming and requires extensive development re-
sources, the challenge is to elaborate a solution to op-
timize the further development of this platform. We
are proposing here a unique system that allows the
automatic creation of specific graphical layouts for
various platforms, including mobile devices, web ap-
plications, etc. To address this challenge, we trans-
formed the EEGBase framework to use object anno-
tations (Jezek et al., 2013a) for creating internal tem-
plates out of the database and convert it into odML.
The odML structure is then transmitted to the mobile
device, which can generate automatically a graphi-
cal layout for rendering the metadata document. The
transformation described here marks a first step to-
wards interoperability of the EEGBase portal with the
G-Node portal and the integration of our work to pro-
pose a unique mobile platform.
3 PROPOSED FRAMEWORK
3.1 System Restrictions and
Requirements
According to the considerations mentioned in Section
1 we decided to design and implement a tool that fa-
cilitates collection of metadata from experiments by a
unified interface customizable to the data structure. In
FrameworkforCollectionofElectrophysiologyData
559

order to ensure an immediate usage, the user interface
must be easy to generate. In addition, the synchro-
nization of collected data with the electrophysiology
database is crucial in the perspective of their sharing
and management.
These design goals imply several requirements
that the system must fulfill. It must be inherently
client-server oriented. The server end must provide
a well-known API to be integrated with most com-
mon electrophysiology databases. Moreover, server
and client must communicate using a unified data for-
mat for easy migration across different platforms and
to be understandable by various clients.
The client end parses the exported data structure
and generates a graphical layout. Once the layout is
generated, the user can customize it. When the user
has the layout prepared, the client is able to download
data from the server or push data stored locally. When
the client works offline, the data must be stored in
an embedded database and synchronized immediately
when the client gets back online.
The idea initially presented in (Jezek et al., 2013a)
described a client-server system. The server is able to
annotate the database layer of an electrophysiology
database. Such annotated layer is then serialized to
a XML structure and sent to the client. This idea is
extended, implemented and validated in this paper.
3.2 Mapping Annotated Code to Data
Template
Modern systems are coded using high-level virtual
machine programming languages such as Java, .NET
or Python. These languages rely on reflection, an abil-
ity of a computer program to examine and modify the
structure at runtime. Moreover, the code in these lan-
guages can be annotated by supplemented metadata.
These metadata can be processed by reflection at run-
time. Java uses Java Reflection API (Forman and
Forman, 2004) to read Java Annotations (Microsys-
tems, Sun, 2008). These annotations give additional
semantic meaning to common Java classes. When a
database layer in Java programs is represented by PO-
JOs
4
, a suitable approach is their extension by spe-
cially designed annotations. These annotations pro-
vide the means to describe a data template.
As a solution we designed a set of annotations
described in Table 1. The annotation
@Form
repre-
sents one form on the layout. If one form is rep-
resented by more than one entity, the annotation
@Multiform
with the same name is used for each en-
tity included in the form. The
@FormItem
annotation
4
Plain Old Java Object - A Java class with class at-
tributes accessed by get/set methods
Table 1: Supported Annotations.
Annotation Parameter Meaning
@Form label Label of the form.
@FormDescription value The description.
@MultiForm value Identifier of the form (string).
label Label of the form.
@FormItem label Label of the item.
required Determines a required value (default: false)
preview Determines items used in data previews.
@FormItemRestriction minLength Minimum length of the input.
maxLength Maximum length of the input.
minValue Minimum value of numerical item.
maxValue Maximum value of numerical item.
defaultValue The default value.
values Enumeration of possible values.
denotes attributes that appear in a specific form. The
@FormItemRestriction
annotation defines restric-
tions on individual items. These restrictions can be
e. g. minimal and maximal numerical values or max-
imal length of text values.
When the data layer of the database is anno-
tated, it must be mapped to a suitable transport
format. Formats solve description of electrophys-
iology data on different levels of abstraction from
low-level binary formats, through highly abstract
implementation-independent data formats to formats
based on semantic web ontologies. Each approach
has its benefits and drawbacks that need to be over-
come. We require a format that provides a sufficient
level of abstraction to be system independent on one
hand, but it should be easy-to-use without any spe-
cific requirements for user’s knowledge on the other
hand. The most flexible formats are Hierarchical Data
Format (HDF5) (HDF5 Group, 2013) or respectively
epHDF or NIX (G-Node, 2014) that are specialized
HDF5 for electrophysiology. HDF5 and epHDF5 are
the containers for raw binary data and metadata rep-
resented in the JSON
5
format. NIX (Stoewer et al.,
2014) provides a data model for storing experimental
data in HDF5, together with metadata in the odML
format. The odML (Grewe et al., 2011) is an open
format to store and transport metadata more then a
container for data. This format brings the following
advantages: (1) platform-independence, (2) simplic-
ity and human-readability, (3) ability to transfer lay-
outs as well as metadata with the same format. Be-
cause of mentioned benefits of odML we selected it
for the framework.
4 IMPLEMENTATION
4.1 Mapping Implementation
The mapping presented in Section 3.2 shows trans-
ferring persistent Java classes to templates. Then,
5
http://json.org/
HEALTHINF2015-InternationalConferenceonHealthInformatics
560

these templates are mapped to odML representation
and transferred to the client. OdML is based on Sec-
tions that can contain Properties and Values. More-
over, each section can contain subsections (tree-like
structures can be created). In our approach forms are
represented by sections, items by subsections and re-
strictions by properties. Definition 1 formalizes an ex-
traction process from Java Annotations to correspond-
ing odML representation.
Definition 1. (Java annotation extraction process)
The process is the transformation of a set of Java an-
notations JA to an odML Section S in the odML doc-
ument D that satisfies:
∀ JA
i
∈ (@Form) ∃ odML Section S
i
∈ D ⇒ JA
i
∈ a class annotation.
∀ JA
j
∈ (@FormItem) ∃ odML Section S
j
⊂ S
i
∈
D ⇒ JA
j
∈ a property annotation.
∀ JA
k
∈ (@FormRestriction) ∃ odML Property
P
k
∈ S
j
∈ D ⇒ JA
k
∈ a property annotation.
Listing 1 shows an Example of the class Person
containing several attributes givenname, surname and
gender supplemented by metadata annotations (other
annotations, class imports, get/set methods, and other
fields are omitted for simplicity).
Listing 1: Java Class Example.
@Form
public class Person{
@FormItem
private String givenname;
@FormItem
private String surname;
@FormItem(required =
true)
@FormItemRestriction(values = {
"M",
"F", "X"}, defaultValue = "X")
private char gender;
}
An XML serialization of this class to an odML
document is presented in Listing 2. The document
shows one section named Person with two subsection
givenname and gender (note: Because of the XML
serialization is quite long, similar subsections repre-
senting other attributes are omitted). All restrictions
are represented by properties of this section. In ad-
dition, because the Java code is reflective, data types
or names of attributes can be straightforwardly trans-
ferred to odML properties. With respect to commonly
used data types String, Integer, DateTime, Float, Char
or Boolean are supported. In addition, a combobox
data type is used for arrays or lists of values.
4.2 Templates Generator
The template generator is a server part of the pre-
sented tool. Figure 1 shows a block diagram of the
Listing 2: odML Representation.
<odML>
<
section>
<
type>form</type>
<
name>Person</name>
<
property>
<
name>label</name>
<
value>
Person
<
type>string</type>
</
value>
</
property>
<
property>
<
name>layoutName</name>
<
value>
Person-generated
<
type>string</type>
</
value>
</
property>
<
section>
<
type>textbox</type>
<
name>givenname</name>
<
property>
<
name>cardinality</name>
<
value>
1
<
type>int</type>
</
value>
</
property>
<
property>
<
name>required</name>
<
value>
false
<
type>boolean</type>
</
value>
</
property>
<
property>
<
name>datatype</name>
<
value>
string
<
type>string</type>
</
value>
</
property>
</
section>
<
section>
<
type>combobox</type>
<
name>gender</name>
<
property>
<
name>label</name>
<
value>
Gender
<
type>string</type>
</
value>
</
property>
<
property>
<
name>cardinality</name>
<
value>
1
<
type>int</type>
</
value>
</
property>
<
property>
<
name>required</name>
<
value>
true
<
type>boolean</type>
</
value>
</
property>
<
property>
<
name>datatype</name>
<
value>
string
<
type>string</type>
</
value>
</
property>
<
property>
<
name>defaultValue</name>
<
value>
X
<
type>string</type>
</
value>
</
property>
<
property>
<
name>values</name>
<
value>
M
<
type>string</type>
</
value>
<
value>
F
<
type>string</type>
</
value>
<
value>
X
<
type>string</type>
</
value>
</
property>
</
section>
</
section>
</
odML>
template generator. The parsing process contains two
steps. The first parser ”Annotations parser” reads
data entities, parses their names, attributes, data types
and annotations and stores them in an internal model.
The template stored in this model is transferred to an
odML document. The Annotation parser is driven by
user requests that come through a unified API. When
the client generates the layout, the data can be down-
loaded by the second parser, a ”Data parser”. The
Data parser selects requested data and serializes them
into the equally structured odML document. In re-
verse operation, when data are uploaded, they are de-
serialized and transferred to related Java classes by an
object builder implemented inside the Data parser.
4.3 Mobile Client
The mobile client is a framework implemented for the
FrameworkforCollectionofElectrophysiologyData
561
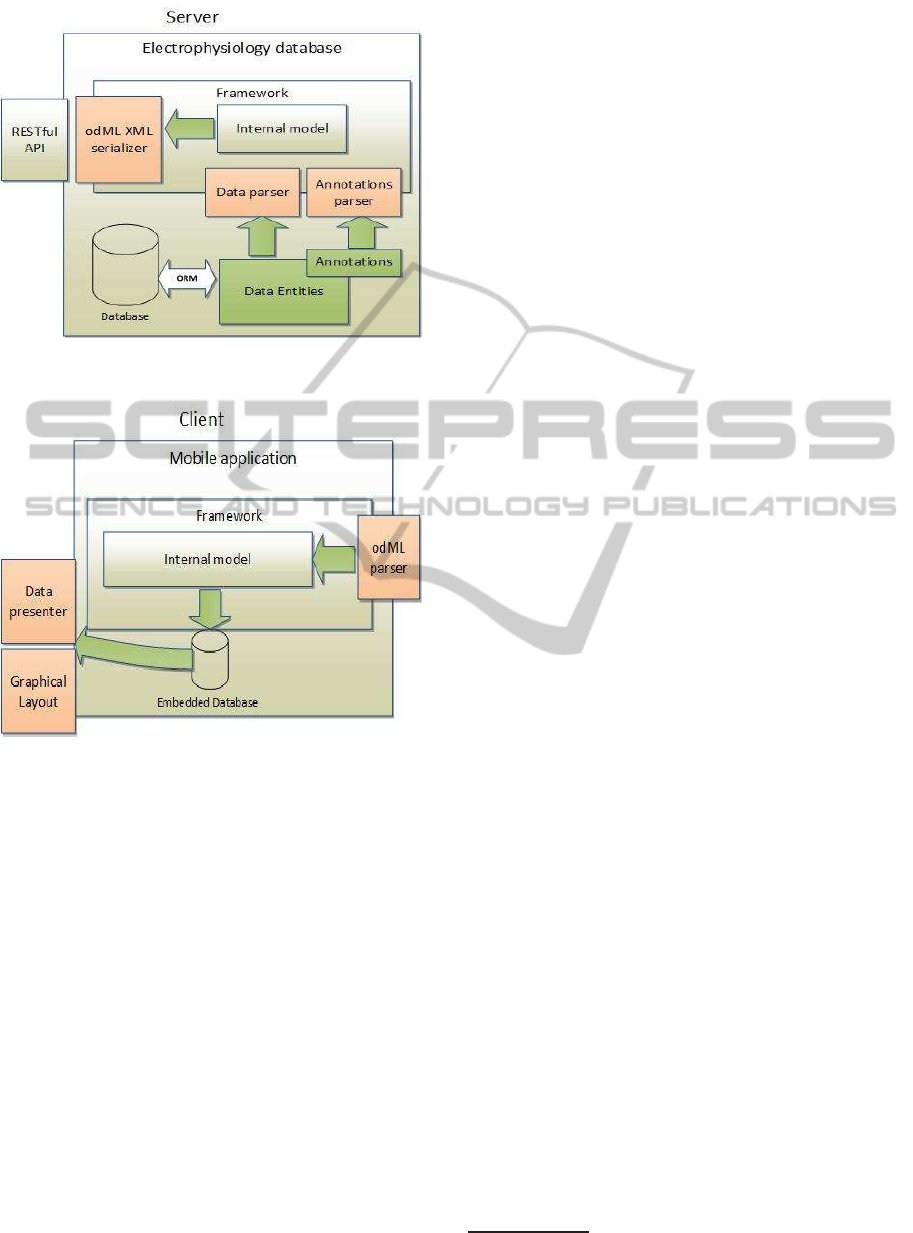
Figure 1: Framework for Generating Templates and Data
Transfer.
Figure 2: Framework Implemented in the Mobile Client.
Android platform. A client block diagram is shown in
Figure 2. A client input point is an odML deserializer
that parses an input odML document into an internal
model. The internal model can contain both data and
templates. When the client requests a template, this
template is transferred to a layout using a layout gen-
erator. When the client is requesting data for a specific
layout, these data are processed by a data parser. Both
layouts and data are stored in an embedded database
(SQLLite). It enables users to work offline.
5 USE CASE
5.1 Domain Specialization
The main focus of the research group are methods
and techniques of electroencephalography (EEG) and
event-related-potentials (ERP). Experiments are con-
ducted in a dedicated laboratory equipped with highly
specialized hardware and software. Besides this lab-
oratory, we are equipped with a mobile laboratory
that enables us to perform experiments outside. In
addition we are collaborating between several INCF
National Nodes, including German, Belgian, Czech,
USA or Japan Nodes. Our equipment and a lot of dis-
cussion with our colleagues motivate us to design and
test the solution described.
Experimental procedure follows severalsteps. Ex-
periments are performed according to defined sce-
nario. Then data and metadata are collected. These
data are analyzed and results are stored and published.
The central point of this infrastructure is EEGBase
(Jezek et al., 2013b) that serves for storing and man-
agement of EEG/ERP experiments.
Although the system uses a web-based interface,
many experiments are conducted outside the labora-
tory where a common computer, or Internet access
are not available. The aim of this use case is to val-
idate the integration of the presented framework and
generation of templates according to data structure.
Generation of templates is controlled by an Android
phone. When the template is generated, new data can
be collected and synchronized with EEGBase.
5.2 EEGBase
EEGBase
6
provides a simple wizard that guides the
user when uploading experiments. The user is in-
structed what metadata have to be filled in. The col-
lected metadata respecting experience of our research
partners with designing and performing experiments
and experimental scenarios. The metadata follow on-
tology described in (Jezek and Moucek, 2011). A user
interface preview is shown in Figure 3.
The core structure contains the followingsemantic
metadata groups:
• Scenario of experiment (name, length, descrip-
tion, ...)
• Experimenters, tested people, experiment owner
(given name, surname, contact, experiences,
handicaps, ...)
• Description of raw data (format, sampling fre-
quency, ...)
• Additional metadata in codebooks (weather,
notes, electrode settings, artifact information, ...)
We prepared a simple proof-of-concept imple-
mentation based on a restricted set of metadata to
demonstrate functionality of the framework. We se-
lected the most important medata and annotate them
by described annotations. Table 2 shows selected en-
tities with their annotated attributes.
6
https://eegdatabase.kiv.zcu.cz
HEALTHINF2015-InternationalConferenceonHealthInformatics
562
Figure 3: EEGBase Preview.
Since EEGBase implements RESTful web ser-
vices (Richardson and Ruby, 2007), control of the
framework API by client requests is ensured. We im-
plemented two methods; one for listing available tem-
plates and the second one for downloading a selected
template
7
.
5.3 Mobile Client
The mobile client uses common design of Android
applications. Main activities of the use-case study are
shown in several screen-shots in Figure 4. Because
the client is intended to work with various electro-
physiological databases, it enables creating separate
workspaces where each workspace works with differ-
ent user credentials. In Figure 4(a) the user creates
a new workspace. Users credentials are optional be-
cause the client can work as a stand alone system as
well. In Figure 4(b) available layouts are listed; the
user can download them. Finally, in Figure 4(c) the
user can download data from the server or add new
data as shown in Figure 4(d).
By following the described steps we created one
workspace connected to EEGBase (see Figure4(a))
and downloaded available layouts (Person, Scenario,
Experiment and DataFile as listed in Figure 4(b)).
When the layouts were downloaded experiments from
the server were listed (see Figure 4(c)). Finally, we
add several new experiments using form listed in Fig-
ure 4(d). These experiments were stored on the server.
6 FUTURE WORK
The proposed solution works satisfactorily and the
presented use-case study validates its functionality on
7
All API methods all described more in detail in a
project wiki page: https://github.com/INCF/eeg-database/
wiki/RESTful-API
Table 2: Entities and their Annotated Fields.
Entity Annotated fields
Person
@FormItem
private String email;
@FormItem(required = true)
private String givenname;
@FormItem(required = true)
private String surname;
@FormItem
private Timestamp dateOfBirth;
@FormItem(required = true)
@FormItemRestriction(values = ”M”, ”F”)
private char gender;
@FormItem(required = true)
@FormItemRestriction(values = ”L”, ”R”, ”X”, defaultValue = ”X”)
private char laterality;
private String phoneNumber;
@FormItem
private String note;
Scenario
@FormItem(required = true)
private Person person;
@FormItem(required = true)
private ResearchGroup researchGroup;
@FormItem(required = true)
private String title;
@FormItem
@FormItemRestriction(minLength =0)
private int scenarioLength;
@FormItem
private boolean privateScenario;
@FormItem
@FormItemRestriction(maxLength = 255)
private String description;
@FormItem
private String scenarioName;
@FormItem
private String mimetype;
Experiment
@FormItem(required = true)
Weather weather;
@FormItem(required = true, label = ”Subject person”)
private Person personBySubjectPersonId;
@FormItem(required = true)
private Scenario scenario;
@FormItem(required = true, label = ”Owner”)
private Person personByOwnerId;
@FormItem(required = true)
private ResearchGroup researchGroup;
@FormItem(required = true)
private Digitization digitization;
@FormItem(required = true)
private SubjectGroup subjectGroup;
@FormItem(required = true)
private Artifact artifact;
@FormItem(required = true)
private ElectrodeConf electrodeConf;
@FormItem(preview = PreviewLevel.MAJOR)
private Timestamp startTime;
@FormItem
private Timestamp endTime;
@FormItem
private int temperature;
@FormItem
private boolean privateExperiment;
@FormItem(preview = PreviewLevel.MINOR)
private String environmentNote;
Stimulus
@FormItemRestriction(maxLength = 255)
String description
DataFile
@FormItemRestriction(maxLength = 255)
String description
real data. However, the presented pilot implementa-
tion has several gaps that are planned to be overcome.
First, we will focus our effort on developing the
mobile client to offer the possibility to create tem-
plates natively on the client, allowing to work of-
fline. For this purpose, we will merge efforts be-
tween the EEGBase client and the mobile odML ed-
itor (Le Franc et al., 2014) to propose a unique mo-
bile client that would work on both database. With
this work, we will be able to extend 1- the interoper-
ability between these two resources and 2- extend the
functionalities of the mobile client presented here to
FrameworkforCollectionofElectrophysiologyData
563

(a) The user can create a new
workspace
(b) Then select available layout (c) Finally list data (d) He/she can add a new item
when clicks to a + button
Figure 4: Mobile Application Use Case Preview.
iOS and ultimately to more platforms
8
. Secondly, we
will address the problem of data and metadata stor-
age on both client and server sides. As electrophysi-
ological data are heterogeneous, the usage of NoSQL
databases seems to be more practical. Our first tests
prove (Jeˇzek et al., 2014) that a shift from a relational
database to a NoSQL one brings a lot of benefits han-
dling metadata. The major step is to propose a suit-
able mapping of heterogeneous metadata stored in a
NoSQL database to a suitable data template. Finally,
we are planning to extend the server part of the frame-
work to work with a larger diversity of servers than
Java-based servers.
7 CONCLUSION
Experiments in electrophysiology domain produce a
lot of collections of semi-structured data and meta-
data. To organize these metadata, various data for-
mats implemented in electrophysiological databases
have been introduced. Data in these databases are ac-
cessed from laboratory computers using fixed user in-
terfaces. Therefore these databases are unreachable
where a common computer is not available. With ex-
pansion of mobile devices users would like to manage
data using their smart phones. Because the dual devel-
opment of user interfaces for both common comput-
ers and mobile devices increases demands to develop-
ers and reduces users experience with these systems,
we proposed an annotation framework that enables to
generate graphical layouts for mobile devices auto-
matically. This framework enables users to map a data
layer of an electrophysiological database and create
the template representing the data structure. This tem-
8
several frameworks as http://phonegap.com/ enables
development of platform independent mobile applications
plate is serialized to an odML document and trans-
ferred to the client system. The client system parses
this template and generates a graphical layout dis-
played in the device. In addition, when the layout is
prepared, the experimental data can be synchronized
with the server.
ACKNOWLEDGEMENT
This work was supported by the European Regional
Development Fund (ERDF), Project ”NTIS - New
Technologies for Information Society”, European
Centre of Excellence, CZ.1.05/1.1.00/02.0090, the
UWB grant SGS-2013-039 Methods and Applica-
tions of Bio- and Medical Informatics, and the Ger-
man Federal Ministry of Education and Research
grant 01GQ1302.
REFERENCES
Davison, A. P., Brizzi, T., Guarino, D., Manette, O. F.,
Monier, C., Sadoc, G., and Fr´egnac, Y. (2013).
Helmholtz: a customizable framework for neurophys-
iology data management. Frontiers in Neuroinformat-
ics, (25).
Forman, I. R. and Forman, N. (2004). Java Reflection in
Action (In Action series). Manning Publications.
G-Node (2014). Nix – neuroinformatics exchange format.
Garcia, S., Guarino, D., Jaillet, F., Jennings, T. R., Pr¨opper,
R., Rautenberg, P. L., Rodgers, C., Sobolev, A.,
Wachtler, T., Yger, P., and Davison, A. P. (2014). Neo:
an object model for handling electrophysiology data
in multiple formats. Frontiers in Neuroinformatics,
8(10).
Grewe, J., Wachtler, T., and Benda, J. (2011). A bottom-up
approach to data annotation in neurophysiology. Fron-
tiers in Neuroinformatics, 5(16).
HEALTHINF2015-InternationalConferenceonHealthInformatics
564

HDF5 Group (2013). Hierarchical data format.
INC Working Group (2013). Incf dataspace.
Jeˇzek, P., Mouˇcek, R., and Danˇek, J. (2014). Mongodb for
electrophysiology experiments. pages 422–427. cited
By (since 1996)0.
Jezek, P. and Moucek, R. (2011). Semantic web in eeg/erp
portal: Ontology development and nif registration.
In Biomedical Engineering and Informatics (BMEI),
2011 4th International Conference on, volume 4,
pages 2058 –2062.
Jezek, P. and Moucek, R. (2012). SYSTEM FOR
EEG/ERP DATA AND METADATA STORAGE
AND MANAGEMENT. NEURAL NETWORK
WORLD, 22(3):277–290.
Jezek, P. and Moucek, R. (2013). Eeg/erp portal for android
platform. Frontiers in Neuroinformatics, (46).
Jezek, P., Moucek, R., Le Franc, Y., Wachtler, T., and
Grewe, J. (2013a). Framework for automatic genera-
tion of graphical layout compatible with multiple plat-
forms. In Visual Languages and Human-Centric Com-
puting (VL/HCC), 2013 IEEE Symposium on, pages
193–194.
Jezek, P., Stebet´ak, J., Bruha, P., and Moucek, R. (2013b).
Model of software and hardware infrastructure for
electrophysiology. In Stacey, D., Sol´e-Casals, J., Fred,
A. L. N., and Gamboa, H., editors, HEALTHINF,
pages 352–356. SciTePress.
Le Franc, Y., Gonzalez, D., Mylyanyk, I., Grewe, J., Jezek,
P., Moucek, R., and Wachtler, T. (2014). Mobile meta-
data: bringing neuroinformatics tools to the bench.
Frontiers in Neuroinformatics, (53).
Microsystems, Sun (2008). Annotations (jdk 5.0 java pro-
gramming language-related apis & developer guides).
Moucek, R., Bruha, P., Jezek, P., Mautner, P., Novotny, J.,
Papez, V., Prokop, T., Rond´ık, T.,
ˇ
Stebetˇak, J., and
Vareka, L. (2014). Software and hardware infrastruc-
ture for research in electrophysiology. Frontiers in
Neuroinformatics, 8(20).
Richardson, L. and Ruby, S. (2007). Restful Web Services.
O’Reilly, first edition.
Sobolev, A., Stoewer, A., Leonhardt, A. P., Rautenberg,
P. L., Kellner, C. J., Garbers, C., and Wachtler, T.
(2014). Integrated platform and api for electrophys-
iological data. Frontiers in Neuroinformatics, 8(32).
Stoewer, A., Kellner, C., Benda, J., Wachtler, T., and Grewe,
J. (2014). File format and library for neuroscience
data and metadata. Front. Neuroinform. Conference
Abstract: Neuroinformatics 2014.
Teeters, J., Harris, K., Millman, K., Olshausen, B., and
Sommer, F. (2008). Data sharing for computational
neuroscience. Neuroinformatics, 6(1):47–55.
Watson, P., Jackson, T., Pitsilis, G., Gibson, F., Austin, J.,
Fletcher, M., Liang, B., and Lord, P. (2007). The
CARMEN Neuroscience Server. In Proceedings of
the UK e-Science All hands Meeting, pages 135–141.
FrameworkforCollectionofElectrophysiologyData
565
