
A SYSTEMATIC REVIEW OF OUTLIERS DETECTION
TECHNIQUES IN MEDICAL DATA
Preliminary Study
Juliano Gaspar
1,2
, Emanuel Catumbela
1,2
, Bernardo Marques
1,2
and Alberto Freitas
1,2
1
Department of Biostatistics and Medical Informatics, Faculty of Medicine, University of Porto, Porto, Portugal
2
CINTESIS - Center for Research in Health Technologies and Information Systems
University of Porto, Porto, Portugal
Keywords: Outliers detection, Data mining, Medical data.
Abstract: Background: Patient medical records contain many entries relating to patient conditions, treatments and lab
results. Generally involve multiple types of data and produces a large amount of information. These
databases can provide important information for clinical decision and to support the management of the
hospital. Medical databases have some specificities not often found in others non-medical databases. In this
context, outlier detection techniques can be used to detect abnormal patterns in health records (for instance,
problems in data quality) and this contributing to better data and better knowledge in the process of decision
making.
Aim: This systematic review intention to provide a better comprehension about the techniques used to
detect outliers in healthcare data, for creates automatisms for those methods in the order to facilitate the
access to information with quality in healthcare.
Methods: The literature was systematically reviewed to identify articles mentioning outlier detection
techniques or anomalies in medical data. Four distinct bibliographic databases were searched: Medline, ISI,
IEEE and EBSCO.
Results: From 4071 distinct papers selected, 80 were included after applying inclusion and exclusion
criteria. According to the medical specialty 32% of the techniques are intended for oncology and 37% of
them using patient data. Considering only articles that used administrative medical data, 59% of the
techniques were statistical based.
Conclusion: The area with outliers detection techniques most widely used in medical administrative data is
the statistics, when compared with techniques from data mining such as clustering and nearest neighbor.
1 BACKGROUND
Medical databases generally involve many different
types of data and produces a lot of information
(Kumar et al., 2008). The data typically consists in
records which may have several different types of
features such as patient age, blood group, weight,
clinical images, patient diagnoses, lab test results
and other details from patient treatments (Chandola
et al., 2009).
Patient medical records include many electronic
entries related to patient conditions and treatments
and their laboratory results. These are useful in
providing a better picture about the individual
patient, however, the benefits of such data in
decision support or in discovery of new clinical
knowledge are far from being exhausted
(Hauskrecht et al., 2007).
In latest years, with the exponential development
of information technology in hospitals, the volume
of medical data has increased significantly. At the
same time, new interest in the analysis of this
information has emerged, taking place not only as a
source for clinical decision making and research in
epidemiological studies, but also to support of
hospital management (Silva-Costa et al., 2010).
The value assigned to these data is directly
related to their quality. This way, as higher is the
quality of data, higher is its utility (Arts et al., 2002).
For healthcare organizations, this kind of
information is essential in providing healthcare, as
well as a consistent and careful financial
575
Gaspar J., Catumbela E., Marques B. and Freitas A..
A SYSTEMATIC REVIEW OF OUTLIERS DETECTION TECHNIQUES IN MEDICAL DATA - Preliminary Study.
DOI: 10.5220/0003168705750582
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2011), pages 575-582
ISBN: 978-989-8425-34-8
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

management. On most systems, the quality of data is
completely neglected, this way, could become a
nightmare, when users of the information produced
are not aware about its veracity and quality (Silva-
Costa et al., 2010).
In health, this quality is even more important,
since a wrong decision by a clinician, may even lead
to death of the patient or, in a most extreme case,
may also have a more global reach, when this weak
quality is associated with the exchange of
documents. A professional takes the decision based
on surveys that sometimes and without him aware
that they are not the same patient that he is
addressing. For example, a study on the integration
of hospital information systems (Cruz-Correia et al.,
2006), it stated that about 0.1% (423 in 391,258) of
the documents associated with the process of clinical
patients in a hospital central contain identifying
information wrong.
Data mining a knowledge discovery in medical
databases are not substantially different from mining
in other types of databases. There are some
particularities in medical databases that are absent in
non-medical database (Cios, 2001).
One of these particularities are that the
physician’s interpretation of images, signals, or any
other clinical data, is written in unstructured free-
text, and because of this, standardize is very
difficult. Other feature of medical data mining is that
the underlying data structures of medicine are poorly
characterized mathematically, as compared to many
areas of the physical sciences. Physical scientists
could substitute data into formulas, equations, and
models that reflect the relationships among their data
(Cios, 2001).
1.1 Outliers Detection in Medicine
Outlier detection is very important to medicine,
because this data translate to significant information
and often critical data (Chandola et al., 2009). In
healthcare databases, outlier detection techniques are
used to detect anomalous patterns in patient records
which can contain valuable data as, for instance,
symptoms of a new disease.
There are many definitions for outliers which
differ in words found in different studies (Laurikkala
et al., 2000). According to Barnett and Lewis, an
outlier can be defined as “an observation, or subsets
of observations, appearing to be inconsistent with
the remainder of that set of data” (Barnett and
Lewis, 1994). In other words, an outlier is an
element that deviates from a standard set of data
from which it belongs. However, an outlier is always
an element of a group. An element is said outlier
when compared to a standard, therefore, an element
can be called outlier compared to the standard X and
not an outlier compared to standard Y (Silva, 2004).
1.1.1 Techniques
The outliers detection techniques debated in this
study are:
Statistical: Statistical techniques fit a statistical
model, usually for normal behavior, to the given
data and then a statistical inference test is applied to
determine if an unseen instance belongs to the model
or not. Instances that have a low probability to be
generated from the learnt model, based on the
applied test statistic, are declared as outliers
(Chandola et al., 2009).
Clustering: Is used to group similar data into
clusters. Even though clustering and outliers
detections appear to be fundamentally different from
each other, several clustering based outlier detection
techniques have been developed.
Classification: This technique is used to learn a
model from a set of labeled data instances and, then,
classify a test instance into one of the classes using
the learned model. Classification based anomaly
detection techniques operate in a similar two-phase:
the training phase learns a classifier using the
available labeled training data and the testing phase
classifies a test instance as normal or anomalous
using the classifier (Chandola et al., 2009).
Nearest Neighbor: Require a distance or
similarity measure defined between two data
instances, that can be computed in different ways
(Chandola et al., 2009). Techniques based on this
approach can be broadly grouped into two
categories: techniques that use the distance of a data
instance to its nearest neighbor as the anomaly score
and techniques that compute the relative density of
each data instance to compute its anomaly score.
Mixture Models: Mixture models comprise a
finite or infinite number of components, possibly of
different distributional types, that can describe
different features of data (Marin et al., 2005). In
statistics, a mixture model is a probabilistic model
for density estimation using a mixture distribution.
A mixture model can be regarded as a type of
unsupervised learning or clustering.
Spectral: Try to find an approximation of the
data using a combination of attributes that capture
the bulk of variability in the data (Chandola et al.,
2009). This technique defines subspaces in which
the anomalous instances can be easily identified.
HEALTHINF 2011 - International Conference on Health Informatics
576

1.2 Aim
This systematic review aims to provide a better
comprehension about the used techniques to detect
outliers in administrative healthcare data, to create
automatisms for those methods in order to facilitate
the access to information with quality, to make a
better decision by managers in health.
2 METHODS
2.1 Eligibility Criteria
We defined a pair review, independently, involving
the two reviewers in the study. The selection was
based on the studies titles and abstracts. The pair
review considered eligible articles that contained the
following criteria:
The Inclusion covered the topics outlier
detection, anomaly detection, extreme data or gross
error, and the topics healthcare, health, medical,
medicine, clinical or patient.
The exclusion criteria covered only the articles
that described the technique or method used.
Only selection by both reviewers was considered
adequate. In case of disagreement the decision was
based on a consensus meeting between the
reviewers.
To maximize specificity, a preliminary analysis
was performed with 20 papers, to evaluate and
synchronize the reviewers.
2.2 Review Team
The review team was composed by a Computer
Scientist, Juliano Gaspar, and a Medical Doctor,
Emanuel Catumbela, advised by a Computer
Scientist with expertise in medical informatics,
Professor Alberto Freitas.
The statistical analysis was performed by a
Statistician, Bernardo Marques, with PASW version
18
®
.
2.3 Search Methods
The search for studies was performed between May
and June 2010 in bibliographic databases. Since
there were no specific standardized MeSH terms, we
developed a search string that includes the concepts
of outlier or anomaly detection in healthcare. We
decided to not include restrictions on language or
date of published articles. Four distinct bibliographic
databases were searched: Medline (via Pubmed), ISI
(ISI Web of Knowledge), IEEE (IEEE Xplore) and
EBSCO (EBSCOhost
®
databases). The query search
string used in each database was:
(((outlier or outliers) and (detect or detection or
observation or analysis)) or ((anomalous and
data) and (detect or detection or observation or
analysis)) or “extreme data” or “gross error” or
“anomalous record” or “anomalous register”)
and (medical or medicine or clinical or patient or
care or health).
2.4 Definition of Variables
The variables analyzed in this preliminary review
are:
Outliers Detection Techniques: statistical,
clustering, classification, nearest neighbor, mixture
models and spectral.
Data Type: type of data used in the use of each
technique (eg.: images, biosinal, patient data).
Medical domain: which defines the medical
specialty or domain of medicine used in the study is
employed.
Clinical Stage: phase clinical notes that the
study is applied to diagnosis, prognosis, treatment,
or for assessing outcomes and performance.
Were also collected information if the data used
were primary, secondary or simulated, the country
where the study was conducted and the year of
publication.
3 RESULTS
The design of this systematic review showed the
following results: the search method found 2697
articles in Medline, 1169 in ISI, 185 in IEEE and
414 in EBSCO, a total of 4465 articles. After
eliminating duplicate articles 4071 were selected.
As a result, a total of 177 out of 4071 articles
were selected to be read entirely. In this preliminary
study we present and analyze 80 articles. Figure 1 is
a flowchart illustrating the different stages of paper
selection.
The agreement rate between reviewers during the
phase of studies selection was 72%.
The table 1 lists all outlier detection techniques
considered in this review. In the 80 articles analyzed
were identified 112 techniques, which were
classified into 6 categories. The “statistical”
technique appears with 55.4% of the studies.
A SYSTEMATIC REVIEW OF OUTLIERS DETECTION TECHNIQUES IN MEDICAL DATA - Preliminary Study
577
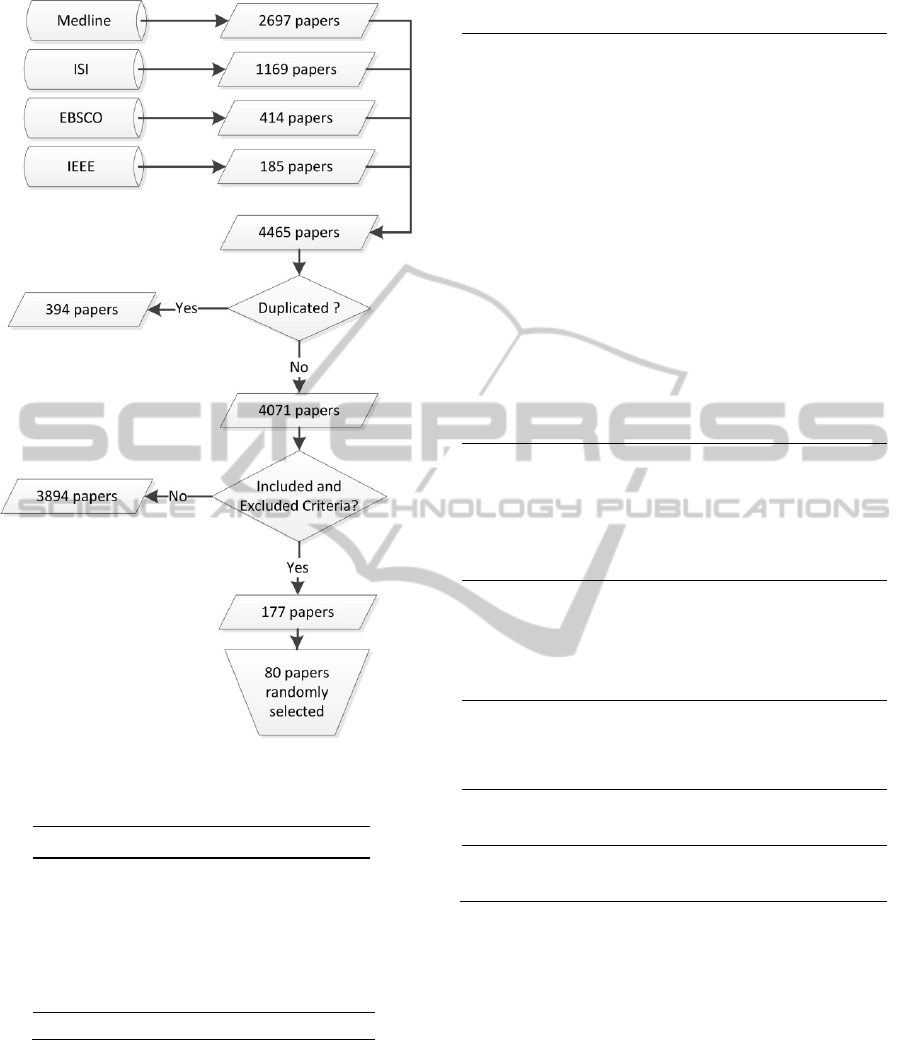
Figure 1: Flowchart illustrating the papers selection.
Table 1: List of outlier detection techniques.
Type N (%)
Statistical 62 55.4
Clustering 17 15.2
Classification 14 12.5
Nearest Neighbor 10 8.9
Mixture Models 6 5.4
Spectral 3 2.7
Total 112 100.0
It can be observed in Table 2 listing the 80
articles reviewed, grouped by techniques that each
presents. As described earlier, an article may appear
more than once, since some studies show more than
one technique.
According to the “clinical stage”, this study
showed that 81.3% of the techniques to detect
outliers for the diagnostic, while only 3.8% for
treatment.
Table 2: List of the articles by techniques.
Statistical
(Aalen et al., 2004; Ahdesmaki et al., 2005; Ahlers and Figg,
2006; Alameda and Suarez, 2009; Allen et al., 2010; Asare et
al., 2009; Bakhshi-Raiez et al., 2007; Beguin and Hulliger,
2004; Bickel, 2003; Booth and Lee, 2003; Branden and
Verboven, 2009; Breen et al., 2002; Cho et al., 2008;
Cluitmans and van de Velde, 2000; Cohen et al., 1996;
Comanor et al., 2006; Commowick and Warfield, 2009;
Cooney et al., 2003; Englesbe et al., 2009; Fomenko et al.,
2006; Freifeld et al., 2009; Ghosh, 2010; Ghosh and
Chinnaiyan, 2009; Glance et al., 2003; Glance et al., 2002;
Glance et al., 2007; Gold and Hoffman, 1976; Grotkjaer et al.,
2006; Hanauer et al., 2007; Hayes et al., 2007; Hojjatoleslami
et al., 1997; Hu, 2008; Hughes et al., 1997; Irigoien and
Arenas, 2008; Jackson et al., 2009; Jacobs, 2001; Kauffmann
and Huber, 2010; Kazmierczak et al., 2007; Liu and Wu, 2007;
Livesey, 2007; MacDonald and Ghosh, 2006; Mahadevan et
al., 2004; Meloun et al., 2004; Model et al., 2002; Nielsen and
Hansen, 2002; Nielsen et al., 2001; Oh and Gao, 2009;
Ohlssen et al., 2007; Penny and Jolliffe, 1999; Penny and
Jolliffe, 2001; Read, 1999; Rochelson et al., 2006; Rubin and
Chinnaiyan, 2006; Ryan, 2009; Song and Wyrwicz, 2009;
Tomlins et al., 2008; Van Leemput et al., 2001;
Vankeerberghen et al., 1995; Vellido and Lisboa, 2006;
Whitley and Ball, 2002; Wu, 2007; Zervakis et al., 2009)
Clustering
(Aggarwal and Yu, 2005; Azmandian et al., 2007; Beguin and
Hulliger, 2004; Bickel, 2003; Duan et al., 2009; Freifeld et al.,
2009; Gold and Hoffman, 1976; Goovaerts and Jacquez, 2004;
Grotkjaer et al., 2006; Hibbs et al., 2005; Irigoien and Arenas,
2008; Jackson et al., 2009; Janeja and Atluri, 2009; Koufakou
and Georgiopoulos, 2010; Mramor et al., 2007; Vellido and
Lisboa, 2006; Yang et al., 2007)
Classification
(Aggarwal and Yu, 2005; Baker and Jackson, 2008; Cardoso et
al., 2007; Commowick and Warfield, 2009; Gold and
Hoffman, 1976; Grotkjaer et al., 2006; Kazmierczak et al.,
2007; Law et al., 2001; Lopes et al., 2003; Mahadevan et al.,
2004; Oh and Gao, 2009; Ohlssen et al., 2007; Whitley and
Ball, 2002; Zervakis et al., 2009)
Nearest Neighbor
(Antao et al., 2008; Beguin and Hulliger, 2004; Chen et al.,
2008; Duan et al., 2009; Freifeld et al., 2009; Goovaerts and
Jacquez, 2004; Irigoien and Arenas, 2008; Jackson et al., 2009;
Janeja and Atluri, 2009; Koufakou and Georgiopoulos, 2010)
Mixture Models
(Ghosh, 2010; Ghosh and Chinnaiyan, 2009; Hu, 2008; Lopes
et al., 2003; Model et al., 2002; Penny and Jolliffe, 2001)
Spectral
(Cohen Freue et al., 2007; Hubert and Engelen, 2004; Song
and Wyrwicz, 2009)
Table 3 lists the data types more frequently
found in this study. They are, respectively, “Patient
Data” (37.5%) and “Genomic Data Sets” (32.1%)
followed by “Images” (15.1%) which includes, for
example, the two-dimensional images, three-
dimensional, electroencephalography, magnetic
resonance imaging and ultrasonography.
Table 4 presents the medical domain data. Found
in the medical specialties, the “oncology” (32.1%)
and “genetics” (15.2%) are the areas of health with
more usage of outlier detection techniques.
However, studies specifically related to
administrative medical data, “healthcare and quality
indicators” (24.1%), are ranked in second when
compared to medical specialties.
HEALTHINF 2011 - International Conference on Health Informatics
578

Table 3: Data Type.
Type N (%)
Patient Data 42 37.5
Genomic Data Sets 36 32.1
Images 17 15.1
Mass Spectrometry 6 5.4
Biosignal 4 3.6
Blood 4 3.6
Others 3 2.7
Total 112 100.0
Table 4: Medical Domain.
Specialties
N (%)
Oncology 36 32.1
Healthcare and Quality Indicator 27 24.1
Genetics 17 15.2
Neurology 13 11.6
Biochemistry 7 6.3
Clinical Analysis 4 3.6
Others 8 7.1
Total 112 100.0
Considering only the administrative medical
data, “healthcare and quality indicator”, is
presented in Table 5 a cross table between
techniques described and the data types used by the
authors of the studies.
Table 5: Administrative Medical Data.
Data Type
Patient
Data
Biosignal Total
Techniques N N N (%)
Statistical 14 2 16 59.3
Clustering 4 0 4 14.8
Nearest Neighbor 4 0 4 14.8
Classification 3 0 3 11.1
Total 25 2 27 100.0
Table 5 one can observe that the data used to
analyze administrative medical data, the main focus
of this review, are “patient data” (92.6%) and
“biosignal” (7.4%).
4 DISCUSSION
This review shows that, considering only
administrative medical data, statistical techniques
are the most commonly used (59%), followed by
clustering (15%) and nearest neighbor (15%). Even
considering only administrative medical data, the
techniques mainly use data such as "patient data".
Methods used to detect outliers in healthcare
databases are mostly applied for diagnosis (81%).
Considering medical specialty, oncology and genetic
studies are the most applied.
Thus, we conclude that statistical techniques are
widely used in administrative medical data, and that
techniques from data mining such as clustering and
nearest neighbor are still little used in this context.
There is a considerable field for developing
techniques to detect outliers in medical data
management, based on clustering and nearest
neighbor, beyond of the statistics techniques already
widespread.
4.1 Limitations
A major difficulty observed in this systematic
review was the variety of names used to define
outliers. The lack of objectivity and clarity to
describe the methods and techniques used in some
articles, demanded the reviewers to have a special
attention to distinguish which outlier detection
technique was used by the author in his study.
4.2 Future Works
In future work, we will implement a prototype for
detection of outliers in medical administrative
databases using clustering techniques, nearest
neighbor techniques, beyond statistical techniques.
The aim is to improve the process of decision
making the department directors and hospital
administrators, providing them with data with more
quality and accuracy.
ACKNOWLEDGEMENTS
The authors would like to thank the support given by
the research project HR-QoD - Quality of data
(outliers, inconsistencies and errors) in hospital
inpatient databases: methods and implications for
data modeling, cleansing and analysis (project
PTDC/SAU-ESA/75660/2006).
A SYSTEMATIC REVIEW OF OUTLIERS DETECTION TECHNIQUES IN MEDICAL DATA - Preliminary Study
579

REFERENCES
Aalen, O. O., Fosen, J., Weedon-Fekjaer, H., Borgan, O.,
and Husebye, E. (2004). Dynamic analysis of
multivariate failure time data. Biometrics 60, 764-773.
Aggarwal, C. C., and Yu, P. S. (2005). An effective and
efficient algorithm for high-dimensional outlier
detection. Vldb J 14, 211-221.
Ahdesmaki, M., Lahdesmaki, H., Pearson, R., Huttunen,
H., and Yli-Harja, O. (2005). Robust detection of
periodic time series measured from biological systems.
BMC Bioinformatics 6, 117.
Ahlers, C. M., and Figg, W. D. (2006). ETS-TMPRSS2
fusion gene products in prostate cancer. Cancer Biol
Ther 5, 254-255.
Alameda, C., and Suarez, C. (2009). Clinical outcomes in
medical outliers admitted to hospital with heart
failure. Eur J Intern Med 20, 764-767.
Allen, D. P., Stegemoller, E. L., Zadikoff, C., Rosenow, J.
M., and Mackinnon, C. D. (2010). Suppression of deep
brain stimulation artifacts from the
electroencephalogram by frequency-domain Hampel
filtering. Clin Neurophysiol.
Antao, T., Lopes, A., Lopes, R. J., Beja-Pereira, A., and
Luikart, G. (2008). LOSITAN: a workbench to detect
molecular adaptation based on a Fst-outlier method.
BMC Bioinformatics 9, 323.
Arts, D., Keizer, N., and Scheffer, G.-J. (2002). Defining
and Improving Data Quality in Medical Registries: A
Literature Review Case Study, and Generic
Framework. J Am Med Inform Assoc 9, 600-611.
Asare, A. L., Gao, Z., Carey, V. J., Wang, R., and Seyfert-
Margolis, V. (2009). Power enhancement via
multivariate outlier testing with gene expression
arrays. Bioinformatics 25, 48-53.
Azmandian, F., Kaeli, D., Dy, J. G., Hutchinson, E.,
Ancukiewicz, M., Niemierko, A., and Jiang, S. B.
(2007). Towards the development of an error checker
for radiotherapy treatment plans: a preliminary study.
Phys Med Biol 52, 6511-6524.
Baker, R., and Jackson, D. (2008). A new approach to
outliers in meta-analysis. Health Care Management
Science 11, 121-131.
Bakhshi-Raiez, F., Peek, N., Bosman, R. J., de Jonge, E.,
and de Keizer, N. F. (2007). The impact of different
prognostic models and their customization on
institutional comparison of intensive care units. Crit
Care Med 35, 2553-2560.
Barnett, V., and Lewis, T. (1994). Outliers in Statistical
Data (England).
Beguin, C., and Hulliger, B. (2004). Multivariate outlier
detection in incomplete survey data: the epidemic
algorithm and transformed rank correlations. J R Stat
Soc Ser A-Stat Soc 167, 275-294.
Bickel, D. R. (2003). Robust cluster analysis of
microarray gene expression data with the number of
clusters determined biologically. Bioinformatics 19,
818-824.
Booth, D. E., and Lee, K. (2003). Robust regression-based
analysis of drug-nucleic acid binding. Anal Biochem
319, 258-262.
Branden, K. V., and Verboven, S. (2009). Robust data
imputation. Comput Biol Chem 33, 7-13.
Breen, H. J., Rogers, P. A., and Johnson, N. W. (2002).
Improvements in methods of periodontal probing:
comparison of relative attachment level data selected
by outlier reduction protocols from Florida disc probe
measurements. J Clin Periodontol 29, 679-687.
Cardoso, F. F., Rosa, G. J., and Tempelman, R. J. (2007).
Accounting for outliers and heteroskedasticity in
multibreed genetic evaluations of postweaning gain of
Nelore-Hereford cattle. J Anim Sci 85, 909-918.
Chandola, V., Banerjee, A., and Kumar, V. (2009).
Anomaly Detection: A Survey. ACM Computing
Surveys 41.
Chen, D., Lu, C.-T., Kou, Y., and Chen, F. (2008). On
Detecting Spatial Outliers. GeoInformatica 12, 455-
475.
Cho, H., Kim, Y. J., Jung, H. J., Lee, S. W., and Lee, J. W.
(2008). OutlierD: an R package for outlier detection
using quantile regression on mass spectrometry data.
Bioinformatics 24, 882-884.
Cios, K. (2001). Medical data mining and knowledge
discovery (Physica-Verlag).
Cluitmans, P. J. M., and van de Velde, M. (2000). Outlier
detection to identify artefacts in EEG signals. In
Proceedings of the 22nd Annual International
Conference of the Ieee Engineering in Medicine and
Biology Society, Vols 1-4, J. D. Enderle, ed. (New
York, Ieee), pp. 2825-2826.
Cohen Freue, G. V., Hollander, Z., Shen, E., Zamar, R. H.,
Balshaw, R., Scherer, A., McManus, B., Keown, P.,
McMaster, W. R., and Ng, R. T. (2007). MDQC: a
new quality assessment method for microarrays based
on quality control reports. Bioinformatics 23, 3162-
3169.
Cohen, Y. C., Olmer, L., and Mozes, B. (1996). Two-
dimensional outcome analysis as a guide for quality
assurance of prostatectomy. Int J Qual Health Care 8,
67-73.
Comanor, W. S., Frech, H. E., 3rd, and Miller, R. D., Jr.
(2006). Is the United States an outlier in health care
and health outcomes? A preliminary analysis. Int J
Health Care Finance Econ 6, 3-23.
Commowick, O., and Warfield, S. K. (2009). A
Continuous STAPLE for Scalar, Vector, and Tensor
Images: An Application to DTI Analysis. IEEE
Transactions on Medical Imaging 28, 838-846.
Cooney, R. N., Haluck, R. S., Ku, J., Bass, T., MacLeod,
J., Brunner, H., and Miller, C. A. (2003). Analysis of
cost outliers after gastric bypass surgery: What can
we learn? Obesity Surgery 13, 29-36.
Cruz-Correia, R., Vieira-Marques, P., Ferreira, A.,
Oliveira-Palhares, E., Costa, P., and Costa-Pereira, A.
(2006). Monitoring the integration of hospital
information systems: How it may ensure and improve
the quality of data. Stud Health Technol Inform 121,
176-182.
HEALTHINF 2011 - International Conference on Health Informatics
580

Duan, L., Xu, L. D., Liu, Y., and Lee, J. (2009). Cluster-
based outlier detection. Annals of Operations
Research 168, 151-168.
Englesbe, M. J., Dimick, J. B., Fan, Z., Baser, O., and
Birkmeyer, J. D. (2009). Case Mix, Quality and High-
Cost Kidney Transplant Patients. American Journal of
Transplantation 9, 1108-1114.
Fomenko, I., Durst, M., and Balaban, D. (2006). Robust
regression for high throughput drug screening.
Comput Methods Programs Biomed 82, 31-37.
Freifeld, O., Greenspan, H., and Goldberger, J. (2009).
Multiple Sclerosis Lesion Detection Using
Constrained GMM and Curve Evolution. Int J Biomed
Imaging 2009, 715124.
Ghosh, D. (2010). Discrete nonparametric algorithms for
outlier detection with genomic data. J Biopharm Stat
20, 193-208.
Ghosh, D., and Chinnaiyan, A .M. (2009). Genomic
outlier profile analysis: mixture models, null
hypotheses, and nonparametric estimation.
Biostatistics 10, 60-69.
Glance, L. G., Dick, A. W., Osler, T. M., and Mukamel,
D. (2003). Using hierarchical modeling to measure
ICU quality. Intensive Care Medicine 29, 2223-2229.
Glance, L. G., Osler, T. M., and Dick, A. W. (2002).
Identifying quality outliers in a large, multiple-
institution database by using customized versions of
the Simplified Acute Physiology Score II and the
Mortality Probability Model II0. Crit Care Med 30,
1995-2002.
Glance, L. G., Osler, T. M., Mukamel, D. B., and Dick, A.
W. (2007). Use of a matching algorithm to evaluate
hospital coronary artery bypass grafting performance
as an alternative to conventional risk adjustment. Med
Care 45, 292-299.
Gold, E. M., and Hoffman, P. J. (1976). Flange detection
cluster analysis. Multivariate Behavioral Research 11,
217-235.
Goovaerts, P., and Jacquez, G. M. (2004). Accounting for
regional background and population size in the
detection of spatial clusters and outliers using
geostatistical filtering and spatial neutral models: the
case of lung cancer in Long Island, New York.
International Journal of Health Geographics 3, 14-23.
Grotkjaer, T., Winther, O., Regenberg, B., Nielsen, J., and
Hansen, L. K. (2006). Robust multi-scale clustering of
large DNA microarray datasets with the consensus
algorithm. Bioinformatics 22, 58-67.
Hanauer, D. A., Rhodes, D. R., Sinha-Kumar, C., and
Chinnaiyan, A. M. (2007). Bioinformatics approaches
in the study of cancer.
Curr Mol Med 7, 133-141.
Hauskrecht, M., Valko, M., Kveton, B., Visweswaran, S.,
and Cooper, G. F. (2007). Evidence-based anomaly
detection in clinical domains. AMIA Annu Symp
Proc, 319-323.
Hayes, K., Kinsella, A., and Coffey, N. (2007). A note on
the use of outlier criteria in Ontario laboratory quality
control schemes. Clin Biochem 40, 147-152.
Hibbs, M. A., Dirksen, N. C., Li, K., and Troyanskaya, O.
G. (2005). Visualization methods for statistical
analysis of microarray clusters. BMC Bioinformatics
6, 115.
Hojjatoleslami, A., Sardo, L., and Kittler, J. (1997). An
RBF based classifier for the detection of
microcalcifications in mammograms with outlier
rejection capability. Paper presented at: Neural
Networks,1997, International Conference on.
Hu, J. (2008). Cancer outlier detection based on
likelihood ratio test. Bioinformatics 24, 2193-2199.
Hubert, M., and Engelen, S. (2004). Robust PCA and
classification in biosciences. Bioinformatics 20, 1728-
1736.
Hughes, S. L., Ulasevich, A., Weaver, F. M., Henderson,
W., Manheim, L., Kubal, J. D., and Bonarigo, F.
(1997). Impact of home care on hospital days: a meta
analysis. Health Serv Res 32, 415-432.
Irigoien, I., and Arenas, C. (2008). INCA: new statistic for
estimating the number of clusters and identifying
atypical units. Stat Med 27, 2948-2973.
Jackson, M. C., Huang, L., Luo, J., Hachey, M., and
Feuer, E. (2009). Comparison of tests for spatial
heterogeneity on data with global clustering patterns
and outliers. Int J Health Geogr 8, 55.
Jacobs, R. (2001). Outliers in Statistical Analysis: Basic
Methods of Detection and Accommodation.
Janeja, V. P., and Atluri, V. (2009). Spatial outlier
detection in heterogeneous neighborhoods. Intell Data
Anal 13, 85-107.
Kauffmann, A., and Huber, W. (2010). Microarray data
quality control improves the detection of differentially
expressed genes. Genomics 95, 138-142.
Kazmierczak, S. C., Leen, T. K., Erdogmus, D., and
Carreira-Perpinan, M. A. (2007). Reduction of multi-
dimensional laboratory data to a two-dimensional
plot: a novel technique for the identification of
laboratory error.
Clinical Chemistry & Laboratory
Medicine 45, 749-752.
Koufakou, A., and Georgiopoulos, M. (2010). A fast
outlier detection strategy for distributed high-
dimensional data sets with mixed attributes. Data Min
Knowl Discov 20, 259-289.
Kumar, V., Kumar, D., and Singh, R. K. (2008). Outlier
Mining in Medical Databases: An Application of Data
Mining in Health Care Management to Detect
Abnormal Values Presented In Medical Databases.
IJCSNS International Journal of Computer Science
and Network Security.
Laurikkala, J., Juhola, M., and Kentala, E. (2000).
Informal Identification of Outilers in Medical Data.
Intelligent Data Analysis in Medicine and
Pharmacology.
Law, G. R., Cox, D. R., Machonochie, N. E., Simpson, J.,
Roman, E., and Carpenter, L. M. (2001). Large tables.
Biostatistics 2, 163-171.
Liu, F., and Wu, B. (2007). Multi-group cancer outlier
differential gene expression detection. Comput Biol
Chem 31, 65-71.
Livesey, J. H. (2007). Kurtosis provides a good omnibus
test for outliers in small samples. Clin Biochem 40,
1032-1036.
A SYSTEMATIC REVIEW OF OUTLIERS DETECTION TECHNIQUES IN MEDICAL DATA - Preliminary Study
581

Lopes, H. F., Müller, P., and Rosner, G. L. (2003).
Bayesian Meta-analysis for Longitudinal Data Models
Using Multivariate Mixture Priors. Biometrics 59, 66-
75.
MacDonald, J. W., and Ghosh, D. (2006). COPA--cancer
outlier profile analysis. Bioinformatics 22, 2950-2951.
Mahadevan, V., Narasimha-Iyer, H., Roysam, B., and
Tanenbaum, H. L. (2004). Robust model-based
vasculature detection in noisy biomedical images.
IEEE Trans Inf Technol Biomed 8, 360-376.
Marin, J. M. M., Kerrie, L., and Robert, C. (2005).
Bayesian modelling and inference on mixtures of
distributions, Vol 25 (Elsevier).
Meloun, M., Hill, M., Militký, J., Vrbiková, J., Škrha, J.,
and Stanická, S. (2004). New methodology of
influential point detection in regression model
building for the prediction of metabolic clearance rate
of glucose. Clinical Chemistry & Laboratory Medicine
42, 311-322.
Model, F., Konig, T., Piepenbrock, C., and Adorjan, P.
(2002). Statistical process control for large scale
microarray experiments. Bioinformatics 18 Suppl 1,
S155-163.
Mramor, M., Leban, G., Demsar, J., and Zupan, B. (2007).
Visualization-based cancer microarray data
classification analysis. Bioinformatics 23, 2147-2154.
Nielsen, F. A., and Hansen, L. K. (2002). Modeling of
activation data in the BrainMap (TM) database:
Detection of outliers. Human Brain Mapping 15, 146-
156.
Nielsen, F. A., Hansen, L. K., and Kjems, U. (2001).
Modeling of locations in the BrainMap database:
Detection of outliers. NeuroImage 13, S211-S211.
Oh, J. H., and Gao, J. (2009). A kernel-based approach for
detecting outliers of high-dimensional biological data.
BMC Bioinformatics 10 Suppl 4, S7.
Ohlssen, D. I., Sharples, L. D., and Spiegelhalter, D. J.
(2007). A hierarchical modelling framework for
identifying unusual performance in health care
providers. Journal of the Royal Statistical Society:
Series A (Statistics in Society) 170, 865-890.
Penny, K. I., and Jolliffe, I. T. (1999). Multivariate outlier
detection applied to multiply imputed laboratory data.
Statistics In Medicine 18, 1879-1895.
Penny, K. I., and Jolliffe, I. T. (2001). A Comparison of
Multivariate Outlier Detection Methods for Clinical
Laboratory Safety Data. Journal of the Royal
Statistical Society: Series D (The Statistician) 50, 295.
Read, R. J. (1999). Detecting outliers in non-redundant
diffraction data. Acta Crystallogr D Biol Crystallogr
55, 1759-1764.
Rochelson, B., Vohra, N., Krantz, D., and Macri, V.J.
(2006). Geometric morphometric analysis of shape
outlines of the normal and abnormal fetal skull using
three-dimensional sonographic multiplanar display.
Ultrasound in Obstetrics & Gynecology 27, 167-172.
Rubin, M. A., and Chinnaiyan, A.M. (2006).
Bioinformatics approach leads to the discovery of the
TMPRSS2:ETS gene fusion in prostate cancer. Lab
Invest 86, 1099-1102.
Ryan, A. M. (2009). Effects of the Premier Hospital
Quality Incentive Demonstration on Medicare Patient
Mortality and Cost. Health Services Research 44, 821-
842.
Silva-Costa, T., Marques, B., and Freitas, A. (2010).
Problemas de Qualidade de Dados em Bases de
Dados de Internamentos Hospitalares. Paper
presented at: 5ª Conferência Ibérica de Sistemas e
Tecnologias de Informação (Santiago de Compostela).
Silva, F. R. (2004). Uma abordagem para detecção de
outliers em dados categoricos. In Instituto de
Computação (Campinas, SP Universidade Estadual de
Campinas).
Song, X., and Wyrwicz, A. M. (2009). Unsupervised
spatiotemporal fMRI data analysis using support
vector machines. NeuroImage 47, 204-212.
Tomlins, S. A., Rhodes, D. R., Yu, J., Varambally, S.,
Mehra, R., Perner, S., Demichelis, F., Helgeson, B. E.,
Laxman, B., Morris, D. S., et al. (2008). The role of
SPINK1 in ETS rearrangement-negative prostate
cancers. Cancer Cell 13, 519-528.
Van Leemput, K., Maes, F., Vandermeulen, D.,
Colchester, A., and Suetens, P. (2001). Automated
segmentation of multiple sclerosis lesions by model
outlier detection. IEEE Transactions on Medical
Imaging 20, 677-688.
Vankeerberghen, P., Smeyersverbeke, J., Leardi, R., Karr,
C. L., and Massart, D.L. (1995). Robust Regression
and Outlier Detection for NonLinear Models Using
Genetic Algorithms. Chemometrics Intell Lab Syst 28,
73-87.
Vellido, A., and Lisboa, P. J. (2006). Handling outliers in
brain tumour MRS data analysis through robust
topographic mapping. Comput Biol Med 36, 1049-
1063.
Whitley, E., and Ball, J. (2002). Statistics review 1:
presenting and summarising data. Crit Care 6, 66-71.
Wu, B. (2007). Cancer outlier differential gene expression
detection. Biostatistics 8, 566-575.
Yang, S., Guo, X., Yang, Y. C., Papcunik, D., Heckman,
C., Hooke, J., Shriver, C. D., Liebman, M. N., and Hu,
H. (2007). Detecting outlier microarray arrays by
correlation and percentage of outliers spots. Cancer
Inform 2, 351-360.
Zervakis, M., Blazadonakis, M. E., Tsiliki, G., Danilatou,
V., Tsiknakis, M., and Kafetzopoulos, D. (2009).
Outcome prediction based on microarray analysis: a
critical perspective on methods. BMC Bioinformatics
10, 53.
HEALTHINF 2011 - International Conference on Health Informatics
582
