
PATIENT-BASED LITERATURE RETRIEVAL AND
INTEGRATION
A Use Case for Diabetes and Arterial Hypertension
Ana Jimenez-Castellanos
1,2
, Izaskun Fernández
3
, David Perez-Rey
1
, Elisa Viejo
2
Francisco Javier Díez
3
, Xabier García de Kortazar
3
, Miguel García-Remesal
1
, Víctor Maojo
1
,
Antonio Cobo
4,2
and Francisco del Pozo
2,4
1
Dept Inteligencia Artificial, Facultad de Informática, Universidad Politécnica de Madrid
Campus de Montegancedo S/N, 28660 Boadilla del Monte, Madrid, Spain
2
Centre for Biomedical Technology, Technical University of Madrid (CTB – UPM)
Campus de Montegancedo – UPM Autopista M-40, km 38.Pozuelo de Alarcón, 28223, Madrid, Spain
3
Tekniker-IK4,Av Otaola, 20, 20600 Eibar, Guipuzcoa, Spain
4
Biomedical Research Networking Center in Bioengineering, Biomaterials and Nanomedicine (CIBER –BBN)
Madrid, Spain
Keywords: Electronic Health Record, Search engines, Literature retrieval, Integration, Federated search.
Abstract: Specialized search engines such as PubMed, MedScape or Cochrane have increased dramatically the
visibility of biomedical scientific results. These web-based tools allow physicians to access scientific papers
instantly. However, this decisive improvement had not a proportional impact in clinical practice due to the
lack of advanced search methods. Even queries highly specified for a concrete pathology frequently retrieve
too many information, with publications related to patients treated by the physician beyond the scope of the
results examined. In this work we present a new method to improve scientific article search using patient
information. Two pathologies have been used within the project to retrieve relevant literature to patient data
and to be integrated with other sources. Promising results suggest the suitability of the approach,
highlighting publications dealing with patient features and facilitating literature search to physicians.
1 INTRODUCTION
Physicians attending daily a high number of patients
hamper their availability to keep up to date with the
latest research news in their field. They frequently
lack the time to locate relevant information related
to the patient Electronic Health Record (EHR).
Physicians need specific information rather than
large amounts of information, however the amount
of data generated nowadays and stored both, within
EHR and research literature, is overwhelming. The
more data the more challenging is to find relevant
information (for research and training of the
physician) and to integrate it with current resources.
Search engines and other Web 2.0 technologies such
as RSS feeds facilitate these tasks, but advanced
methods are required to retrieve research
information related to, not only a pathology, but also
a specific demographic group.
Scientific research papers regarding clinical
practice deals with population groups rather than
specific patients. Physicians must generalize patient
data in order to find patient-related clinical
literature. They should analyze the EHR of the
patient to include relevant keywords in the query
that filter the results from several information
sources. After that, the physician reviews the
scientific articles retrieved and selects the relevant
information for the patient, discarding the majority
of the results. This is particularly interesting for
patients who do not respond to therapies or
treatments, who need special attention to avoid
further complications. These kind of patients are
those for whom physicians need more specific
research information about their disease.
33
Jimenez-Castellanos A., Fernández I., Perez-Rey D., Viejo E., Javier Díez F., García de Kortazar X., García-Remesal M., Maojo V., Cobo A. and del
Pozo F..
PATIENT-BASED LITERATURE RETRIEVAL AND INTEGRATION - A Use Case for Diabetes and Arterial Hypertension.
DOI: 10.5220/0003132900330041
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2011), pages 33-41
ISBN: 978-989-8425-34-8
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

In this work we present a new method,
implemented by the authors, aiming to provide an
advanced method to retrieve biomedical literature
based on EHRs. Diabetes and arterial hypertension
are the two use cases that have been tested, within
the framework of “Treatment 2.0”, a research and
development project aiming to create a generic
middleware platform that serves as the basis for the
development of management services and intelligent
application for treatment for patients, especially
chronic (Tratamiento 2.0 2010 June).
2 BACKGROUND
Patient information inclusion in search queries for
biomedical literature, requires techniques and
technologies with a high research activity. Those
used to store the required information, such as
Electronic Health Records and Decision Support
Systems, and those used to extract information, such
as Natural Language Processing and Federated
Searching described in this section.
Electronic Health Records (EHR), defined as
“electronic objects that contain data, evaluations and
information of any kind, on the status and the
clinical course of one patient through the care
process” (Healthcare Information and Management
Systems Society 2010 June), have been a
fundamental advance in clinical practice. Electronic
information can be automatically analyzed and there
exist research efforts to generate knowledge from
EHRs such as (Weaver et al. 2005), where the data
generated from care delivery and captured in the
EHR systems, is used for being analyzed to discover
and then inform about best practice. In (Antolik
2005), how to transfer knowledge from a medical
record written in a free text form into a structured
format represented by the EHR is analyzed. Finally,
(Natarajan et al. 2010) is a study aiming to
understand user needs as captured by their search
queries in an EHR system.
EHR are the essential infrastructure to other
software elements in clinical practice such as
Decision Support Systems, a kind of information
systems that supports decision-making activities
(Keen, Morton 1978). Specifically, Clinical
Decision Support Systems (CDSS) are software
programs that assist the physicians in the decision
making process (Kawamoto et al. 2005, Sim et al.
2001). Some methods to integrate CDSS and EHR
are: (van der Weijden et al. 2010) and (Seidling et
al. 2010), systems dealing with CDSS adaptation, to
facilitate the integration of individual patients
preferences and characteristics and improve decision
making.
To retrieve further clinical information, the EBM
(Evidence-Based Medicine Working Group 1992)
recommends physicians to formulate clinical
questions in terms of the problem/population,
intervention, comparison, and outcome for searching
clinical reports efficiently. These elements comprise
the PICO frame: P represents problem/population; I
intervention’s information; C the comparison; and
finally O the outcome. The construction of this kind
of query is not an easy task as it is shown in (Huang,
Lin & Demner-Fushman 2006). For instance,
defining P requires an exhaustive reading of the
patient EHR and the selection of the most relevant
characteristics for the current context.
Information extraction dealing with free text
instead of structured data is treated with Natural
Language Processing (NLP) techniques, that have
been applied to many tasks of Biomedicine such as
bio-entity recognition, protein/gene normalization,
interaction, extraction and many others. As there are
many fields where NLP techniques can be used,
various NLP tools are available. There are domain
specific tools resolving an specific task like
GoAnnotatorTool (Couto et al. 2006), but also we
can find more generic and flexible NLP tools, like
Freeling (Atserias et al. 2006) or GATE
(Cunningham 2002) which can be used for many
tasks and domains. The fundamental problem of
NLP analysis is the fact, that a particular meaning
may be expressed using different synonymous
expressions. Lexical repositories recording meaning
and lexical forms relations are frequently used in
NLP techniques for interpreting texts. MeSH (Díaz-
Galiano et al. 2008) or SNOMED (Spackman,
Reynoso 2004) are two well known lexical
repositories in the medicine domain which can be
exploited for scientific article interpretation.
Finally, heterogeneous data sources stored in
different locations require distributed or federated
methods to extract information. The federated search
paradigm was thus created, evolving in response to
the vast number of information resources. As
defined in (Jacso 2004), federated searching consists
of firstly transforming a query and broadcasting it to
a group of different sources; merging the results
obtained from the different asked sources;
presenting it in a unified format without
duplications; and providing the option of sorting the
result set. Search engines like Sphinx (Lee 1989)
help in the development of this kind of systems,
HEALTHINF 2011 - International Conference on Health Informatics
34
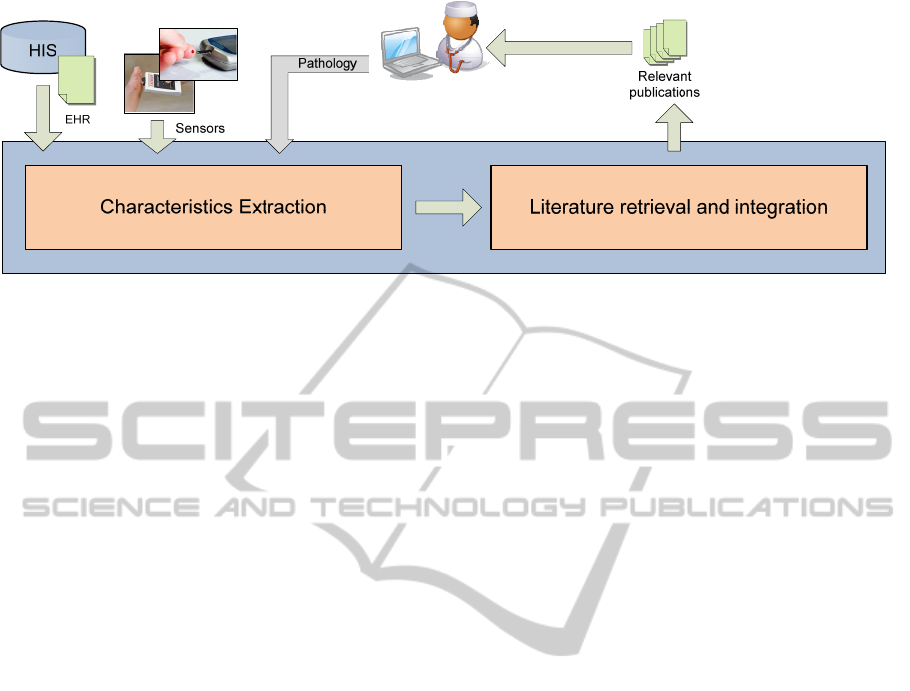
Figure 1: Patient-based literature retrieval architecture.
indexing the information and providing a fast
information retrieval.
These techniques have been used in projects such
as Parallel IE for bio-medical text mining and
indexation at Merck kGaA, Darmstadt and Medline
Analysis (Karopka et al. 2004) at Institute for
Medical Informatics and Biometry, University of
Rostock, Germany for extracting causal functional
relations on MedLine abstracts using GATE. There
are also tools like MeSHMap (Srinivasan 2001) and
MedMeSH Summarizer (Kankar et al. 2002) that
exploits MeSH ontology for document indexation
and summarization, respectively.
3 PATIENT-BASED LITERATURE
RETRIEVAL AND
INTEGRATION
The approach proposed in this paper aims to provide
an improved method to search biomedical literature
based on patient data. Data used to identify the
relevant information for literature retrieval is
collected from two sources: sensors on the patient’s
home — described elsewhere (Tratamiento 2.0 2010
June) — and data contained in the EHR of the
patient. Both sources are stored in the platform of
the project and then feed the services explained
below.
Figure 1 presents the two main phases performed
to retrieve relevant publications. Firstly, the system
should receive both data sources and locate relevant
characteristics of the patient and then looking for
and integrating the most relevant publications based
on these characteristics.
To automatically query current biomedical
literature search engines for relevant publications
based on EHRs and sensor info, a web services
architecture was proposed. Three main web services
were identified to cover the required functionality:
patientCharacterictics, receiving a set of
parameters from the EHR and sensors to
identify the relevant characteristics.
federatedSearch, receiving the relevant
characteristics, generates and launches queries
against different data sources.
resultAggregation, that collect, integrates and
present the results to the user.
These web services are explained within the next
subsections, 3.1, 3.2 and 3.3 respectively.
3.1 Characteristics Extraction
The patientCharacteristics web service receives a
set of parameters in XML format from the platform
of the project. This platform stores data contained in
the patient's EHR in addition to other data resulting
from the monitoring of the patient at home — e.g.
certain data is needed in real time (like glucose
levels before and after eating). This was
implemented using sensors within the patient
environment. Figure 2 shows the structure of the
web service which extracts the characteristics of a
patient.
Two separated phases can be identified: (i)
parameter processing and (ii) curation. Within the
first phase, parameters are processed to extract the
relevant characteristics. Afterwards, during the
curation phase, an expert may evaluate the
characteristics extracted to filter those relevant for
the publication search.
PATIENT-BASED LITERATURE RETRIEVAL AND INTEGRATION - A Use Case for Diabetes and Arterial
Hypertension
35
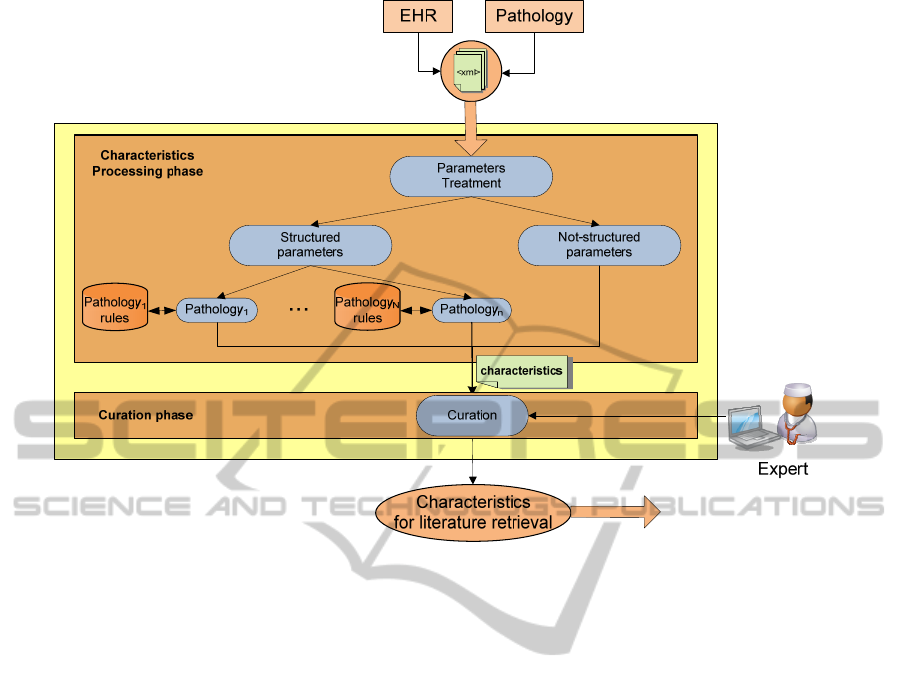
Figure 2: Characteristics Extraction.
Parameters from EHR or sensors are integrated
into a single XML parameter list:
<patient_parameters>
<pathology>diabetes</pathology>
<parameters_list>
<parameter>
<name>age</name>
<value>62</value>
</parameter>
...
</parameters_list>
</patient_parameters>
For each parameter its name and its value are
required. The patientCharacteristics web service
extracts the significant characteristics focused on a
given pathology.
The set of parameters is not closed, the service
can receive any type of parameters, i.e. that the
application will analyze only the parameters that can
be treated according to certain rules, discarding
those that are not recognized.
After receiving the parameters, the web service
separates among structured and non-structured
parameters. Structured data are those which have a
numeric or enumerated value (E.g.: the numeric
value of the patient’s age, or the values “male” or
“female” identifying the sex of the patient). Not-
structured data are free text and they are directly
addressed to the curation phase as characteristics of
the patient.
Depending on the pathology, structured
parameters are selected for its relevance. For each
disease there is a knowledge base of rules that is
responsible for processing related parameters. Thus,
when the characteristics of a patient have to be
extracted based on a new disease, only the
corresponding rules are required. The knowledge
base of each pathology is built based on standards of
CDSSs.
Relevant data for the pathology is analyzed
following rules contained within the knowledge
bases, which are associated with the parameters.
Those rules deal with patient stratification. Most of
the rules are constructed based on expert approved
thresholds. Depending on parameters values,
characteristics are extracted and mapped to one or
several MeSH terms.
All parameters are evaluated to extract
significant characteristics related to one patient. In
the curation phase, the physician (using a simple
interface) may choose characteristics to be used to
build the search query. Relevant characteristics
selected by the physicians will be sent to the next
web service for retrieving the corresponding
literature.
HEALTHINF 2011 - International Conference on Health Informatics
36
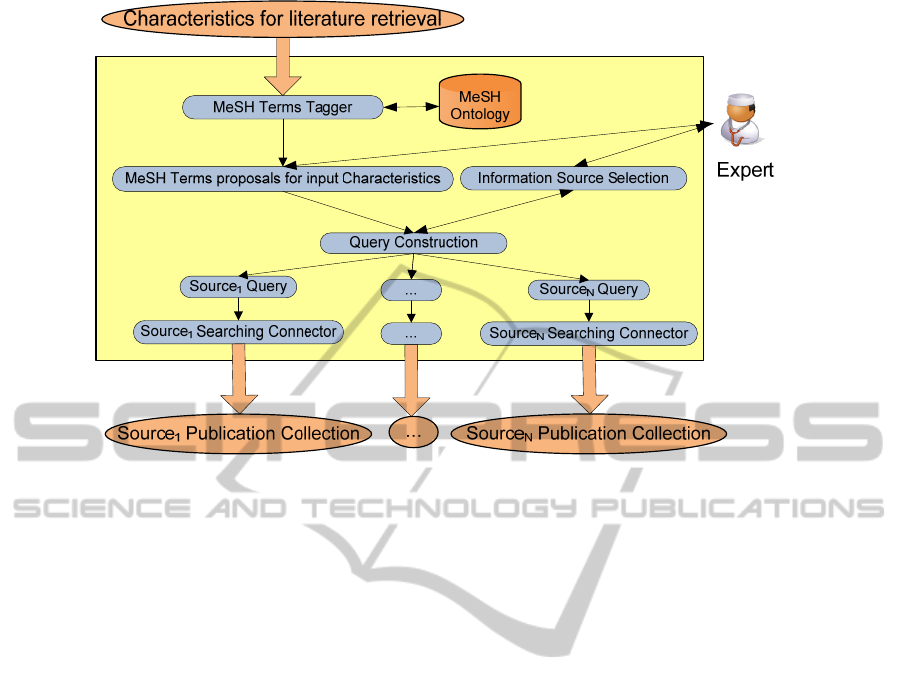
Figure 3: Query Generation and Information Retrieval.
3.2 Query Generation
Search federation intends to aggregate information
units from heterogeneous sources, presenting the
results on a unified way, without duplicates and
ordered by relevance. This architecture enables a
high scalability and stability, since adding new
sources is only a matter of new connectors’
configuration, and system remains working even if
some sources are down.
So the federatedSearch web service gets as input
the XML characteristics generated by
patientCharacteristics service as context and a query
defined by the expert, which expresses the searching
topic. In order to represent efficiently those
characteristics and the free query, the service
exploits MeSH ontology combined with NLP tools,
trying to find corresponding MeSH terms both, for
the characteristics and on the input free text. The
MeSH lexicon as a flexible Gazetteer on GATE tool
has been integrated in the service. In this way MeSH
terms on any text are tagged, first parsing it with the
morphological module of GATE and then
identifying MeSH terms with the gazetteers module.
The characteristics are extracted from the patient
EHR, so it can be considered that the selected MeSH
terms define the problem (P) of the PICO query.
Intervention (I) and outcome (O) will be completed
with the identified and after selected terms on the
expert query. At this moment, the system does not
consider the comparison (C) parameter.
In order to get the federated search, the web
service queries different information sources. Each
registered expert has associated a collection of
information sources. For each new query the expert
selects a set of those sources to search. Each
information source encloses its own query format.
Before searching, it is necessary to transform the
PICO query to the each source query format. For the
transformation process, regardless of the source, the
system applies always the same methodology: taking
PICO query as input, it applies an xslt based
transformation. The xslt object is a source dependent
element where the source specific query syntax is
considered for the final query representation.
Together with query representations, the service
launches different queries, getting one result set
from each source in XML format. This procedure is
graphically represented at figure 3.
3.3 Integration and Presentation
The publication sets obtained from the previous web
service are the input of the resultAggregation
service. The aim of this service is to aggregate all
the result sets presenting to the expert the
publications as a unique collection. The final
collection should not have duplicated items and must
be sorted by relevance regardless of the source. The
web service processes the publications, tagging
MeSH terms and storing both the item itself and the
tagged MeSH terms in a MySQL database in order
to index the entire collection with Sphinx and
PATIENT-BASED LITERATURE RETRIEVAL AND INTEGRATION - A Use Case for Diabetes and Arterial
Hypertension
37
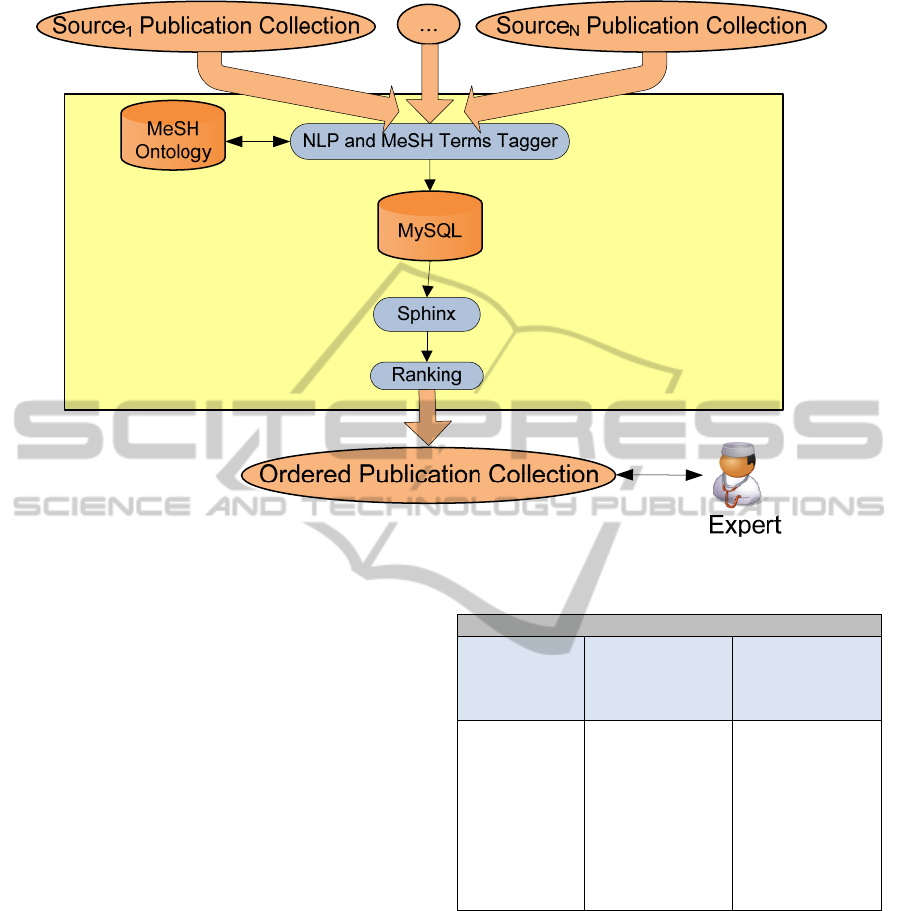
Figure 4: Information Aggregation.
improve the efficiency of the sorting search.
Once the collection is indexed, the service ranks
the publications based on the initial query using
Sphinx. The result is a sorted collection based on the
relevance of each publication respect the initial
query as it is shown in figure 4.
4 USE CASES: DIABETES AND
ARTERIAL HYPERTENSION
Two use cases were developed to test the proposed
method: (i) diabetes and (ii) arterial hypertension. A
knowledge base to analyze the parameters related
with them has been implemented to test the system.
Information needed to evaluate each parameter
received by the patientCharacteristics web service is
stored in different knowledge (one for each
pathology). Structured parameters were selected by
experts within the project using a focus group
methodology. These parameters are shown in table
1.
Depending on the pathology, structured
parameters are selected by its relevance. Due to the
characteristics of the diseases, there are some
parameters that are common to both and others are
specific for each pathology (table 1).
Table 1: Structured data recognized.
Structured parameters
Common
parameters
Diabetes specific
parameters
Arterial
hypertension
specific
parameters
age
race
sex
body_mass_inde
x
diabetic_familiar
neuropathy
basal_glycemia
gestational_diabetes
retinopathy
nephropathy
cardiopathy
glucose_intolerance
glycated_hemoglobi
n
diabetic_foot
hypertensive_familia
r
total_cholesterol
triglycerides
systolic_pressure
diastolic_pressure
HDL
abdominal_wall_size
smoker
cLDL
Finally, for each structured parameter, the Web
Service applies several rules, extracting the related
characteristic. For example, treatment of the
glycated hemoglobin (HbA1c) and related
characteristics are:
HbA1c < 6.5 Æ "normal HbA1c"
HbA1c < 7 Æ "high HbA1c"
HbA1c < 8 Æ "high HbA1c AND glycemic
control"
HbA1c >= 8) Æ "high HbA1c AND
intensive glycemic control"
HEALTHINF 2011 - International Conference on Health Informatics
38

Figure 5: Interface to select the parameters and sources of
the search.
Once all the characteristics are extracted went
through the curation phase, the system constructs a
searching query with the pathology, the relevant
characteristics previously gathered and the MeSH
terms identified with the expert’s input query (with
the searching topic)
With queries close to the mentioned pathologies,
the system searches on expert defined sources using
their corresponding connectors to obtain the most
relevant publications and evidences. Nowadays,
sources covered by the system are PubMed,
Cochrane and MedScape, where the most relevant
publications about diabetes and hypertension are
published.
Finally, the results of each source are integrated,
indexed and presented to the expert as a unique
collection as shown in the following section.
5 RESULTS
Characteristics are extracted according to the
pathology and the parameters introduced. From this
output, the expert will select the most relevant
features to be launched to the search.
A preliminary test set of ten patient data and fifty
queries were used to check the functionality of the
system. The retrieved characteristics were correct for
all the cases. The results correctly identified the 90%
of the relevant papers, according to assessment of
experts of the project.
As it is shown in figure 5, the expert can choose
a more specific term for the search, i.e. optimizing
the query that will be launched against the selected
sources. The expert can also specify the number of
results obtained from the query.
With the optimized query, the system accesses
the different sources and aggregates the results,
filtering those fulfilling the initial requirements and
finally presenting them ordered by relevance, as it is
shown on figure 6.
Two filters were applied to the parameters in
order to extract the most relevant literature referred
to a patient: (i) a transformation of the parameters
into Mesh terms and (ii) an optimization of the
characteristics, using the MeSH ontology and the
medical knowledge of the expert. Through the final
query and the federated search the expert will
retrieve biomedical literature based on patient data.
6 CONCLUSIONS AND FUTURE
LINES
In this work, a new method for search clinical
literature has been proposed and implemented within
the framework of a research project for the
personalization of treatments and therapeutic
strategies. Promising results of the system suggest
that EHR-based literature retrieval may facilitate the
work of physicians obtaining biomedical patient-
related research publications. Also, within the
project, a set of tests with real patients and experts
from the “Hospital Universitario de Valencia” will
be conducted.
As work in progress, extensions of services
described within this paper are being implemented:
further non-structured data processing, clustering
techniques to group the results, open of generic
connectors and source weighting for final ranking.
First, the service will use Natural Language
Processing techniques to process non-structured data
of the patient for characteristic extraction.
Clustering techniques are being used to present
results not only as a sorted collection but also
grouped by their semantic similarity, providing a
more intuitive representation for the navigation.
Regarding information sources, connector
designing process is being generalized to facilitate
new source integration within the system. This
PATIENT-BASED LITERATURE RETRIEVAL AND INTEGRATION - A Use Case for Diabetes and Arterial
Hypertension
39

Figure 6: Aggregated Results.
service will add a weighting functionality for
sources, to adjust the final publication relevance not
only based on the initial query but also considering
the relevance of the source.
The system presented in this work has proven the
suitability of a patient-based literature retrieval
approach. And we are confident that current
extensions will improve the location of relevant
publications for physicians, facilitating more
relevant results when searching at main biomedical
literature search engines.
ACKNOWLEDGEMENTS
The present work has been funded by the Spanish
Ministry of Industry, Tourism and Trade in the
framework of the "Avanza I+D" programme under
the project “Tratamiento 2.0” (project number TSI-
020301-2008-15). We would like to express our
gratitude to the “Hospital Universitario de Valencia”
for their collaboration in the election of the
parameters used in this work.
REFERENCES
Antolik, J. 2005, "Automatic annotation of medical
records", Studies in health technology and
informatics, vol. 116, pp. 817-822.
Atserias, J., Casas, B., Comelles, E., González, M., Padró,
L. & Padró, M. 2006, "FreeLing 1.3: Syntactic and
semantic services in an open-source NLP library",
Proceedings of the 5th International Conference on
Language Resources and Evaluation (LREC’06), pp.
48–55.
Couto, F. M., Silva, M. J., Lee, V., Dimmer, E., Camon,
E., Apweiler, R., Kirsch, H. & Rebholz-Schuhmann,
D. 2006, "GOAnnotator: linking protein GO
annotations to evidence text", Journal of biomedical
discovery and collaboration, vol. 1, pp. 19.
Cunningham, H. 2002, "GATE, a general architecture for
text engineering", Computers and the Humanities, vol.
36, no. 2, pp. 223-254.
HEALTHINF 2011 - International Conference on Health Informatics
40

Díaz-Galiano, M., García-Cumbreras, M., Martín-
Valdivia, M., Montejo-Ráez, A. & Ureña-López, L.
2008, "Integrating MeSH Ontology to Improve
Medical Information Retrieval", Advances in
Multilingual and Multimodal Information Retrieval,
pp. 601-606.
Evidence-Based Medicine Working Group 1992,
"Evidence-based medicine. A new approach to
teaching the practice of medicine", JAMA : the journal
of the American Medical Association, vol. 268, no. 17,
pp. 2420-2425.
Healthcare Information and Management Systems Society
2010 June, Consensus definition of an Electronic
Health Record. Available: http://www.himss.org/
ASP/topics_ehr.asp.
Huang, X., Lin, J. & Demner-Fushman, D. 2006,
"Evaluation of PICO as a knowledge representation
for clinical questions", AMIA ...Annual Symposium
proceedings / AMIA Symposium.AMIA Symposium, ,
pp. 359-363.
Jacso, P. 2004, "Thoughts about federated searching",
Information Today, vol. 21, pp. 17-17.
Kankar, P., Adak, S., Sarkar, A., Murari, K. & Sharma, G.
2002, "MedMeSH summarizer: text mining for gene
clusters", Proceedings of the Second SIAM
International Conference on Data MiningCiteseer, pp.
11.
Karopka, T., Scheel, T., Bansemer, S. & Glass, A. 2004,
"Automatic construction of gene relation networks
using text mining and gene expression data", Medical
informatics and the Internet in medicine, vol. 29, no.
2, pp. 169-183.
Kawamoto, K., Houlihan, C.A., Balas, E.A. & Lobach, D.
F. 2005, "Improving clinical practice using clinical
decision support systems: a systematic review of trials
to identify features critical to success", BMJ (Clinical
research ed.), vol. 330, no. 7494, pp. 765.
Keen, P. G. W. & Morton, M.S.S. 1978, Decision support
systems: an organizational perspective, Addison
Wesley Publishing Company.
Lee, K. F. 1989, Automatic speech recognition: the
development of the SPHINX system, Kluwer
Academic Pub.
Natarajan, K., Stein, D., Jain, S. & Elhadad, N. 2010, "An
analysis of clinical queries in an electronic health
record search utility", International journal of medical
informatics
Seidling, H. M., Schmitt, S. P., Bruckner, T., Kaltschmidt,
J., Pruszydlo, M. G., Senger, C., Bertsche, T., Walter-
Sack, I. & Haefeli, W. E. 2010, "Patient-specific
electronic decision support reduces prescription of
excessive doses", Quality & safety in health care
Sim, I., Gorman, P., Greenes, R. A., Haynes, R. B.,
Kaplan, B., Lehmann, H. & Tang, P.C. 2001, "Clinical
decision support systems for the practice of evidence-
based medicine", Journal of the American Medical
Informatics Association : JAMIA, vol. 8, no. 6, pp.
527-534.
Spackman, K. A. & Reynoso, G. 2004, "Examining
SNOMED from the perspective of formal ontological
principles: Some preliminary analysis and
observations", KR-MEDCiteseer, pp. 81.
Srinivasan, P. 2001, "MeSHmap: a text mining tool for
MEDLINE", Proceedings / AMIA ...Annual
Symposium.AMIA Symposium, pp. 642-646.
Tratamiento 2.0 2010 June, Available:
http://www.tratamiento20.com.
van der Weijden, T., Legare, F., Boivin, A., Burgers, J. S.,
van Veenendaal, H., Stiggelbout, A. M., Faber, M. &
Elwyn, G. 2010, "How to integrate individual patient
values and preferences in clinical practice guidelines?
A research protocol", Implementation science : IS, vol.
5, pp. 10.
Weaver, C. A., Warren, J. J., Delaney, C., International
Medical Informatics Association, Nursing Informatics
Special Interest Group (IMIA-NI) & Evidence-Based
Practice Working Group 2005, "Bedside, classroom
and bench: collaborative strategies to generate
evidence-based knowledge for nursing practice",
International journal of medical informatics, vol. 74,
no. 11-12, pp. 989-999.
PATIENT-BASED LITERATURE RETRIEVAL AND INTEGRATION - A Use Case for Diabetes and Arterial
Hypertension
41
