
Automatic Determining of Vertebrae from CT Images
B. A. Zalesky, A. M. Nedzved, S. V. Ablameiko and P. V. Lukashevich
United Institute of Informatics Problems of the National Academy of Sciences of Belarus
Surganova 6, Minsk, Belarus
Abstract. Position of the spinal column and vertebrae on the CT images is one
of the main features to determine position of a patient and his organs. In this
paper we propose the algorithm to extract the spinal column and vertebrae with
ribs from CT images and to estimate their coordinates as well as coordinates of
human body in the coordinate system of a CT scanner.
1 Introduction
In recent decades remarkable improvements in diagnostic managements of oncology
patients have been achieved. We became witnesses of a technological breakthrough
in the field of diagnostic imaging, including the computerized tomography (CT),
magnetic resonance imaging (MRI), angiography etc.
Over the past decade, the increasing technological sophistication of CT equipment
has permitted excellent images with a limited dose of radiation which is currently
equal to a plain chest X-ray (0,001 Gy). Modern CT scanners were equipped with
special programs in order to save the information in DICOM files, to analyze images
and to provide interactive measurements. The programs disclose internal object struc-
tures and compute a great number of organ parameters. They make easier interactive
measurements of organ size and volume and identification of the character of the
pathological mass.
At present there are a few special systems and complexes for the automatic diag-
nosis in medicine using imaging techniques. These systems, in general, belong to the
diagnostic management of brain pathology. Image technologies also play the key role
in the diagnosis of Pediatric malignancies. CT imaging more than any other imaging
modality provides documentation of the tumor areas, and topography characteristics.
It gives together with additional intravenous infusion of contrast (CT-angiography)
extra data about the blood vessels of the tumor and surrounding tissue.
Now medical experts use vertebrae and ribs to figure out disposition of regions and
organs of human body. The tasks of extraction of spine column, distinguishing and
counting of vertebrae on CT images are usually carried out by medical specialists
manually. This routing work takes significant time and efforts. Algorithms, which
solve or even partly solve mentioned problems, seriously facilitate the work with CT
datasets having from several dozen to hundreds images. A dataset of CT images of a
patient is stored as 2D grayscale scans of body axial section written in the DICOM
format.
A. Zalesky B., M. Nedzved A., V. Ablameiko S. and V. Lukashevich P. (2010).
Automatic Determining of Vertebrae from CT Images.
In Proceedings of the Third International Workshop on Image Mining Theory and Applications, pages 85-91
DOI: 10.5220/0002963000850091
Copyright
c
SciTePress

Medical experts very often need localization of parts of human body and human
organs shown in 2D scans. For this purpose numbering of vertebrae is traditionally
used.
We offer an approach to extract the spine column from CT images and distinguish
vertebrae. It enables automatic segmentation of spine column region and automatic or
semiautomatic separation of vertebrae. We offer our solution in the traditional deci-
sion making form as a prompt for the medical expert who can easily correct the result
of automatic separation of vertebrae.
2 Formulation of Problem
Usually a dataset of CT images of one patient consists of 20÷100 scans. Distance
between scans can vary from 0.1 to 1 centimeter. All images are stored as files in the
DICOM format. Each DICOM file contains 3D coordinates of the top left corner of
the 2D scan image relative to the coordinate system of the CT scanner.
One of difficulties for experts to work with the CT dataset consists in absence of
interconnection of successive numbers of files and their real 3D coordinates. A CT
scan can have arbitrary Z-coordinate relative to the human body Therefore, 2D scans
should be ordered in regard to their space disposition before one begins to segment
human organs.
DICOM pictures are represented as 16-bit grayscale images. Brightness of
DICOM images of 2D CT scans corresponds to the Hounsfield units that characterize
organ densities. Unfortunately, it is not referred to topograms. An appropriate trans-
formation of DICOM images, especially topograms, into the standard 8-bit gray scale
ones can help to get maximum accuracy of further steps. All images are provided
with their 3D coordinates according to the coordinate system of the CT scanner.
Our task is to estimate position of vertebrae and vertebra in a 2D scan. The main
steps of the offered decision rely on finding characteristic vertebra for comparing
different investigation of one patient.
3 Vertebra Segmentation
Vertebra segmentation from CT pictures is an urgent task for many medical applica-
tions. It is widely used to control dynamic of conditions of the spine, to recognize its
deceases and to treat them. Besides, this procedure is an intermediate step for seg-
mentation of abdominal organs, such as the liver, kidneys, and spleen, from CT scan
imagery [1]. Among semiautomatic and automatic approaches to the problem are
model-based, discrete optimization, neural network, active contours, morphologic
methods or their combinations [2-4] with or without of use a prior information etc.
One of the difficulties of vertebra segmentation, often mentioned by authors of algo-
rithms, consists in discrimination between the spine and ribs.
We propose a new automatic algorithm, which allows both: reliable automatic de-
tection of the vertebrae on 2D CT images in DICOM format, and separation of ribs
touching the spine. Also, detection of vertebrae on 256-color gray scale CT images is
86
possible, as well. The algorithm does not need learning. It contains five main steps:
image preprocessing, finding body section, reliable detection of round part of the
vertebra, accurate segmentation of the bone, which image can break up into several
parts and be touched by ribs, and discrimination between the spine and ribs.
Image preprocessing is performed by version of the region growing algorithm
leaving only the largest connected component of the CT image, which always is the
body section, and removing all other artifacts and noisy clusters. After, the region of
interest (ROI) is found as the bounding box of the body section.
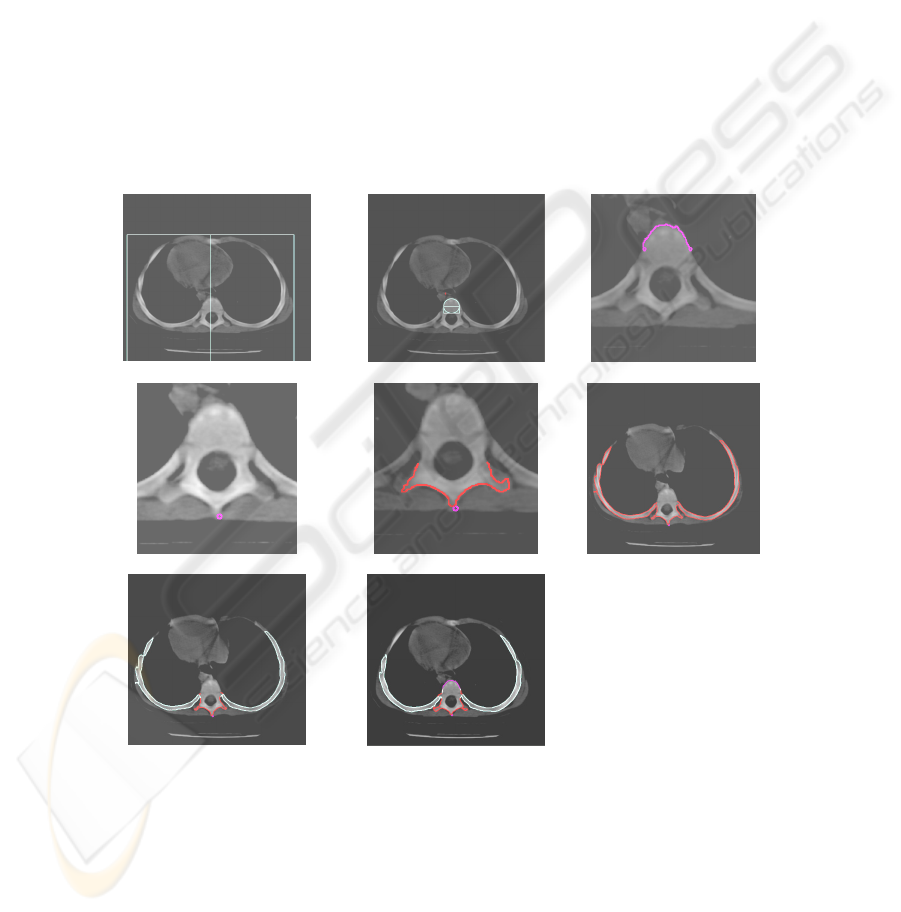
Simple prior information on round shape of a vertebra frontal is used to detect its
location in the image. The shape of matching window is drawn in Fig.1b. Despite of
the simple form of prior information practically all vertebrae were detected by the
chosen window.
Then, top boundary of the vertebrae is determined (Fig.1c). Further outlining of
the vertebra boundary is started from the lowest pixel of the bone (Fig.1d), since 2D
slice CT image of this one piece bone can contain different number of separate piec-
es.
a
b
c
d
e
f
g
h
Fig.1. Steps of performance of the algorithm.
The lower boundary of the vertebrae is found after binaryzation of the CT image
at values 1000 -1200 of Hounsfield units corresponding bones of human body. It
allows outlining bottom part of the vertebra, possibly, with touching ribs. Outlined
87

part of the vertebra without touching rigs is depicted in Fig.1e, and with them – in
Fig.1f.
The algorithm analyzes bottom part of the outlined boundary and recognizes
whether it contains rib frontiers.
In case of recognized touching ribs the algorithm estimates their contact points
with the vertebrae and removes rib frontiers from the bottom boundary. Removed rib
frontiers are shown in Fig. 1g by aquamarine color. The final segmented image with
removes ribs colored in aquamarine can be seen in Fig.1h.
This algorithm was tested by CT-images of children and included in software for
monitoring of mediastinal and retroperitoneal tumors. It is allow to detect description
of organs position by vertebrae geometrical coordinates. This information is very
usefulness for extraction 3D patterns of organs by module-based segmentation algo-
rithms.
4 Definition of Vertebra Orientation
One of the basic feature of analysis CT Images is a topology of organs in body. For
quality topology analysis It is necessary to define body orientation. For solving this
task the algorithm of orientation definition was developed.
Orientation is determined on bone elements and the vertebra by using threshold
segmentation. For the CT image histogram of brightness is divided into three gauss-
like shape classes. On their position we can determine the location of the bodies
(fig.2).
Fig. 2. Histogram of brightness for the CT images.
Position of the last local minimum is determined by the watershed thresholding me-
thod with smoothing of the histogram. Bone fragments carved this thresholding.
However, there is a geometric noise on binary image. These noises correspond to the
soft tissue and acquisition errors. Removal of such errors is performed through sever-
al steps. The first step is to remove small noise, which is performed by analyzing the
size of objects and thin clamped to the edge of objects. This removal is realized by
analyzing the image boundaries (Fig.3).
88
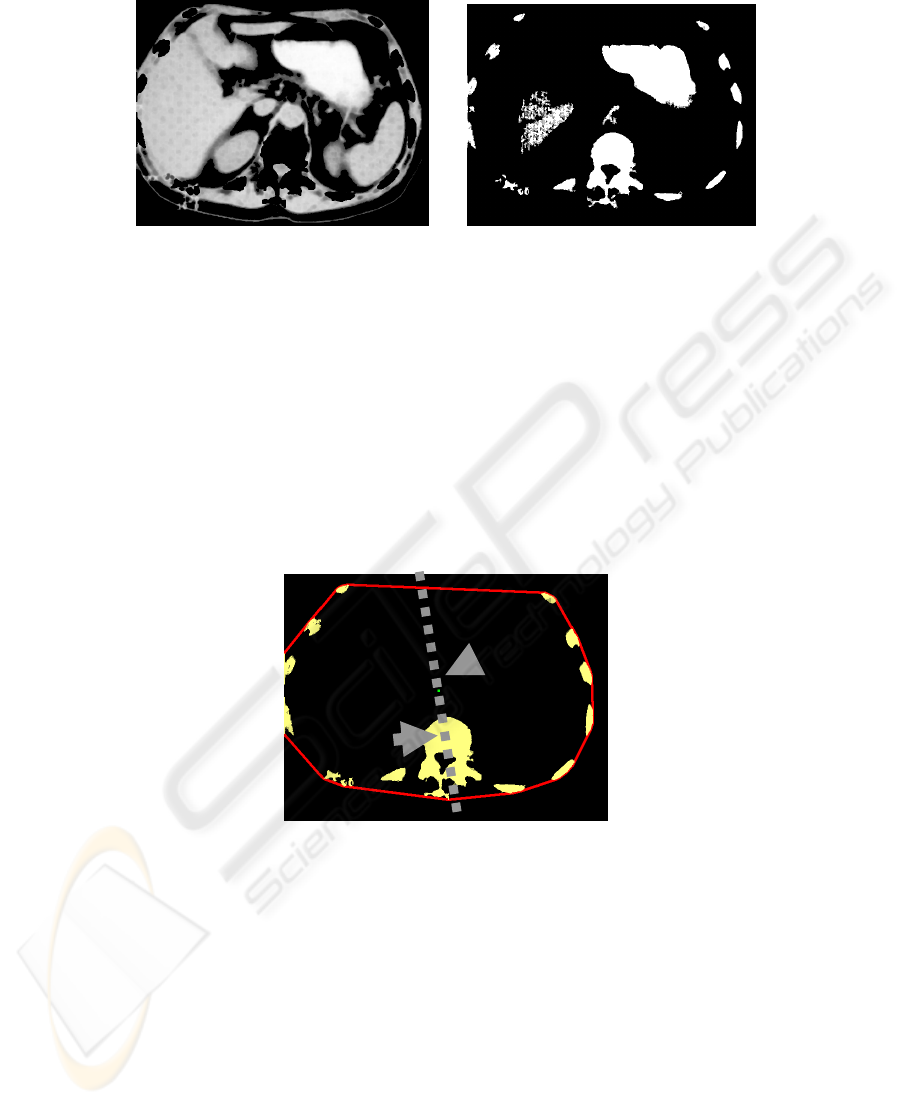
Fig. 3. Source CT image and Image with remote small object.
Spatial position of bone on the slice is used for definition of bone regions. Practically
all bone elements are placed along the edges. So the overall operation with convex
contour on all elements of the image is used for them to determine. Objects are saved,
if they cross the convex contour.
To determine the remaining elements of the bone dilatation increases convex con-
tour. Analysis of the factor of shape for all elements of the bone tissue determines the
element that corresponding to the vertebra. Then the morphological open operation is
performed with a high depth, which allows to get a round object. Center of gravity is
calculated as a binding element.
Another element of the anchor is the center of gravity for the convex object.
Based on these coordinates calculated line corresponding to the slice orientation
(Fig.4)
Fig. 4. Images from the orientation axis, the arrows indicate the centers of mass of a convex
region and the vertebra.
Further detection of organs regions is carried out based on the watershed. On the
basis of the subtraction of very smoothed watershed lines from the original image and
regions binarization separate organs are defined.
Such analysis allow to define topology properties organs and body that can be
used for monitoring and diagnostic task.
89

Fig. 5. Regions corresponding to different organs.
5 Conclusions
The presented algorithms allows automatic and semiautomatic extraction of the spine
column and thoracic vertebrae from datasets of CT images stored in the medical
DICOM format. It enables of reliable extraction and separation of thoracic vertebrae
in order to number them relative to the bottom human vertebra with ribs.
In turn, it gives possibility for the medical expert to know the number of the vertebra
he sees in the current 2D scan image.
A new unsupervised algorithm to segment vertebrae in 2D CT images has been pre-
sented. It allows reliable finding this bone and its automatic extraction. The algorithm
does not use prior information on the spine shape. The results of tests showed possi-
bility of automatic extraction of vertebra from CT images. In order to be applicable to
practical extraction the algorithm needs following feasible improvements in order to:
process correctly images, which do not contain the vertebra; process in special way
CT images of patients that took contrast agents; test shapes of outlines contours.
The drawback of the algorithm is its real applicability to separate and number only
thoracic but not lumbar and sacral vertebrae.
Acknowledgements
This work was supported by ISTC project #B-1489.
References
1. Serlie I. W. O., Vos F.M., Truyen R., Post, F.H. van Vliet, L.J., Classifying CT Image Data
Into Material Fractions by a Scale and Rotation Invariant Edge Model, IP(16), No. 12, De-
cember 2007, pp. 2891-2904.
2. Wang L. I., Greenspan M., Ellis, R.E., Validation of bone segmentation and improved 3-D
registration using contour coherency in CT data, MedImg(25), No. 3, March 2006, pp. 324-
334.
90

3. Seise M., McKenna S.J., Ricketts I.W., Wigderowitz C.A., Learning Active Shape Models
for Bifurcating Contours, MedImg(26), No. 5, May 2007, pp. 666-677.
4. Yao W., Abolmaesumi P., Greenspan M., Ellis R.E., An Estimation/Correction Algorithm
for Detecting Bone Edges in CT Images, MedImg(24), No. 8, August 2005, pp. 997-1010.
5. Jackowski M. and Goshtasby A., "A computer-aided design system for segmentation of vo-
lumetric images," in Handbook of Biomedical Image Analysis, Volume III: Registration
Models, J. S. Suri, D. L. Wilson, and S. Laxminarayan, Kluwer Academic, New York,
2005, pp. 251-272
6. Zagorchev L., Goshtasby A., Satter M., Bernstein T. Segmentation and Registration of Spi-
nal MR and CT Images ( http://www.cs.wright.edu/~agoshtas/wkni_spine.html)
91
