
A MOLECULAR CONCEPT OF MANAGING DATA
Christoph Schommer
Department of Computer Science, University Luxembourg, 6 Coudenhove-Kalergi, 1359 Luxembourg, Luxembourg
Keywords:
Artificial life, Relational data management, Pattern recognition, Bio-inspired computing.
Abstract:
The following (position) paper follows the concept of the field of Artificial Life and argues that the (relational)
management of data can be understood as a chemical model. Whereas each data itself is consistent with atomic
entities, each combination of data corresponds to a (artificial) molecular structure. For example, an attribute
D inside a relational system can be represented by a nucleus α
D
sharing a cloud of values, which consists
of so-called valectrons (the values for the column D). By using reaction rules like the selection of tuples or
projection of attributes, a retrieve of molecules can be achieved quite easily. Advantages of the chemical model
are no data types, a fast data access, and the associative nature of the molecules: this automatically supports a
direct identification of patterns in the sense of data mining. A disadvantage is the need for restructuring that
must eventually be done, because the incoming data stream is allowed to influence the chemical model. With
this position paper, we present our basic concept.
1 INTRODUCTION
Since more than 30 years, relational database sys-
tems are successful, being the most important data
management system worldwide. Based on the theory
on sets, a relational database system takes advantage
from the concepts of the relational algebra, which has
led – among other functionalities – to today’s stan-
dard query language SQL. Although in recent past,
some alternative database architectures have been de-
veloped (object-oriented, object-relational, and XML
databases, etc.), they have never received the desired
breakthrough. And although the relational systems
suffer from an efficient management (often, the more
complex the system is the more time and capacity is
needed to guarantee the data consistency), the rela-
tional architecture still proves reliability, consistency,
and precision.
With this position paper, we foster on a com-
pletely different approach of data management and
try to figure out that data is unlike a data value inside
a well-structured environment but even more a fluid
(dynamic) and molecular concern. With respect to
the natural example, we understand data as an atomic
structure and combinations of data as a molecule –
invoked on the field of Artificial chemistry (Dittrich
et al., 2001). Commonly, Artificial chemistry is un-
derstood as a theoretical model following the natural
example, which is used to simulate types of systems
in the spirit of chemical reactions (Leach, 2001). It
originates in the field of Artificial Life (Kelemen and
Sos
´
ık, 2001) and has proven to be a manifold and
powerful pathway of modeling (Skusa et al., 2000),
(Ziegler and Banzhaf, 2001), (Schommer, 2009).
In general, an artificial chemistry is defined as
a triple (M, R, A), where M refers to the set of
molecules {m
1
. . . ,m
n
}, which is possibly of infinite
size, R to the a set of n-ary operations/reaction rules
{r
1
. . . ,r
n
} on the molecules, and A, which denotes an
algorithm describing how to apply the rules R to a
subset P⊂M. Each reaction rule r
i
∈ R is written as a
chemical reaction like
(x
1
, x
2
) →
r
i
x
∗
1
x
∗
2
With that, we firstly introduce the molecular
model, present several reaction rules to explain its
depth, and demonstrate its strength on an example.
2 A SET OF MOLECULES
We understand each attribute D
i
inside a database ta-
ble D as a nucleus α
i
that owns a cloud of values
e
1
, . . . e
k
at distance ε
i
. Each e
i
corresponds to a data
411
Schommer C. (2010).
A MOLECULAR CONCEPT OF MANAGING DATA.
In Proceedings of the 2nd International Conference on Agents and Artificial Intelligence - Artificial Intelligence, pages 411-415
DOI: 10.5220/0002758304110415
Copyright
c
SciTePress
value d
i
∈ D
i
that might be e.g. of type string or num-
ber (integer, real, dots), but not a list of values (first
normal form is valid). A nucleus α
i
owns a name (=
the attribute name) and shares a higher valency ν
D
i
,
the more dense the cloud of values is. The distance
between the nucleus α
i
and each valectron e
i
gives the
strength of existence, meaning that if the occurrence
of e
i
increases the occurrence of e
j
, the distance to
the nucleus is shorter. If the nucleus owns only one
e
i
, then α
i
= e
i
.
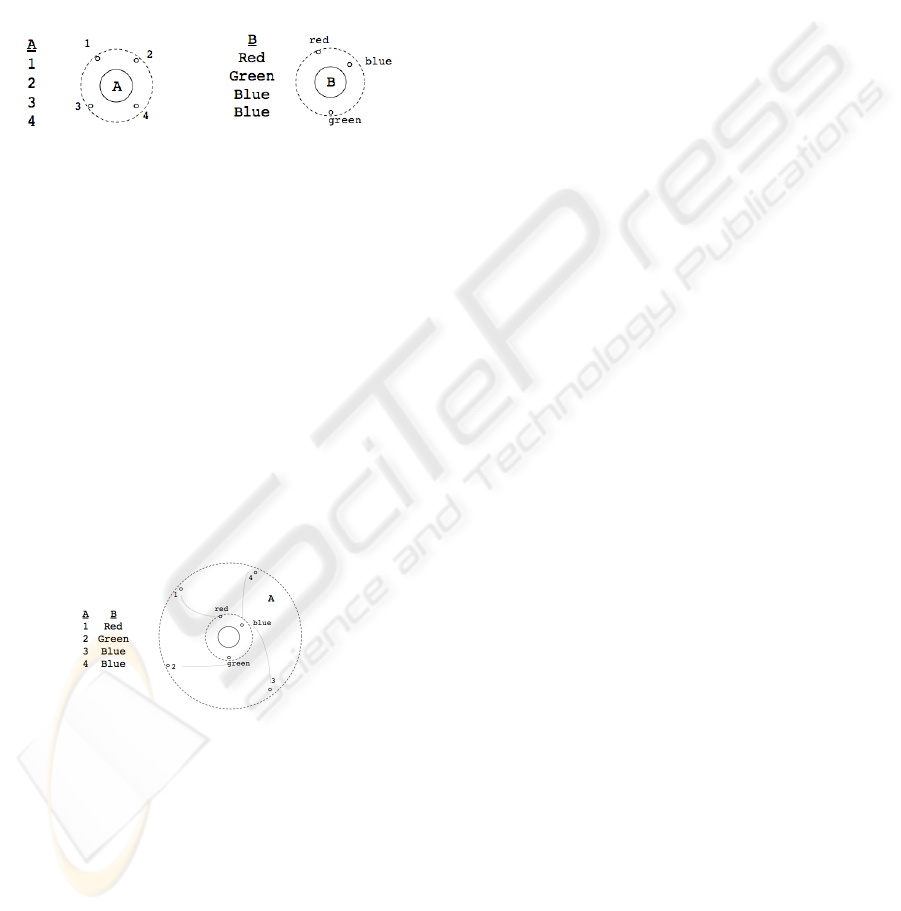
a) b)
Figure 1: Database attributes D
A
and D
B
with the corre-
sponding nuclei α
A
and α
B
.
Figure 1 presents two database attributes D
A
and D
B
with the corresponding nuclei α
A
and α
B
.
Whereas all valectrons of nucleus α
A
shares the same
distance, the distance of e
blue
of nucleus α
B
is shorter
than for e
green
and e
red
.
In opposite to its atomic basis, a database table of
≥ 2 attributes D
1
, . . . , D
k
is consequently a set of nu-
clei α
D
1
, . . . , α
D
k
. The nuclei, however, are not organ-
ised in an arbitrary way, but keep themselves ordered:
• The lower the valency ν
D
i
is the more centric the
nucleus α
i
will be.
• In case that some nuclei share the same valency,
we may randomly select one of them.
Figure 2: Simulation of two database attributes D
A
and
D
B
of the database table D with the corresponding three-
dimensional molecule m
A,B
(ordered by their valency).
The principle of ordering is unlike the ordering
of a set of numbers but more the arrangement of
the nucleus including their cloud of values. With
respect to this, a chemical structure of size ≥ 2 –
which is said as to be a molecule m∈M – therefore
can not be a two-dimensional model anymore: the
cloud of values embraces each previously selected nu-
cleus and associates each valectron e
i
with its cor-
responding partner of the other nucleus. Figure 2
shows a simulation of two database attributes D
A
and
D
B
of the database table D with the corresponding
three-dimensional molecule m
A,B
(ordered by their
valency). As presented in Figure 2, the merge of nu-
clei is as follows:
• Assume that ν
D
i
< ν
D
i+1
< . . . ν
D
k
, then ν
D
i
has
the highest priority and therefore takes over the
innermost position, followed by ν
D
i+1
, and so on.
• The nuclei α
i
, . . . α
k
are nested and represented by
their cloud of values only.
• Originally associated tuples inside D =
{D
i
, . . . D
k
} are connected by molecular bridges
γ
i,k
of a certain strength, which may vary.
3 ENZYMATIC REACTIONS
An artificial enzyme is a protein that is able to execute
reactions. Whereas in the natural example an enzyme
takes over the responsibility of many functions that
concern the metabolism of an individual, the simula-
tion of enzyme in a database environment can be un-
derstood as the adequate to reaction rules. Enzymatic
reactions work in one or two ways
• the targeted nuclei α
i
can be copied.
• the molecular bridges γ
i,k
between the valectrons
may be destroyed.
but the enzymatic instruction decides if both or
only the latter action takes place. For example, an
enzyme that simply has to read existing molecules
surely copies the existent structure and then keeps
only those connections that satisfy the enzymatic in-
struction. On the other side, a permanent delete of
data in the original molecule does not afford a copy
but only the delete of the molecular bridges.
With respect to a retrieval, fundamental enyzmes
concern the selection and projection enzyme. Given
a molecule as presented in Figure 2, then the reac-
tion rule σ
A,B
characterizes a chemical reaction of the
original molecule – which consists of the two cloud
of values A and B – to another molecule A
∗
B
∗
. The
density and the valency change, since for example
ν
A,B
> ν
A
∗
B
∗
of the new molecule:
A, B →
σ
A
∗
B
∗
Similarly, the reaction rule π characterizes a
chemical reaction as well, but in contrast to the reac-
tion σ, the valency remains stable, whereas the num-
ber of resulting nuclei α
i
changes:
A, B →
π
A
∗
B
∗
ICAART 2010 - 2nd International Conference on Agents and Artificial Intelligence
412
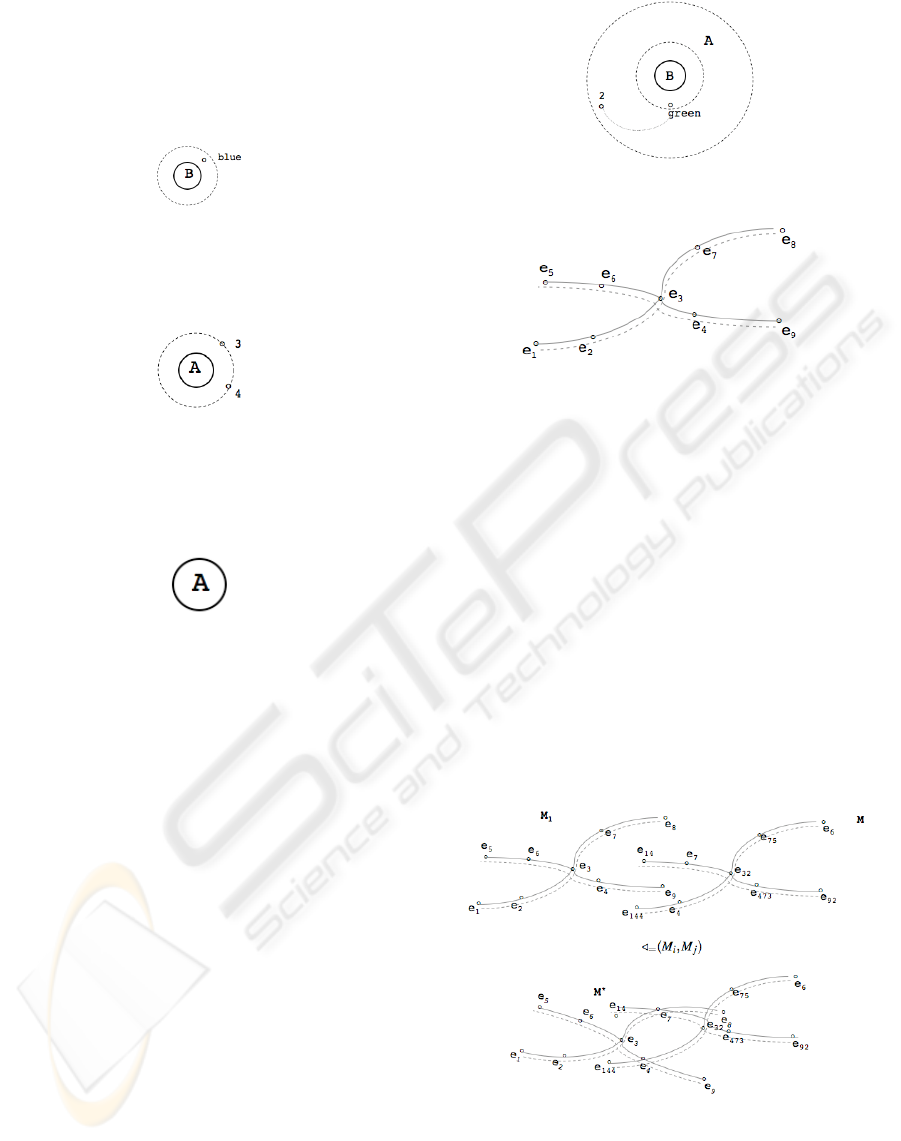
As an example, Figure 3 shows an enzymatic reac-
tion π
B
(σ
B=‘blue
0
(D)), where the original molecule is
copied and all molecular bridges and valectrons (ex-
cept ‘blue’) are removed. Please note that the distance
of ε
blue
remains unchanged, i.e., the valectron remains
at the same position.
Figure 3: Examples of the Reactor Rule σ: select B from D
where B=‘Blue’
Similarly, Figure 4 shows an enzymatic reaction
π
A
(σ
A>2
(D)), where two valectrons occur.
Figure 4: Examples of the Reactor Rule σ: select A fom D
where A > 2
The enzymatic reaction π
A
(σ
A>4
(D)) given in
Figure 5, however, gives only the nucleus α
A
, but no
valetrons.
Figure 5: Examples of the Reactor Rule σ: select A from D
where A > 4
Finally, Figure 6 shows an enzymatic reaction
π
A,B
(σ
A=2
(D)) where both nuclei α
A
and α
B
occur
and the molecular bridge γ
‘green
0
,2
between the corre-
sponding valectrons still exist.
With an enzymatic reaction ∈
m,M
, the insert of
a new molecule m into an existing molecular struc-
ture M takes place by a simple addition. In case that
valectrons are already present, these become merged.
As an example, Figure 7 shows the merge of two
molecules where the valectron e
3
is common. How-
ever, such an insert may violate the correctness of
the existing data landscape, because it allows the cre-
ation of molecules that do not exist. The insert of the
molecule
∈
(e
5
,e
6
,e
3
,e
4
,e
9
),M
seems to be safe, but the existence of another
molecule (e
1
, e
2
, e
3
, e
7
, e
8
) causes an error, since in-
herently the molecules (e
5
, e
6
, e
3
, e
7
, e
8
) and (e
1
, e
2
,
e
3
, e
4
, e
9
) might untruly be present as well. By using
just one molecular bridge γ
i, j
, we therefore risk the
inconsistency of the whole molecule.
An insert, and moreover the presence of collec-
tions of valectrons must not have a single molecular
bridge but a double one. With this, the dashed bridge
Figure 6: Examples of the Reactor Rule σ: select B from D
where A = 2
Figure 7: The reaction rule ∈ (m, M): a molecular bridge
(dashed) characterizes the connections of the valectrons (=
the β-helix β
e
i
...e
e
j
) whereas the solid line the situation of
the molecule after insertion (γ-helix).
characterizes the connections of the valectrons. This
called the β-helix β
e
i
...e
e
j
. The solid line the situa-
tion of the molecule while insertion, representing val-
ues between the associated nuclei α
i
. The molecular
string is therefore called the α-helix. And with that,
a molecule (e
5
, e
6
, e
3
, e
7
, e
8
) does not exist since no
α-helix is from e
5
– e
6
– e
3
to e
7
.
As a third operation, an (equi-)join operation of
molecules may be represented by the reaction rule
/
=
(M
i
, M
j
). As for the insert reaction ∈, those valec-
trons, which occur both in molecule M
i
and in M
j
,
are merged. All original valectrons keep their helix
structure (see Figure 8).
Figure 8: Reaction Rule /
=
(M
i
, M
j
)
With an enzymatic reaction 6∈
m,M
, we denote the
delete of a molecule m within M. The helix β
e
i
...e
e
j
guarantees that only those valectrons, which belong
together, are deleted.
A MOLECULAR CONCEPT OF MANAGING DATA
413

In addition, the composition of several reaction
rules like
A, B →
σ,π
A
∗
B
∗
is possible and appears in that order the reaction
rules are given. The composition is commutative.
Beside the given reaction rule, the authorization
of valectrons might be interesting as well. With au-
thorization, we identify the an enzyme’s right to ac-
cess to a nuclei and it’s cloud of values. This is not
really a reaction rule as the enzymatic reaction does
not results in a chemical reaction; it is more a feature
of the nuclei itself that allows or disallows a permitted
access. We therefore note a disallowed access by
¬α
A
meaning that the nucleus α
A
rejects any kind of
reaction. Instead of delivering a valectron, the result
could be an empty element.
4 DISCUSSION
The idea of understanding data within an artificial
chemical system is potentially unlike the relational
system but offers a variety of characteristics. First,
no data type specification is needed. The presence
of a data item within the chemical database model is
per se self-explaining and does not need any further
specification concerning its type. The consequence
then is that data (of different data type – from a rela-
tional point of view) is being identical. This is not of
disadvantage because the expression of strength be-
tween valectrons through the molecular bridges γ
i, j
is very present. In fact, this is the second point as
strong relationships among valectrons do inherently
exist. If a combination of valectrons e
i
− e
j
occurs
often enough, then its molecular bridge γ
i, j
becomes
stronger as if it occurs only “a few times”. Third,
the consideration of the molecular model towards a
molecular-associative construct offers the identifica-
tion of molecular clumps that are connected with each
other and that represent a symbol, such that they may
form a higher-related (cognitive) construct like a men-
tal image or simply a thought. Assuming, that “tree”
(for nucleus α
1
), “green” (for nucleus α
2
), and “rain”
(for nucleus α
3
) exist, it would certainly be possible
to think of a “staying in the forest on a cold and rainy
day”. As a last point, the molecular data manage-
ment model as described above is open for the input
of data streams. Whereas the relational model lacks
from high administrative efforts, a stream of data may
be handled more effective in the proposed model.
⇒
Figure 9: Restructuring the molecule: the left molecule
refers to the situation where the number of years (α
C
) is
significantly less than the colour (α
A
) and the amount of
(α
B
), whereas the right molecule refers to the more stable
molecule.
On the other side, some kind of efforts is to be
done in keeping the molecules in a stable and con-
sistent form. Stability refers to a general claim that
such nuclei α
i
with a minor valency ν
i
do more con-
tribute to a general model consistency and therefore
to the stability as well as those nuclei with a more
densecloud of values. In consequence of a delete or
an insert of molecules, a restructuring reaction must
take place in order to guarantee stability and con-
sistency. With respect to this, assume that an insert
of a new data leads to a change of the valency with
ν
D
i
> ν
D
i+1
< . . . ν
D
k
. Then, the enzymatic restructur-
ing ψ is as follows:
• A copy α
0
i
of the nuclei α
i
is created; it is then set
on its new place, depending on its valency ν
D
i
.
• All valectrons e
i
of α
i
walk on the β-helix β
i
and
finally reach their cloud of values.
• At each point, a connection of each valectron re-
mains.
On the other side, a continuous change of the num-
ber of values may become counter-productive and fi-
nally refer to a continuous and repeating restructur-
ing of the molecule, such that nuclei are more con-
cerned with internal configurations than with the man-
agement of data. An alternative therefore is to prefer
those nuclei whose cloud of values do not or even less
changes in size. Once the molecule is created (first
approach) and once a certain information about stable
nuclei have been got, the second solution seems to be
more appropriate.
5 CONCLUSIONS
With the presented proposition, we follow the con-
cept of understanding data and information as an (ar-
tificial) chemical model. Each data is consistent with
ICAART 2010 - 2nd International Conference on Agents and Artificial Intelligence
414

an atomic entity but each combination of data corre-
sponds to a molecular structure. An attribute D in-
side a relational system is represented as a nucleus
α
D
sharing a cloud of values, which consists of so-
called valectrons. The nucleus satisfies the first nor-
mal form (atomic values). By using reaction rules like
the selection of tuples σ or projection of attributes π,
a retrieve of molecules can be achieved quite easily.
As mentioned in chapter 4, one of the major advan-
tages is the associative nature of the molecule. The
generation of mental images (or thoughts), beside the
implementation of the given system, will then be next
steps.
ACKNOWLEDGEMENTS
This work is currently been done within the Interna-
tional Laboratory for Intelligent and Adaptive Sys-
tems of the University of Luxembourg. We thank the
members of the MINE research group for their sup-
port.
REFERENCES
Dittrich, P., Ziegler, J., and Banzhaf, W. (2001). Artificial
chemistries-a review. Artificial Life, 7(3):225–275.
Fern
´
andez-Baiz
´
an, M. C., Garc
´
ıa, A., Gonz
´
alez, M. M.,
P
´
erez-Llera, C., Portaencasa, R., and Santos, E.
(1996). Analysis and design of a relational database
management system and implementation of its nu-
cleus. Computers and Artificial Intelligence, 15(4).
Gerrilsan, R. (1975). The application of artificial intelli-
gence of data base management. In IJCAI, pages 521–
527.
Hutton, T. J. (2002). Evolvable self-replicating molecules in
an artificial chemistry. Artificial Life, 8(4):341–356.
Kelemen, J. and Sos
´
ık, P., editors (2001). Advances in Ar-
tificial Life, 6th European Conference, ECAL 2001,
Prague, Czech Republic, September 10-14, 2001, Pro-
ceedings, volume 2159 of Lecture Notes in Computer
Science. Springer.
Leach, A. (2001). Molecular Modelling - Principles and
Applications. Prentice Hall, 2nd edition.
Schommer, C. (2009). An artificial molecular model to fos-
ter communities. In Knowledge Discovery and Infor-
mation Retrieval (KDIR). IEEE Computer Society.
Skusa, A., Banzhaf, W., Busch, J., Dittrich, P., and Ziegler,
J. (2000). K
¨
unstliche chemie. KI, 14(1):12–19.
Tominaga, K., Watanabe, T., Kobayashi, K., Nakamura, M.,
Kishi, K., and Kazuno, M. (2007). Modeling molec-
ular computing systems by an artificial chemistry -
its expressive power and application. Artificial Life,
13(3):223–247.
von Luck, K. and Marburger, H., editors (1994). Man-
agement and Processing of Complex Data Structures,
Third Workshop on Information Systems and Artifi-
cial Intelligence, Hamburg, Germany, February 28 -
March 2, 1994, Proceedings, volume 777 of Lecture
Notes in Computer Science. Springer.
Ziegler, J. and Banzhaf, W. (2001). Evolving control
metabolisms for a robot. Artificial Life, 7(2):171–190.
A MOLECULAR CONCEPT OF MANAGING DATA
415
