
BIOSIG
Standardization and Quality Control in Biomedical Signal Processing using the
BioSig Project
A. Schlögl, C. Vidaurre
Fraunhofer FIRST-IDA, Kekulestrasse 7, 12489, Berlin, Germany
Ernst Hofer, Thomas Wiener
Institute of Biophysics, Medical University of Graz, Harrachgasse 21, 8010, Graz, Austria
Clemens Brunner, Reinhold Scherer
Institute for Knowledge Discovery, University of Technology Graz, 8010, Graz, Austria
Franco Chiarugi
Institute of Computer Science (ICS), Foundation for Research and Technology - Hellas (FORTH) P.O. Box 1385, Vassilika
Vouton, GR 711 10 Heraklion, Crete, Greece
Keywords: Standardization, Quality control, electroencephalogram, Brain Computer Interfaces, Free/libre open source
software (FLOSS).
Abstract: Biomedical signal processing is an important but underestimated area of medical informatics. In order to
overcome this limitation, the open source software library BioSig has been established. The tools can be
used to compare the recordings of different equipment providers, it provides validated methods for artifact
processing and supports over 40 different data formats (more than any other software in this area). BioSig
provides reference implementations for biomedical signal processing questions and holds the top rank
among all biomedical signal processing projects registered at SourceForge. Thus is provides standardization
and quality control for the field of biomedical signal processing.
1 INTRODUCTION
Biomedical signal processing is an important area of
medical informatics and is used in many subject area
(neurology, psychiatry, cardiology, pulmology,
cognitive neuroscience, psychology, biophysics,
biomedical engineer etc) with many different
applications. Unlike other areas of medical
informatics (Imaging, labor diagnostics, patient
information system etc.), biomedical signals are not
well represented in health information systems.
A likely explanation is the fact that many
different disciplines and many small groups do
biomedical signal processing. The interaction
between the various groups is not well organized;
often the same well-known methods are
implemented again and again, the wheel is re-
invented again and again. In order to overcome this
problem, the open source software project BioSig
was founded with the aim to provide a software
library for biomedical signal processing. Motivated
by the successful development model of the Linux
operating system, it was decided that the library
should be also open source, everyone is invited to
use and to contribute to BioSig, and the GNU
General Purpose License (GPL) ensures that BioSig
will stay open source.
Section 2, presents several subprojects that have
been developed within BioSig. Section 3 provides
some numbers about the success of BioSig, Sections
4 and 5 discuss open issues summarize the project.
403
Schlögl A., Vidaurre C., Hofer E., Wiener T., Brunner C., Scherer R. and Chiarugi F. (2008).
BIOSIG - Standardization and Quality Control in Biomedical Signal Processing using the BioSig Project.
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 403-409
DOI: 10.5220/0001065904030409
Copyright
c
SciTePress

2 METHOD
Software development and programming is an
important aspect of biomedical signal processing. In
order to address different needs, several
programming languages are supported. The Matlab
scripting language is widely used in biomedical
signal processing and engineering. For this reason,
the first part of BioSig has been implemented in
Matlab language. Matlab is an proprietary software
product from “The Mathworks Inc”, short TMW, but
there are also some open source alternatives
available; Octave (http://www.octave.org) is
probably the most widely known alternative. Special
effort was undertaken to make the code also
compatible to Octave. This part is now call “BioSig
for Octave and Matlab”, or short “BioSig4OctMat”.
Moreover, C/C++ is a very flexible
programming language and provides a very efficient
(i.e. fast) software; although the software
development takes more time. There is now a
common C/C++ interface to access various data
formats including the SCP-ECG standard (EN1064),
the HL7aECG, GDF (Schlögl et al. 1999b, Schlögl,
2006b) and several other data formats.
Furthermore, projects for the languages Python
(BioSig4Python) and Java (BioSig4Java) have been
started.
2.1 Converter between SCP-ECG and
HL7 aECG
SCP-ECG (Standard Communications Protocol for
Computer-Assisted Electrocardiography) is a
European standard (EN1064:2005) for interpretive
resting ECG. This ECG Standard is the result of an
EU supported project that European, American and
Japanese Manufacturers and Users have jointly
worked and agreed on (1989-1990). In 1993 it
became a European ENV, later was positively
balloted within AAMI (AAMI EC71), and finally
became a European EN at the beginning of 2005.
In 2002 the FDA launched the need of having
the full disclosure digital waveforms submitted for
the support of clinical trials with a flexible XML
schema and a rich set of annotations. The main
American manufacturers thus defined the so-
called FDA XML Data Format Design Specification
that was an FDA XML-based specification covering
the design for the waveform data format as well as
the relevant submission information. This
specification of rising popularity, known also as
Annotated ECG in XML, also became a part of HL7
V.3 currently balloted (HL7 aECG).
OpenECG is a world-wide network supporting
interoperability in electrocardiography through the
consistent implementation of standards. In 2007 it
has about 850 members from more than 60
countries. The development of open source
converters among ECG formats is supported and
encouraged by OpenECG.
At the beginning of 2006 a international working
group formed by people with different expertise was
created by the OpenECG network, with the support
of IEEE 1073 and CEN and the coordination of TU
Graz and Biosig, for the development of an open
source two way converter in C++ between the SCP-
ECG and the HL7 aECG standards. In the
conversion, GDF, the BioSig format was used as an
intermediate form. ECG data sets available in the
OpenECG portal (http://www.openecg.net) were
used to test the converter in different environments
included Linux and Cygwin.
Once the first version was available, there were
some issues that remained open and most of them
were related to an incomplete mapping between the
two standards. This information was an important
retrofit for the relevant Standard Developing
Organizations and some actions in order to solve
these open issues have already been done. In fact, a
harmonization of the ECG lead standard
terminology between the two standards has already
been done leading to the creation of the SCP-ECG
amendment (EN1064:2005+A1:2007) and a similar
revision for the HL7 aECG standard.
The converter has been released as open source
and is currently available in the Biosig Sourceforge
site (http://biosig.sf.net/). Figure 1 shows SCP data
that has been converted into the GDF v1 data format
(Schlögl et al. 2007b).
2.2 BioSig for BCI Research
Brain computer interfacing is one topic closely
related to EEG processing that needs of techniques
capable to work under on-line conditions. Besides,
efficient methods for artifact rejection and/or
correction, online feature extraction techniques,
classifiers, single trial analysis and performance
measurements are important issues in this field.
A Brain Computer Interface (BCI) consists in
general of 4 modules: EEG pre-processing, feature
extraction, classification and feedback. Biosig
provides useful tools for on-line artefact processing;
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
404
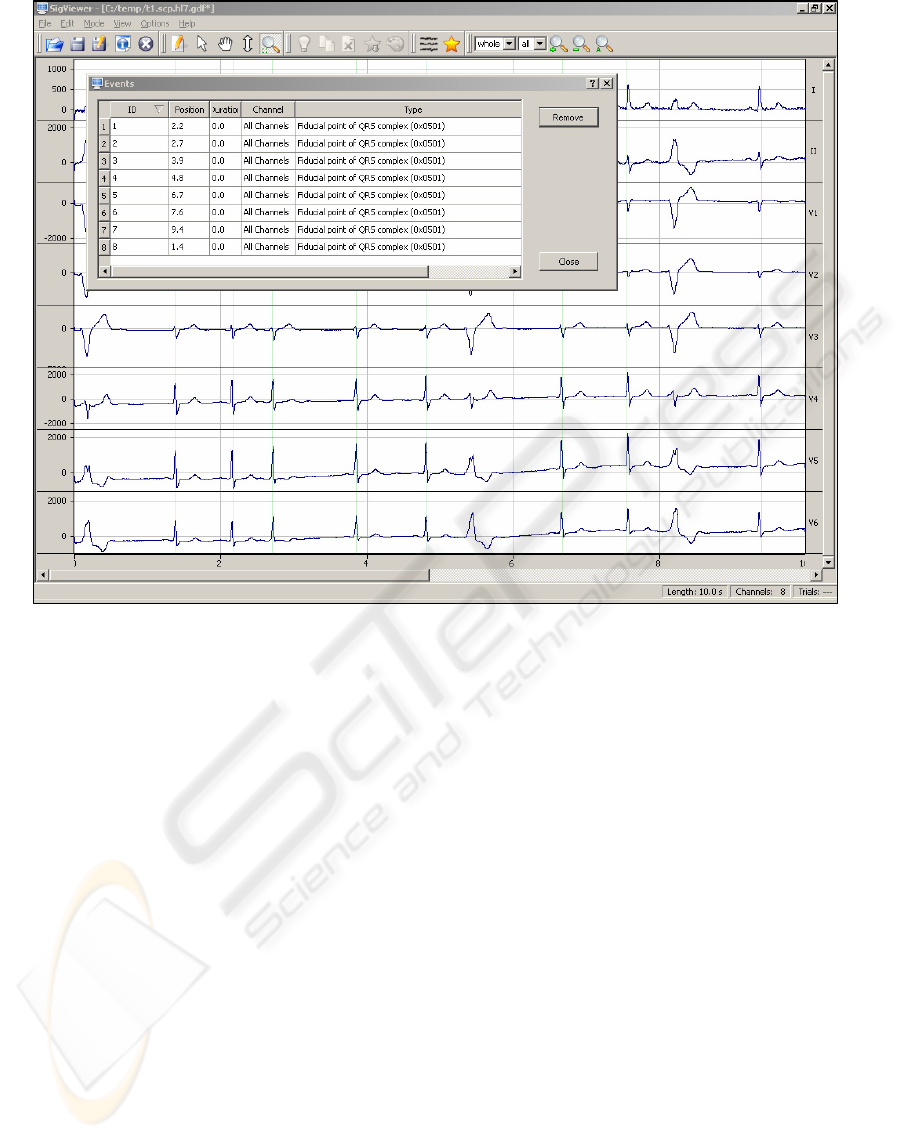
Figure 1: Screenshot of SigViewer showing 10s of 8-channel ECG data. The smaller “window” shows the editable event
table. The data set was converted from the SCP (EN1064) to the GDF v1 (Schögl et al. 1999b) data format using
BioSig4C++.
several on-line feature extractors are also available,
such as adaptive autoregressive parameters or band
power estimates. It also provides many classifiers
(including but not limited to LDA, QDA/MDA,
SVM, NBC, etc) and several single trial analysis
methods to test the performance of systems/subjects.
It can be used to provide initial conditions to all
these modules before starting a BCI on-line session
(see also for the section “rtsBCI below), in which
the system is in general tuned for the subject. Also,
it is especially useful for the analysis of
experimental BCI data. BioSig was the reference
tool for the development of on-line adaptive
classifiers (which were tested in BCI experiments)
(Vidaurre et al. 2006).
2.3 Artifact Processing and Quality
Control
Biomedical signal recordings are often contaminated
by various artifacts. BioSig provides several tools to
address this issue. This include tools for (i) quality
control and determining the saturation values of the
recording systems are provided (Schlögl et al
1999a), (ii) a fully automated reduction methods of
EOG artifacts in EEG recording (Schlögl et al
2007a), and (iii) inverse filtering for detecting
muscle artifacts (Schlögl et al 2000).
2.4 Coupling Analyzing
In order to investigate the interaction and coupling
between brain areas, various coupling measures like
coherency, phase, partial coherence, partial directed
coherence (PDC), directed transfer function (DTF)
etc. can be used. As shown in Schlögl and Supp
(2006), all these coupling measures can be derived
from a multivariate autoregressive (MVAR) model.
The MVAR estimator (Schlögl, 2006a) can be also
applied to data with missing values, thus the various
coupling measures can be obtained from data with
missing values, too. BioSig supports also a non-
parametric statistical analysis using a jackknife
procedure (Efron, 1981) for estimating the
confidence intervals of all the coupling measures.
BIOSIG - Standardization and Quality Control in Biomedical Signal Processing Using the BioSig Project
405

2.5 QRS Detection and HRV Analysis
An important area of biomedical signal processing is
analysing the electrocardiogram (ECG). BioSig
contains several algorithms for QRS detection
(Nygard et al, 1983, Afonso et al 1999), detecting
extrasystoles and other irregular detections (Mateo
and Lugano, 2003) and analyzing the heart rate
variability (Taskforce, 1996). Furthermore, the
efficient algorithm of Berger et al. (1986) for an
equidistant sampling of the heart rate is
implemented.
2.6 rtsBCI
The Graz-BCI open source software package rtsBCI
provides a framework for the development and rapid
prototyping of real-time BCI systems. The software
is based on Matlab/Simulink (The Mathworks, Inc,
Natick, MA, USA) running on Microsoft Windows
(Microsoft Corporation, Redmond, WA, USA) and
licensed under the GNU GPL. For hard real-time
computing and the generation of stand-alone C code
the Real-Time Windows Target (RTWT) and the
Real-Time Workshop (RTW), respectively, are
required. Both toolboxes are extensions of Simulink.
Furthermore, BioSig for Octave and Matlab is
needed for data format handling, and TCP/UDP/IP
toolbox for network communication support.
Additionally to these software requirements, a data
acquisition device is indispensable.
After installation, all rtsBCI modules are listed
in the Simulink Library Browser and can be used to
design (model) the BCI system. Several Matlab
functions and Simulink blocks for (i) data
acquisition and conversion, (ii) storage, (iii) digital
signal processing (e. g. band power feature
estimation, Split-Radix discrete Fourier transform,
adaptive autoregressive parameters (AAR) estimated
with Kalman filtering, linear discriminant analysis,
etc.), (iv) visualization (e. g. signal scope,
presentation of cue information or feedback of a
moving bar), (v) paradigm control (cue-based and
self-paced operation mode) and (vi) network support
(e. g. remote monitoring) are available.
Tunable parameters as well as other information
relevant for the experiment (e.g. subject information,
amplifier settings, electrode setup, paradigm timing)
are stored in an individual configuration file (.INI
file). Before a model is executed, the configuration
is transferred to the model and stored altogether with
the biosignals for further analysis. The division of
model and parameters makes it very easy to deal
with changes: For example, a new classifier requires
only the replacement of the classification block. A
new subject requires only the modification of the
related data in the configuration file.
Modular architecture and rapid prototyping
allow a fast extension and incorporation of new
software as well as hardware components. This
flexibility is a big advantage as is the fact that
Matlab is very popular. The period of vocational
adjustment is reduced, as well as the costs, because
only a reduced number of toolboxes are required.
2.7 SigViewer
SigViewer is a powerful stand-alone viewing and
scoring program for biosignals, originally designed
to process electroencephalogram (EEG) signals.
SigViewer has among its features the ability to load
multi-channel signals such as EEG, ECG, EMG, and
EOG recordings, and display these in various scales.
At the moment, only GDF v1 (Schlögl et al. 1999b)
is supported, but as a workaround, users can convert
other data formats to GDF using the function
“save2gdf” (available in BioSig4OctMat and
BioSig4C++).
Figure 1 shows a screenshot of
SigViewer displaying ECG data.
The other major capability besides the viewing
functions is the scoring of biosignals, which permits
the user to make various annotations to the signals
(e.g. mark segments as artifactious, mark specific
events, like QRS-complexes, etc) and save this
information into a file.
It is also possible to view basic information
about a specific file (e.g. number of channels,
sampling frequency, number of events, time of
recording, and so on). In addition to graphically
scoring the data, the event table is available as a list-
based widget for viewing and deleting events and
annotations (
Figure 1).
SigViewer is written in C++ using the open-
source platform-independent graphical user interface
(GUI) toolkit Qt 4 (Trolltech
®
). SigViewer runs
under many different operating systems such as
Linux, Windows and Mac OS X – in other words, it
is designed to be platform-independent (or more
accurately cross platform). Moreover, it does not
depend on any proprietary software, making it a
truly free program. The source code does not have to
be changed when compiling binaries for specific
platforms, it is enough to take one and the same
source tree and compile it on the target platform.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
406

2.8 Analysis of Cardiac Near-Field
(CNF) Signals
An ongoing research project is the investigation of
the cardiac near-field (CNF) signal. The
spatiotemporal electrical activation at the surface of
heart tissue is assumed to propagate with a smooth
elliptical wave front. Micro-obstacles like embedded
connective tissue may affect the smoothness of the
wave front which results in complex activation
sequences at microscopic size scale. The
investigation of these mechanisms by analysis of the
local activation in microscopic dimensions is
expected to gain deeper insight into structure-related
arrhythmias. For this purpose electrophysiological
in-vitro experiments have been carried out using
autorhythmic or electrically stimulated heart tissue
preparations from Rabbits or Guinea Pigs.
The gradient of electric potential at the cardiac
surface, the Cardiac Near-Field (CNF), can be
computed from four extra-cellular potentials Φ
1
..Φ
4
recorded with ultra-densely placed electrodes
(electrode spacing 50µm). Such a sensor with an
appropriate data acquisition system (sampling rates
of 100 kHz per channel) has been developed
recently (Hofer et al. 2006). It has been shown that
the multivariate signal Φ= [Φ
1
, Φ
2
, Φ
3
, Φ
4
]
T
can be
used to determine local parameters of the
propagating electrical activation, namely velocity
and direction (Plank et al. 2000). Currently, research
is aimed on developing robust procedures for the
calculation of these parameters and the evaluation of
their accuracy (Wiener et al. 2007). In practice, the
two major problems are: First, the acquired signals
are affected by inherent electrode noise and by
stimulus artifacts. Second, in case of structural
discontinuities in the underlying tissue, Φ may be a
composition of multiple local and distal electrical
activation sequences. Therefore, the formation of the
CNF in case of normal and complex activation
sequences in the tissue has been extensively
investigated in computer simulations (Plank et al.
2003).
The BioSig toolbox for Matlab is being used for
off-line analysis of waveforms of Φ obtained from
experiments and computer simulations because it
provides comprehensive procedures for time-
frequency analysis of the multivariate stochastic
process Φ. Moreover the implemented tools can deal
with missing values due to the removal of stimulus
artifacts. Developed signal processing procedures
are being validated by applying them to noise-free
waveforms Φ obtained from computer simulations.
2.9 Miscellaneous
Many smaller algorithms are also included.
Examples are the so-called “Paynter filter” (Bruce et
al. 1977, Platt et al. 1998) to estimate the envelope
of EMG power. There are also many EEG
parameters like Hjorth parameters (Hjorth, 1975),
Barlow parameters (Goncharova and Barlow, 1990),
a global linear descriptor (Wackerman, 1999) and
the brainrate parameter (Pop-Jordanova and Pop-
Jordanov, 2005) supported. Furthermore, methods
for multiple statistical tests for avoiding the problem
of alpha inflation are supported (Hemmelmann et al.
2005), and many different plotting functions for
EEG analysis, like the visualization of Coherence
according to Nolte et al. (2004).
3 RESULTS
BioSig addresses all aspects of biomedical signal
processing, starting with the support for over 40
different data formats, quality control and artifact
processing, methods for signal processing and
feature extraction classification of single trial EEG,
and statistical tests including the multiple
comparison problem. Currently, the main application
areas are research on EEG-based Brain-Computer
Interfaces, coupling analysis of EEG/ECoG/MEG,
processing of EEG artifacts, conversion of different
data formats.
BioSig provides reference implementations of
many biomedical signal processing algorithms and
for many application areas including, EEG, ECoG,
MEG, ECG and HRV analysis, Brain Computer
Interface research, analysing brain connectivity. The
software algorithms can be copied, used, modified
and distributed under the terms of the GNU GPL
(http://www.gnu.org/copyleft/gpl.html). The open
source software library for biomedical signal
processing BioSig is available from
http://biosig.sf.net.
An open source converter between ECG
standardized ECG data formats SCP-ECG (EN1064)
and HL7aECG and several other data formats is
available. Future plans include the development of a
common data format for all biomedical signals.
BIOSIG - Standardization and Quality Control in Biomedical Signal Processing Using the BioSig Project
407
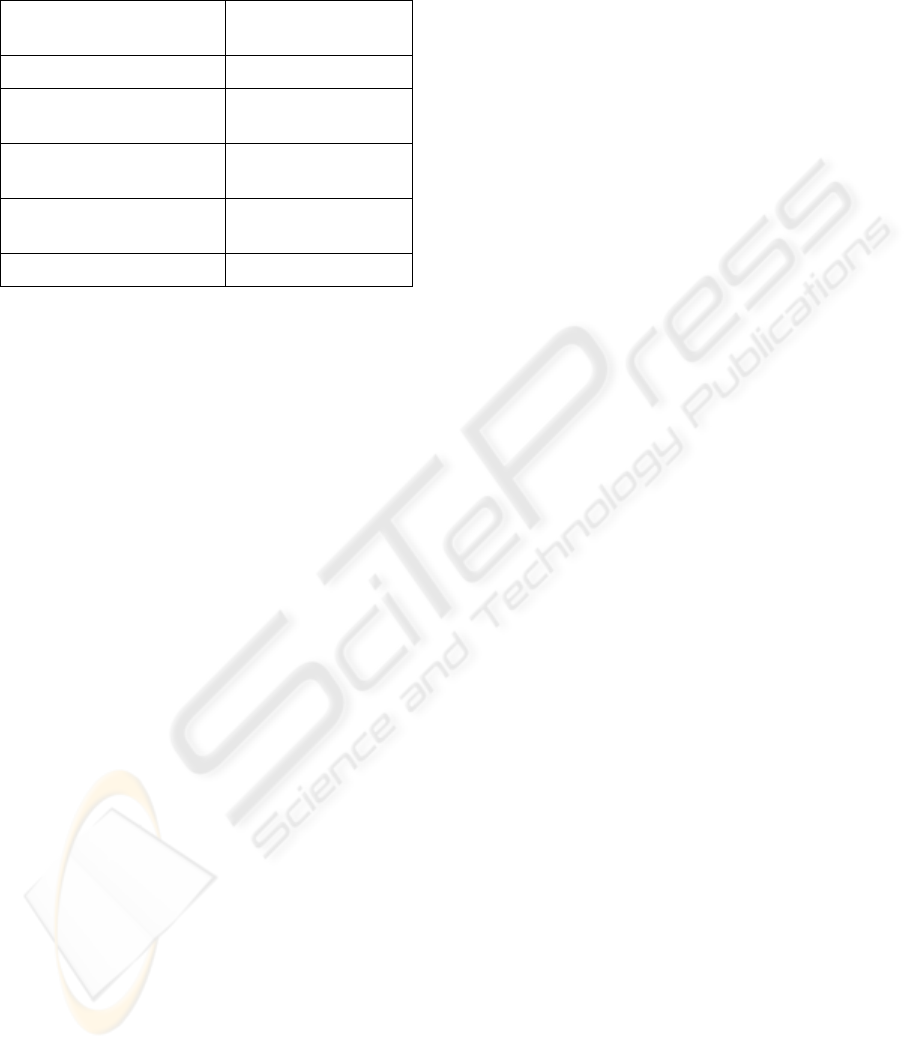
Table 1: Ranking of the Biosig project at SoureForge
among various application areas. The 2
nd
column shows
the ranking of BioSig and the total the number of projects
for each application area.
Topic Ranking 2007-10-22
/ number of projects
SourceForge 456 / 160 049
Biosignals (keyword
search: “EEG, ECG”)
1 / 27
Medical Science
applications
8 / 543
Human Machine
Interfaces
2 / 612
Dataformats 20 / 2139
The BioSig software library is widely adopted.
Currently, the download rate is far beyond 600 per
month and increasing. As of Oct 2007, BioSig is the
highest ranked project for biomedical signal
processing (search term “EEG ECG”) at
SourceForge http://sourceforge.net, a platform that
hosts over 160 000 open source projects. Within the
last two years (Sep 2005 – Sep 2007), the monthly
ranking fluctuated between 2906 (Jan 2007) and 380
(Aug 2007), the overall rank is within the top 2% of
all hosted projects. Besides SourceForge, parts of
BioSig have been incorporated in other projects (e.g.
EEGLab http://www.sccn.ucsd.edu/eeglab/), which
is not considered in the above statistics.
4 DISCUSSION
Although BioSig is routinely used in several
application areas like BCI research, data conversion,
coupling analysis, etc. there are several topics which
are not or only suboptimally supported. Examples
for the current limitations are the following.
(i) Two components (rtsBCI and SViewer)
require proprietary software (Simulink and Matlab).
It would be desirable to have a fully open source
solution without this requirement. SViewer is going
to be replaced by SigViewer, but it is still useful
because SViewer supports more data formats.
Nevertheless, many users do have Matlab anyway,
therefore it is reasonable to distribute these tools.
(ii) The situation on the viewing and scoring
software is not perfect. The SViewer requires the
proprietary Matlab software and is relatively slow,
SigViewer supports only very few data formats.
(iii) The conversion between different data
formats is not always perfect, and can lose some
information (demographic data, annotations, see also
Schlögl et al. 2007b). For this reason, it is important
to unify the various data formats for biomedical
signal processing.
(iv) Support for many specialized application
areas (like advanced ECG analysis, ...) depend on
the contribution and evaluation of expert users. In
order to maintain the growth of BioSig with the aim
to become “the” software library for biomedical
signal processing, participation of users and experts
of the various areas of biomedical signal processing
is crucial. We think of advanced ECG analysis (P-
and T-wave detection, classification of arrhythmias),
the source localization problem in EEG analysis, or
analyzing the activity of spiking neurons.
5 CONCLUSIONS
The BioSig project contains many software tools for
biomedical signal processing. Because BioSig
provides an open source software library, there is no
need to “re-invent the wheel”, but the existing
software algorithms can be used and improved.
These algorithms provide reference
implementations, and can be validated and improved
by everyone. This is an efficient mechanism for
standardization and quality control of software for
biomedical signal processing.
The aim of BioSig providing a software library
for biomedical signal processing has been already
reached, BioSig is also the #1 project for biomedical
signal processing on Sourceforge. The open question
is not whether but how BioSig can help integrating
biosignal analysis into the health information
system.
ACKNOWLEDGEMENTS
This work was supported by the EU grant
“BrainCom” (FP6-2004-Mobility-5 Grant No
024259) and the Austrian Science Fund FWF P-
19993-N15.
REFERENCES
Afonso, V., Tompkins, W., Nguyen, T., Luo S. 1999. ECG
beat detection using filter banks. IEEE Trans. Biomed.
Eng. 46(2):192-202.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
408

Berger R.D., Akselrod S., Gordon D., Cohen R.J. 1986.
An efficient algorithm for spectral analysis of heart
rate variability. IEEE Trans Biomed Eng. 33(9):900-4.
Bruce, E. N., M. D. Goldman, and J. Mead. A digital
computer technique for analyzing respiratory muscle
EMGs. J. Appl. Physiol. 43: 551-556, 1977
Efron, B. 1981. "Nonparametric estimates of standard
error: The jackknife, the bootstrap and other methods",
Biometrika, 68, 589-599.
EN1064 2005. Health informatics. Standard
communication protocol. Computer-assisted
electrocardiography
Goncharova, I.I. and Barlow, J.S. 1990. Changes in EEG
mean frequency and spectral purity during
spontaneous alpha blocking. Electroencephalogr Clin
Neurophysiol. 76(3):197-204.
Hemmelmann C, Horn M, Suesse T, Vollandt R, Weiss S.
2005. New concepts of multiple tests and their use for
evaluating high-dimensional EEG data. J Neurosci
Methods. 142(2):209-17.
Hjorth, B. 1975. Time Domain Descriptors and their
Relation to particulare Model for Generation of EEG
activity. in G. Dolce, H. Kunkel: CEAN Computerized
EEG Analysis, Gustav Fischer, p.3-8.
Hofer, E., Keplinger, F., Thurner, T., Wiener, T., Sanchez-
Quintana, D., Climent, V., Plank, G., 2006. A new
floating sensor array to detect electric near fields of
beating heart preparations. In Biosens. Bioelectron.,
vol. 21, pp 2232-2239.
Mateo, J., Laguna, P. 2003. Analysis of Heart Rate
Variability in Presence of Ectopic Beats Using the
Heart Timing Signal. IEEE Transactions on
biomedical engineering, 50(3): 334-343.
Nolte G, Bai O, Wheaton L, Mari Z, Vorbach S, Hallett
M. 2004. Identifying true brain interaction from EEG
data using the imaginary part of coherency. Clin
Neurophysiol.115(10):2292-307.
Nygards, M.-E. and Sörnmo, L. 1983. Delineation of the
QRS complex using the envelope of the e.c.g.-Med. &
Biol. Eng. & Comput., 21, 538-547.
Plank, G., Hofer, E., 2000. Model study of vector-loop
morphology during electrical mapping of microscopic
conduction in cardiac tissue. In Ann. Biomed. Eng.,
vol. 28, pp 1244-1252.
Plank, G., Vigmond, E., Leon, L.J., Hofer, E., 2003.
Cardiac near-field morphology during conduction
around a microscopic obstacle – a computer
simulation study. In Ann. Biomed. Eng., vol. 31, pp
1206-1212.
Platt, R.S., Hajduk, E.A., Hulliger, M., Easton,P.A. 1998.
A modified Bessel filter for amplitude demodulation
of respiratory electromyograms. J. Appl. Physiol.
84(1): 378-388.
Pop-Jordanova, N. and Jordan Pop-Jordanov J. 2005.
Spectrum-weighted EEG frequency ("Brainrate") as a
quantitative indicator of arousal Contributions, Sec.
Biol. Med. Sci., MASA, XXVI, 2: 35 - 42
Schlögl, A., Kemp, B., Penzel, T., Kunz, D., Himanen, S.-
L., Värri, A., Dorffner, G., Pfurtscheller, G., 1999a.
Quality Control of polysomnographic Sleep Data by
Histogram and Entropy Analysis. Clin. Neurophysiol.
110(12): 2165 - 2170.
Schlögl, A., Filz, O., Ramoser, H., Pfurtscheller, G.
1999b. GDF version 1 - A general dataformat for
biosignals, available at:
http://www.dpmi.tugraz.at/schloegl/matlab/eeg/gdf4/T
R_GDF.pdf
Schlögl, A. 2000. The electroencephalogram and the
adaptive autoregressive model: theory and
applications. Shaker Verlag, Aachen, Germany.
Schlögl, A. 2006a. Comparison of Multivariate
Autoregressive Estimators. Signal processing. 86(9):
2426-9.
Schlögl, A. 2006b. GDF version 2 - A general dataformat
for biosignals. http://arxiv.org/abs/cs.DB/0608052
Schlögl, A. and Supp, G. 2006. Analyzing event-related
EEG data with multivariate autoregressive parameters.
(Eds.) C. Neuper and W. Klimesch, Event-related
Dynamics of Brain Oscillations. Analysis of dynamics
of brain oscillations: methodological advances.
Progress in Brain Research 159: 135 – 147.
Schlögl, A., Keinrath, C., Zimmermann, D., Scherer, R.,
Leeb, R., Pfurtscheller, G. 2007a. A fully automated
correction method of EOG artifacts in EEG
recordings. Clin. Neurophys. 118(1):98-104.
Schlögl, A., Chiarugi, F., Cervesato, E Apostolopoulos,
E., Chronaki, C.E.. 2007b. Two-Way Converter
between the HL7 aECG and SCP-ECG Data Formats
Using BioSig.
Conference on Computers In Cardiology,
Taskforce Heart Rate Variability: Standards of
Measurement, physilogical interpretation and clinical
use. Taskforce of the European Society for Cardiology
and the North American Society of Pacing and
Electrophysiology. European Heart Journal (1996)
17, 354-381.
Vidaurre, C., Schlögl, A.,Cabeza, R., Scherer, R.
Pfurtscheller, G. 2006. A fully on-line adaptive BCI,
IEEE Trans. Biomed. Eng., 53(8): 1214–1219.
Wackermann, J. 1999. Towards a quantitative
characterization of functional states of the brain: from
the non-linear methodology to the global linear
descriptor. International Journal of Psychophysiology,
34: 65-80.
Wiener, T., Thurner, T., Prassl, A.J., Plank, G., Hofer, E.,
2007. Accuracy of local conduction velocity
determination from non-fractionated cardiac activation
signals. In EMBC’07, 29th Annual International
Conference of the IEEE Engineering in Medicine and
Biology Society.
BIOSIG - Standardization and Quality Control in Biomedical Signal Processing Using the BioSig Project
409
