
ESTIMATING LARGE LOCAL MOTION IN LIVE-CELL IMAGING
USING VARIATIONAL OPTICAL FLOW
Towards Motion Tracking in Live Cell Imaging Using Optical Flow
Jan Huben
´
y, Vladim
´
ır Ulman and Pavel Matula
Centre for Biomedical Image Analysis, Faculty of Informatics Masaryk University, Botanick
´
a 68a, Brno 602 00, Czech Republic
Keywords:
Live-cell imaging, motion tracking, 3D imaging, variational optical flow.
Abstract:
The paper studies the application of state-of-the-art variational optical flow methods for motion tracking of
fluorescently labeled targets in living cells. Four variants of variational optical flow methods suitable for
this task are briefly described and evaluated in terms of the average angular error. Artificial ground-truth
image sequences were generated for the purpose of this evaluation. The aim was to compare the ability of
those methods to estimate local divergent motion and their suitability for data with combined global and local
motion. Parametric studies were performed in order to find the most suitable parameter adjustment. It is shown
that a selected optimally tuned method tested on real 3D input data produced satisfactory results. Finally, it is
shown that by using appropriate numerical solution, reasonable computational times can be achieved even for
3D image sequences.
1 INTRODUCTION
There is a steadily growing interest in live cell stud-
ies in modern cell biology. The progress in staining
of living cells together with advances in confocal mi-
croscopy devices has allowed detailed studies of the
behaviour of intracellular components including the
structures inside the cell nucleus. The typical num-
ber of investigated cells in one study varies from tens
to hundreds because of statistical significance of the
results. One gets time-lapse series of three or two di-
mensional images as an output from the microscope.
It is very inconvenient and annoying to analyze such
data sets by hand (especially for 3D series). More-
over, there is no guarantee on the accuracy of the re-
sults. Therefore, there is a natural demand for com-
puter vision methods which can help with analysis of
these time-lapse image series. Estimation or correc-
tion of global as well as local motion belongs to main
tasks in this field. The suitability of the state-of-the-
art optical flow methods for correction of local motion
will be studied in this article.
The live-cell studies are mainly performed using
the confocal microscopes these days. The confocal
microscopes are able to focus on selected z-plane of
the specimen in the same way as the standard wide-
field (non-confocal) microscopes. However, they are
based on principle of suppression of light from planes
which are out of focus. Therefore, they provide far
better 3D image data (less blurred) than wide-field
microscopes. The main disadvantage of confocal mi-
croscopes is their lower light throughput. This causes
larger exposure times as compared to the wide-field
mode. Several optical setups suitable for live-cell
imaging as well as their optimization and automation
are discussed in detail in (Kozubek et al., 2004).
Transparent biological material is visualized with
fluorescent proteins in live-cell imaging. Living spec-
imen usually does not contain fluorescent proteins.
Therefore, the living cells are forced to produce those
proteins in the specimen preparation phase (Chalfie
et al., 1994). The image of the living cells in the
specimen on the microscope stage is acquired peri-
odically. The cells can move or change their internal
structure in the meantime. The interval between two
consecutive acquisitions varies in range from frac-
tions of second up to tens of minutes. It would be
convenient to acquire snapshots frequently in order
to have only small changes between two consecutive
frames. But, the interval length cannot be arbitrary
542
Hubený J., Ulman V. and Matula P. (2007).
ESTIMATING LARGE LOCAL MOTION IN LIVE-CELL IMAGING USING VARIATIONAL OPTICAL FLOW - Towards Motion Tracking in Live Cell Imaging
Using Optical Flow.
In Proceedings of the Second International Conference on Computer Vision Theory and Applications - IU/MTSV, pages 542-548
Copyright
c
SciTePress

small mainly because of photo-toxicity (the living
specimen is harmed by the light) and photo-bleaching
(the intensity of fluorescent markers fades while be-
ing exposed to the light). However, it is usually pos-
sible to find a reasonable compromise between those
restrictions and adjust the image acquisition so that
the displacement of objects between two consecutive
snapshots is not more than ten pixels.
There are two types of tasks to be solved in this
field. First, the global movement of objects should
be corrected before subsequent analysis of an intra-
cellular movement. This goal is often achieved us-
ing common rigid registration methods (Zitov
´
a and
Flusser, 2003). A fast 3D point based registration
method (Matula et al., 2006) was recently proposed
for the global alignment of cells.
The second task is to estimate local changes in-
side the objects. This task is more complex. The ob-
jects inside the cells or nuclei can move in different
directions. One object can split into two or more ob-
jects and vice versa. Moreover, an object can appear
or disappear during the experiment. Therefore, this
task requires computation of dense motion field be-
tween two consecutive snapshots. Manders et. al. has
used block-matching (BM3D) algorithm (de Leeuw
and van Liere, 2002) for this purpose in their study
of chromatin dynamics during the assembly of inter-
phase nuclei (Manders et al., 2003). Their BM3D
algorithm is rather slow. It is similar to basic optic
flow methods but it does not comprise any smooth-
ness term.
We study latest optical flow methods (Bruhn,
2006) for estimation of intracellular movement in this
paper. Up to our best knowledge, nobody investi-
gated the application of these state-of-the-art meth-
ods in live-cell imaging. The simple ancestors of
these methods, which can reliably estimate one pixel
motion, were successfully used for lung motion cor-
rection (Dawood et al., 2005). The examined meth-
ods are able to reliably estimate the flow larger than
one pixel. They can produce piece-wise smooth flow
fields which preserve the discontinuities in the flow
on object boundaries. These properties are needed for
estimation of local divergent motion which occur in
live-cell imaging. We have extended state-of-the-art
optical flow methods into three dimensions. Espe-
cially, we focused on 3D extension of recently pub-
lished optical flow methods for large displacements
(Papenberg et al., 2006). We tested these methods on
synthetic as well as real data and compared their be-
haviour and performance. Our experiments identify
the optical flow methods which can be used in live cell
imaging. We used the efficient numeric techniques for
the optical flow computations (Bruhn, 2006). This al-
lows us to get reasonable computational times even
for 3D image sequences.
The rest of the paper is organized as follows: The
variational optical flow methods are described in Sec-
tion 2. Section 3 is devoted to the experiments and re-
sults obtained for synthetic and real biomedical data.
2 OPTICAL FLOW
In this section, we describe the basic ideas of varia-
tional optical flow methods and in particular the meth-
ods which will be tested in Section 3.
Let two consecutive frames of image sequence be
given. Optical flow methods compute the displace-
ment vector field which maps all voxels from first
frame to their new position in the second frame. Al-
though several kinds of strategies exist for optical
flow computation (Barron et al., 1994), we take only
the so-called variational optical flow (VOF) methods
into our considerations. They currently give the best
results (in terms of error measures) (Papenberg et al.,
2006; Bruhn and Weickert, 2005) and come out from
transparent mathematical modeling (the flow field is
described by energy functional). Furthermore, they
produce dense flow fields and are invariant under ro-
tations.
The first prototype of VOF method was proposed
in (Horn and Schunck, 1981). Horn and Schunck
used the grey value constancy assumption which as-
sumes that the grey value intensity of the moving ob-
jects remains the same and homogenous regulariza-
tion which assumes that the flow is smooth. We will
describe their method first, because even the most so-
phisticated methods available are based on the funda-
mental ideas of Horn and Schunck method.
Let Ω
4
⊂ R
4
denote the 4-dimensional spatial-
temporal image domain and f(x
1
, . . . , x
4
) : Ω
4
→ R
a gray-scale image sequence, where (x
1
, x
2
, x
3
)
⊤
is a
voxel location within a image domain Ω
3
⊂ R
3
and
x
4
∈ [0, T] denotes the time. Moreover, let’s assume
that ∆x
4
= 1 and u = (u
1
, u
2
, u
3
, 1)
⊤
denotes the un-
known flow. The grey value constancy assumption
says
f(x
1
+u
1
, . . . , x
3
+u
3
, x
4
+1)− f(x
1
, . . . , x
4
) = 0 (1)
Optic flow constraint (OFC) is obtained by approxi-
mation of (1) with first-order Taylor expansion
f
x
1
u
1
+ f
x
2
u
2
+ f
x
3
u
3
+ f
x
4
= 0, (2)
where f
x
i
is partial derivative of f. Equation (2) with
three unknowns has obviously more than one solu-
tion. Horn and Schunck assumed only smooth flows
and they therefore penalized the solutions which have

large spatial gradient ∇
3
u
i
where i ∈ 1, 2, 3 and ∇
3
de-
notes the spatial gradient. Thus, the sum
∑
3
i=1
|∇
3
u
i
|
for every voxel should be as small as possible. We
get following variational formulation of the problem
if we combine these two considerations together:
E
HS
(u) =
Ω
( f
x
1
u
1
+ f
x
2
u
2
+ f
x
3
u
3
+ f
x
4
)
2
+ α
3
∑
i=1
|∇
3
u
i
|
2
dx
(3)
The optimal displacement vector field minimizes en-
ergy functional (3). The OFC and the regularizer are
squared, the α parameter has the influence on the
smoothness of the solution. The two terms which
form the functional are called data and smoothness
term. Following the calculus of variations (Gelfand
and Fomin, 2000), the minimizer of (3) is a solution
of Euler-Lagrange equations
0 = f
2
x
1
u
1
+ f
x
1
f
x
2
u
2
+ f
x
1
f
x
3
u
3
+ f
x
1
f
x
4
+ αdiv(∇
3
u
1
)
0 = f
x
1
f
x
2
u
1
+ f
2
x
2
u
2
+ f
x
2
f
x
3
u
3
+ f
x
2
f
x
4
+ αdiv(∇
3
u
2
)
0 = f
x
1
f
x
3
u
1
+ f
x
2
f
x
3
u
2
+ f
2
x
3
u
3
+ f
x
3
f
x
4
+ αdiv(∇
3
u
3
)
(4)
with reflecting Neumann boundary conditions. div(x)
is the divergence operator. The system (4) is usu-
ally solved with common numerical methods like
Gauss-Seidel or SOR. The bidirectional full multigrid
(Briggs et al., 2000) framework for computations of
VOF methods was proposed in (Bruhn et al., 2005).
The computations with multigrid methods are by or-
ders of magnitude faster than the classic Gauss-Seidel
or SOR methods.
Now we describe the VOF methods which will be
tested in Section 3. The current state-of-the-art VOF
methods are still similar to their Horn-Schunck pre-
cursor. Their energy functional consists of data and
smoothness term. The combined local-global (CLG)
method for large displacements proposed in (Papen-
berg et al., 2006) is the first method which we have
tested. This method produces one among the most
accurate results (Bruhn, 2006). We assume that it will
be suitable for our data, because it produces smooth
flow fields and simultaneously flow fields with dis-
continuities. The energy functional of CLG method
is defined as:
E
CLG
(u) =
Ω
Ψ
D
(| f(x+ u) − f(x)|
2
)
+ αΨ
S
3
∑
i=1
|∇
3
u
i
|
2
!
dx
(5)
where
Ψ
D
(s) =
q
s
2
+ ε
2
D
Ψ
S
(s) =
q
s
2
+ ε
2
S
and ε
D
, ε
S
are reasonably small numbers (e.g. ε
D
=
0.01). Note that the data term consists of non-
linearized grey value constancy assumption (1). This
allows to correct estimate of the large displacements.
Moreover, the CLG method uses the non-quadratic
penalizers Ψ
D
(s) and Ψ
S
(s) and therefore it is ro-
bust with respect to noise and outliers. Nevertheless,
these concepts make the minimization of (5) quite
complex. We use the multi-scale warping based ap-
proach, which proposed in (Papenberg et al., 2006),
for minimization of (5). The multigrid numerical
framework for this task was extensively analyzed in
(Bruhn, 2006; Bruhn and Weickert, 2005).
The second tested method consists of robust data
term and anisotropic image driven smoothness term
(Nagel and Enkelmann, 1986). We denote this
method RDIA. Its energy functional is defined:
E
RDIA
(u) =
Ω
Ψ
D
(| f(x+ u) − f(x)|
2
)
+ α
3
∑
i=1
(∇
3
u
⊤
i
P
NE
(∇
3
f)∇
3
u
i
) dx
(6)
where P
NE
(∇
3
f) is projection matrix perpendicular to
∇
3
f defined as
P
NE
=
1
2|∇
3
f|
2
+ 3ε
2
a b c
b d e
c e f
(7)
where
a = f
2
x
2
+ f
2
x
3
+ ε
2
d = f
2
x
1
+ f
2
x
3
+ ε
2
b = − f
x
1
f
x
2
e = − f
x
2
f
x
3
c = − f
x
1
f
x
3
f = f
2
x
1
+ f
2
x
2
+ ε
2
The ε is reasonably small number (e.g. ε = 0.01).
The only difference between CLG and RDIA method
is the smoothing term. RDIA method smoothes the
flow with respect to underlying image. The image
data in live-cell imaging are often low contrast due
to the limitations of the optical setup. We assume that
this smoothing term can help with processing of such
data. The same minimization approach can be used as
for the CLG method.
The third and fourth tested method are variants of
the previous two. We add the gradient constancy as-
sumption to the data term. This should provide us bet-
ter results on image sequences which fade out with in-
creasing time. The energy functional of CLG method

with gradient constancy assumption is defined as
E
CLG
G
(u) =
Ω
Ψ
D
(| f(x+ u) − f(x)|
2
+ γ(|∇ f (x+ u) − ∇ f (x)|
2
))
+ αΨ
S
3
∑
i=1
|∇
3
u
i
|
2
!
dx
(8)
The variant of RDIA method with gradient constancy
assumption is defined as
E
RDIA
G
(u) =
Ω
Ψ
D
(| f(x+ u) − f(x)|
2
+ γ(|∇ f (x+ u) − ∇ f (x)|
2
))
+ α
3
∑
i=1
(∇
3
u
⊤
i
P
NE
(∇
3
f)∇
3
u
i
) dx
(9)
where γ is the parameter which controls the influence
of gradient constancy assumption. Note that the gra-
dient constancy assumption is again included in the
non-linearized form.
3 RESULTS AND DISCUSSION
In this section, we test the behaviour of CLG, RDIA,
CLG
G
and RDIA
G
methods on artificial and real im-
age data. We measure their performance on live-
cell image sequences with large local displacements.
Moreover, we present the results of the methods on
image sequences with combined global rigid (trans-
lation, rotation) and local displacements. We present
the results on both two and three dimensional data.
Finally, we discuss the computational time and stor-
age demands.
The ground-truth flow fields are needed for the
evaluation purposes. Obviously, they are not avail-
able for real image sequences. Therefore, we gener-
ate the artificial data with artificial flow fields from
the real image data. We use the two-layered approach
in which user-selected foreground is locally moved
and inserted into an artificially generated background.
Hence, our generator requires one real input frame,
the mask of the cell (the background) and the mask
of the objects (the foreground). Both first and sec-
ond frames are generated in two steps. First, the
foreground objects are extracted, an artificial back-
ground is generated and the rigid motion is applied
on background and foreground separately. Second,
every foreground object is translated independently
on each other and finally inserted into the generated
background. The movements were performed by us-
ing backward registration technique (Lin and Barron,
Figure 1: Generation of artificial data. We use two-layered
approach. Both artificial frames are generated from the real
input frame. The second frame is almost identical to the real
one. The first frame is the backward registered copy of sec-
ond frame. The artificial ground-truth flow field is used for
the backward registration. The background and foreground
movements are independent. (top left). Real input frame.
(top right) Cell nucleus mask (background) in white, object
mask (foreground) in red. (bottom left) Second artificial
frame. (bottom right) First artificial frame (red channel)
superimposed over second frame (green channel).
1994) according to the generated flow field. This flow
determines the movement and becomes the ground-
truth flow. The flow field determines the movement
uniquely. Thus, it becomes the ground-truth flow field
between these two frames and can be used for testing
purposes. Owing to the property of the backward reg-
istration technique, the first frame represents the in-
put real image before the movement while the second
frame represents it after the movement. Therefore, the
second frame is similar to the real input image. The
generation process is illustrated in Fig. 1.
The first experiment was performed on artificial
two dimensional data. We prepared a data set of arti-
ficial images with large local displacements. The data
set consisted of six different frame couples. The size
of input frames was 400× 400 pixels. The number of
objects which moved inside the nucleus varies from
seven to eleven. The size of the individual translations
vectors varies from 1.2 to 11.3 pixels. According to
the literature, the input frames were filtered with gaus-
sian blur filter with standard deviation σ = 1.5. We
performed a parametric study over α parameter for
four tested methods over whole data set. The results
were compared with respect to average angular error
(AAE) (Fleet and Jepson, 1990) where angular error
is defined as
arccos
(u
1
)
gt
(u
1
)
e
+ (u
2
)
gt
(u
2
)
e
+ 1
q
((u
1
)
2
gt
+ (u
2
)
2
gt
+ 1)((u
1
)
2
e
+ (u
2
)
2
e
+ 1)
(10)

where (u
i
)
gt
, (u
i
)
e
denote the i-component of ground-
truth and estimated vector, respectively. There were
two coupled goals behind this experiment. We wanted
to identify the best method by the mean of AAE and
at the same time find suitable setting of α parameter.
The results are presented in Fig. 2. The AAE was
computed for each run of particular method with par-
ticular α. Then, the averages of AAE over the frame
pairs in the data set were computed. These averages
are depicted in the graphs in Fig. 2. The AAE was
computed only inside the moving objects. We can see
that all methods perform reasonably well. The CLG
method is outperformed by the RDIA method. The
gradient variant of CLG provided better results than
the simple CLG method. The RDIA
G
method is not
depicted in the graph because we noticed that its re-
sults depended on data and the average value was bi-
ased. It was slightly better than simple RDIA method
in some cases. But, it was slightly worse in other
cases.
2
4
6
8
10
12
14
16
5 10 15 20
AAE
α parameter
AAE of CLGg method
AAE of CLG method
2.16
2.18
2.2
2.22
2.24
2.26
2.28
2.3
2.32
2.34
2.36
2.38
50 100 150 200 250 300
AAE
α parameter
AAE of RDIA method
Figure 2: The dependency of average angular error on α pa-
rameter. The methods were tested on artificial image data
with large local displacements up to 11.3 pixels. (top).
CLG and CLG
G
method. (bottom) RDIA method.
The goal of the second experiment was to examine
the performance of the tested methods on sequences
with combination of global and local movement. We
again prepared a dataset with artificial frame pairs.
Image size and other input sequence properties were
the same as in the previous experiment. The cell nu-
clei were transformed with global translation (up to
5 pixels) and rotation (up to 4 degrees). After that
the local displacement were applied on the foreground
object inside. It becomes clear from our experiments
that we should divide the data into two groups. The
results of the computations were influenced by the
following fact. If the majority of objects inside the
cell nucleus moved in the direction similar to the
12
14
16
18
20
22
24
26
2 4 6 8 10 12 14
AAE
α parameter
AAE of CLGg method
AAE of CLG method
11
12
13
14
15
16
17
18
50 100 150 200 250 300
AAE
α parameter
AAE of RDIAg method
AAE of RDIA method
8
9
10
11
12
13
14
4 6 8 10 12 14
AAE
α parameter
AAE of CLGg method
AAE of CLG method
8.5
9
9.5
10
10.5
11
11.5
12
12.5
50 100 150 200 250 300
AAE
α parameter
AAE of RDIAg method
AAE of RDIA method
Figure 3: The dependency of average angular error on α
parameter. The methods were tested on artificial image
data with global movement (translation up to 5 pixels, ro-
tation up to 4 degrees) and large local displacements up to
11.3 pixels. (top). CLG and CLG
G
method. (upper cen-
ter) RDIA and RDIA
G
method. Majority of local displace-
ments have the same direction as compared to global move-
ment (top, upper center). (lower center) CLG and CLG
G
method. (bottom) RDIA and RDIA
G
method. Majority
of local displacements has different direction than global
movement (lower center, bottom).
global translation the results fell into the first group
and vice versa. The AAE was computed on the cell
nucleus mask (see the Fig. 1) in this experiment. The
results for both groups are illustrated in Fig. 3.
We tested the methods with best parameter set-
tings on real three dimensional data in the third ex-
periment. We computed the displacement field of two
input frames of human HL-60 cell nucleus with mov-
ing HP1 protein domains. There are global as well as
local movements in the frame pair (see Fig. 4). We es-
timated the flow with RDIA method, α was set to 100.
The size of the input frames was 276×286×106. The
results are illustrated in Fig. 4.
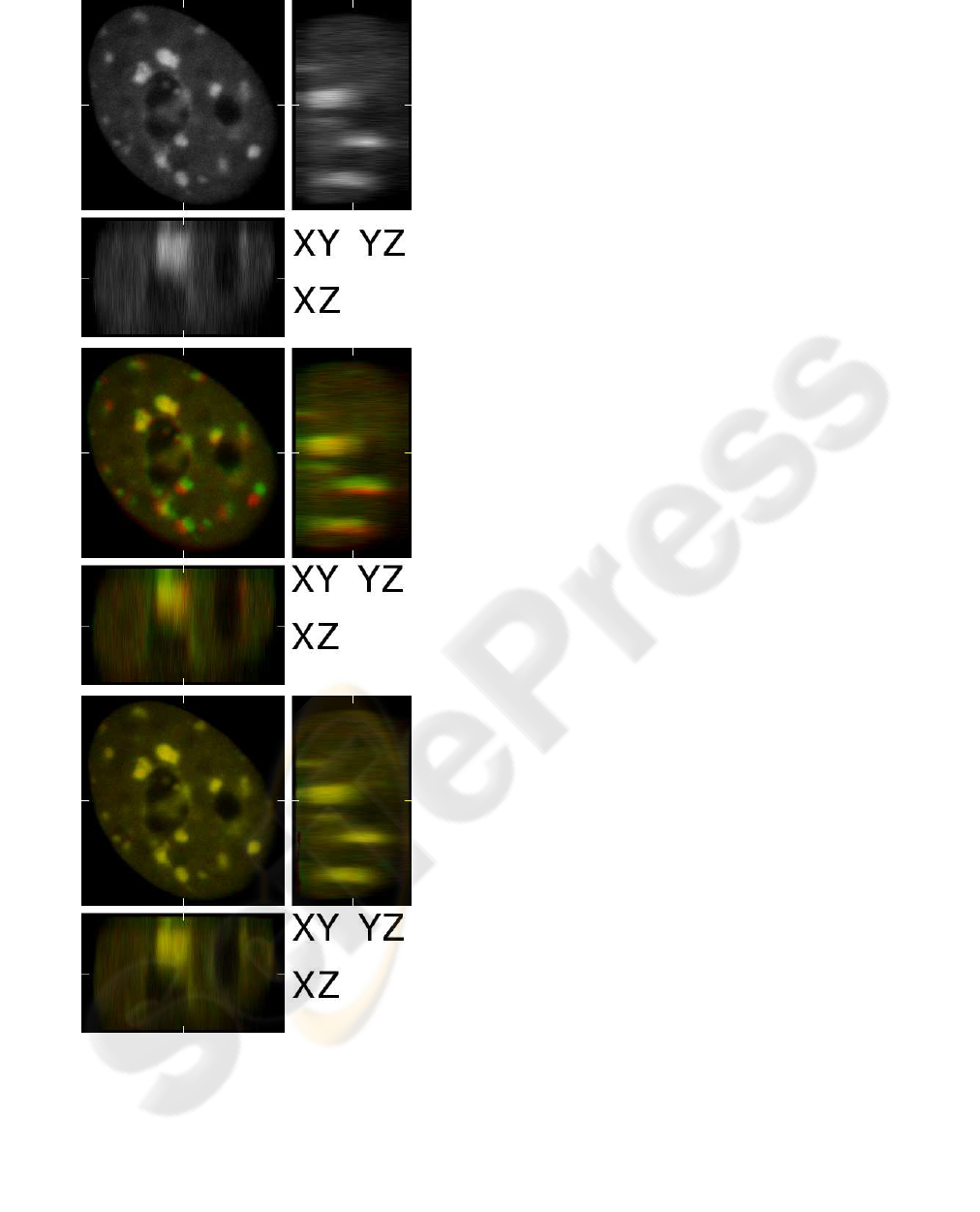
Figure 4: Experiment with real 3D data. Frame
size 276× 286×106. xy, xz and yz cuts on position
(138, 143, 53) are shown. (top). First input frame. (cen-
ter) First input frame (red channel) superimposed on sec-
ond frame (green channel). Correlation is 0.901. (bottom)
The RDIA method with α = 100 computes the flow field.
Backward registered second frame (green channel) is su-
perimposed onto first frame (red channel). Correlation is
0.991.
The VOF methods were implemented in
C++
lan-
guage and tested on common workstation (Intel Pen-
tium 4 2.6 GHz, 2 GB RAM, Linux 2.6.x). We use the
multigrid framework (Bruhn, 2006) for numerical so-
lution of tested VOF methods. The computations on
two 3D frames of size 276× 286× 106 took from 700
to 920 seconds and needed 1.5 GB of RAM. Compu-
tations on two 2D frames of size 400× 400 took from
13 to 16 seconds and needed 13 MB of RAM.
3.1 Discussion
We found out that the RDIA methods produce slightly
better results than the CLG methods (with respect to
AAE) in both experiments on synthetic live-cell im-
age data. Moreover, RDIA methods are less sensi-
tive to α parameter setting. It became clear that the
smoothing term of RDIA method is more suitable for
the low contrast image sequences. The computed flow
field can be easily oversmoothed by the CLG meth-
ods, because they consider the moving objects to be
outliers in the data by particular parameter settings
(larger α).
The use of “gradient” variants of tested method
can slightly improve their performance on live-cell
image data. On the other hand, there is no warranty
that the result will be always better. Actually, the
RDIA method seems to be more sensitive to α param-
eter when using its “gradient” variant. Surprisingly,
the expected improvement of “gradient” variants was
not significant even for fading out sequences. We sup-
pose that the gradient constancy assumption does not
help a lot, because the decrease of the intensities be-
tween two consecutive frames is not big. To sum it up,
the 2D tests show that the RDIA method is the most
suitable for our data. Therefore, we used it for the ex-
periment on the 3D real image data. The backward
registered results show perfect match on xy-planes.
The match in xz and yz planes is a little bit worse.
This is caused by the lower resolution of the micro-
scope device in the z axis.
The bleeding-edge multigrid technique allowed us
to get the flow fields in reasonable times even for 3D
(up to 15 minutes for one frame pair). Small data sets
which consist of only tens of frames of several cells
can be analyzed in order of days on one common PC.
Larger data sets as well as parametric studies in 3D
should be analyzed on a computer cluster.
4 CONCLUSION
We studied state-of-the-art variational optical flow
methods for large displacement for motion tracking

of fluorescently labeled targets in living cells. We fo-
cused on 2D as well as 3D images. Up to our best
knowledge, we tested those methods first time in the
literature for three dimensional image sequences.
We showed that these methods can reliably es-
timate large local divergent displacements up to ten
pixels. Moreover, the methods can estimate the global
as well as local movement simultaneously. The vari-
ants of CLG and RDIA method with gradient con-
stancy assumptions did not bring significant improve-
ment for our data. The RDIA method produced the
best results in our experiments. We achieved reason-
able computation times (even for three dimensional
image sequences) using the full bidirectional multi-
grid numerical technique.
We plan to perform larger parametric studies on
three dimensional data. This studies need to be per-
formed on computer cluster or grid because of com-
putational demands. Owing to the achieved results,
we also feel confident in building a motion tracker as
an application based on tested methods. By analyz-
ing computed flow field one can extract important bi-
ological data regarding the movement of intracellular
structures.
ACKNOWLEDGEMENTS
This work was partly supported by the Ministry of
Education of the Czech Republic (Grants No. MSM-
0021622419 and LC-535) and by Grant Agency of the
Czech Republic (Grant No. GD102/05/H050).
REFERENCES
Barron, J. L., Fleet, D. J., and Beauchemin, S. S. (1994).
Performance of optical flow techniques. Int. J. Com-
put. Vision, 12(1):43–77.
Briggs, W. L., Henson, V. E., and McCormick, S. F. (2000).
A multigrid tutorial: second edition. Society for In-
dustrial and Applied Mathematics, Philadelphia, PA,
USA.
Bruhn, A. (2006). Variational Optic Flow Computation:
Accurate Modelling and Efficient Numerics. PhD the-
sis, Department of Mathematics and Computer Sci-
ence, Universit
¨
at des Saarlandes, Saarbr
¨
ucken.
Bruhn, A. and Weickert, J. (2005). Towards ultimate motion
estimation: Combining highest accuracy with real-
time performance. In Proc. 10th International Confer-
ence on Computer Vision, pages 749–755. IEEE Com-
puter Society Press.
Bruhn, A., Weickert, J., and Schn
¨
orr, C. (2005). Variational
optical flow computation in real time. IEEE Transac-
tions of Image Processing, 14(5).
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., and
Prasher, D. C. (1994). Green fluorescent protein as a
marker for gene-expression. Science, 263(5148):802–
805.
Dawood, M., Lang, N., Jiang, X., and Sch
¨
afers, K. P.
(2005). Lung motion correction on respiratory gated
3d pet/ct images. IEEE Transactions on Medical
Imaging.
de Leeuw, W. and van Liere, R. (2002). Bm3d: Motion es-
timation in time dependent volume data. In Proceed-
ings IEEE Visualization 2002, pages 427–434. IEEE
Computer Society Press.
Fleet, D. J. and Jepson, A. D. (1990). Computation of com-
ponent image velocity from local phase information.
Int. J. Comput. Vision, 5(1):77–104.
Gelfand, I. M. and Fomin, S. V. (2000). Calculus of Varia-
tions. Dover Publications.
Horn, B. K. P. and Schunck, B. G. (1981). Determining
optical flow. Artificial Intelligence, 17:185–203.
Kozubek, M., Matula, P., Matula, P., and Kozubek, S.
(2004). Automated acquisition and processing of
multidimensional image data in confocal in vivo
microscopy. Microscopy Research and Technique,
64:164–175.
Lin, T. and Barron, J. (1994). Image reconstruction error
for optical flow. In Vision Interface, pages 73–80.
Manders, E. M. M., Visser, A., Koppen, A., de Leeuw,
W., van Driel, R., Brakenhoff, G., and van Driel, R.
(2003). Four-dimensional imaging of chromatin dy-
namics the assembly of the interphase nucleus. Chro-
mosome Research, 11(5):537–547.
Matula, P., Matula, P., Kozubek, M., and Dvo
ˇ
r
´
ak, V. (2006).
Fast point-based 3-d alignment of live cells. IEEE
Transactions on Image Processing, 15:2388–2396.
Nagel, H. H. and Enkelmann, W. (1986). An investigation
of smoothness constraints for the estimation of dis-
placement vector fields from image sequences. IEEE
Trans. Pattern Anal. Mach. Intell., 8(5):565–593.
Papenberg, N., Bruhn, A., Brox, T., Didas, S., and Weick-
ert, J. (2006). Highly accurate optic flow computa-
tion with theoretically justified warping. International
Journal of Computer Vision, 67(2):141–158.
Zitov
´
a, B. and Flusser, J. (2003). Image registration meth-
ods: a survey. 21(11):977–1000.
