
Preliminary Steps towards Efficient Classification in Large Medical
Datasets: Structure Optimization for Deep Learning Networks through
Parallelized Differential Evolution
Ivanoe De Falco
1
, Giuseppe De Pietro
1
, Antonio Della Cioppa
2
, Giovanna Sannino
1
,
Umberto Scafuri
1
and Ernesto Tarantino
1
1
ICAR, National Research Council of Italy (CNR), Via P. Castellino 111, Naples, Italy
2
NCLab, DIEM, University of Salerno, Via Giovanni Paolo II 132, Salerno, Italy
Keywords:
Medical Databases, Deep Learning, Optimization, Deep Neural Network Structure, Differential Evolution.
Abstract:
Deep Neural Networks are being more and more widely used to perform several tasks over highly-sized
datasets, one of them being classification. Finding good configurations for Deep Neural Network structures is
a very important problem in general, and particularly in the medical domain. Currently, either trial-and-error
methodologies or sampling-based ones are considered. This paper describes some preliminary steps towards
effectively facing this task. The first step consists in the use of Differential Evolution, a kind of an Evolution-
ary Algorithm. The second lies in using a parallelized version in order to reduce the turnaround time. The
preliminary results obtained here show that this approach can be useful in easily obtaining structures that allow
increases in the network accuracy with respect to those provided by humans.
1 INTRODUCTION
Nowadays, thanks to the ever-increasing use of sen-
sors in the medical field, a huge amount of datasets are
being created starting from the continuous monitoring
of bio-signals. In many cases, these datasets consist
of a very high amount of data, even tens of millions
of items or more, where each item, on its turn, could
be composed by dozens of parameters.
One of the main and most frequent tasks that
should be carried out on such datasets makes refer-
ence to the classification of all the items making up
the dataset, especially in a supervised way. This lat-
ter involves the division of the available items into a
training set and a test one.
Already several years ago, as long as the amount
of sensors that were applied to a subject showed an
increasing trend, and the duration of their use in-
creased as well, it became evident that the classical
tools that were applied at that time to perform classifi-
cation tasks could be no longer suitable for these new,
very large datasets. For example, among Artificial
Neural Networks (Hertz et al., 1991), the classically
used Multi Layer Perceptron (MLP) models (Rumel-
hart et al., 1985), showing good performance over the
small-sized datasets typically gathered in the nineties,
could no longer effectively face this burden of data.
Consequently, new structures, as auto-encoders and
Restricted Boltzmann Machines, were put forward.
Later on, it was noted that those structures can be
stacked so as to obtain networks with a high num-
ber of internal layers, referred to as Deep Neural Net-
works (DNNs) (LeCun et al., 2015). The good news
was that these latter can be trained one layer at a time,
which helps strongly reducing the problems of van-
ishing gradient and over-fitting. Structures as stacked
auto-encoders, deep belief networks, and convolu-
tional networks have become very popular in these
last years.
As a consequence, classification over largely-
sized datasets can be effectively performed by taking
advantage of the DNNs, that are nowadays the stan-
dard de facto in many fields and for many applica-
tion problems (Najafabadi et al., 2015), with excel-
lent results being obtained in, e.g., computer vision,
speech recognition, and machine translation, apart
from, of course, classification. TensorFlow (Abadi
et al., 2016) is probably the most widely used open-
source software library for machine learning, and it is
extremely popular to experiment with DNNs.
Yet, these powerful methods suffer from one im-
portant drawback. Namely, given a dataset onto
De Falco I., De Pietro G., Della Cioppa A., Sannino G., Scafuri U. and Tarantino E.
Preliminary Steps towards Efficient Classification in Large Medical Datasets: Structure Optimization for Deep Learning Networks through Parallelized Differential Evolution.
DOI: 10.5220/0006730006330640
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (HEALTHINF 2018), pages 633-640
ISBN: 978-989-758-281-3
Copyright
c
2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

which classify, the user has to manually configure
the DNN. This means to perform the choice of a set
of parameters, as the number of hidden layers, the
configuration of each layer, and the setting of some
more parameters related to the learning. Finding good
ways to effectively face this multivariable optimiza-
tion problem is a far-from-trivial task, and may highly
impact the results that can be obtained in the anal-
ysis of Big Data. As concerns possible ways to ef-
fectively make this set of choices, the state of the art
is quite simple, as it relies on approaches based ei-
ther on user ability with a trial-and-error approach, or
on just slightly more sophisticated algorithm-driven
approaches as the so-called grid search and random
search. In the former, for each configuration parame-
ter a suitable range is chosen, so that a uniform grid
is obtained in the search space of the possible con-
figurations, and uniform sampling takes place in the
grid. In the latter, instead, the search is not uniform,
rather some parameters are considered as more im-
portant and the search samples more thoroughly the
configuration space along these parameters. The ac-
tivity involved in all of the three above methods is
very laborious for the user, and there is no guarantee
that the obtained structure is satisfactory.
In this paper we move some preliminary steps to
help users overcome this tricky step. Namely, we
propose the use of an Evolutionary Algorithm (EA)
(B
¨
ack et al., 1997) to find a good parameter setting
for the DNN. EAs are stochastic heuristic optimiza-
tion algorithms based on mimicking in a computer
the behavior of a population subject to the pressure of
the environment. Although EAs do not provide any
formal proof for convergence, they have frequently
proved their ability to find good sub-optimal solutions
to multivariable optimization problems in many dif-
ferent areas. Actually, many EAs exist. Here we con-
sider Differential Evolution (Price et al., 2006), one
of the most recent and successful EAs.
One well-known drawback of EAs is that they
need time to evolve a good solution, because many
iterations should be performed, in each of which new
solutions must be obtained from the currently avail-
able ones, and each of them should be evaluated in
terms of its quality in solving the given problem. This
drawback becomes of much higher relevance when-
ever the time needed to evaluate the quality of a so-
lution is high, as it is the case of classification over
big datasets. In fact, for such a problem, the evalua-
tion of a possible configuration requires for the related
DNN a learning phase over an extremely large train-
ing set, and this task can require minutes, and even
hours. Consequently, the whole evolution requires
an amount of time ranging from hours to days, up to
weeks. This is not a problem for the classification task
itself, because, once a good model is found, its use to
classify a new, previously unseen, item requires just
an extremely small amount of time, so it is a real-time
activity, yet the turnaround time to obtain the model
can be excessive.
To relieve the search from this drawback, the de-
sign of parallel models for EAs and their implemen-
tation and utilization on parallel machines is of great
help. As here the main problem is related to the
computation of the quality of each proposed DNN
model, the idea is to run a parallelized master-slave
model (Tomassini, 2006) in which the DE population
evolves on a master node of the parallel architecture,
while at each iteration the evaluation of the quality
of each individual is delegated to one of the available
slave nodes. In this way, if there are N slave nodes,
the search can show a speed up of a factor up to about
N, being the time for all the evolutionary operations in
the order of, say, a few seconds, thus negligible with
respect to the evaluation activities.
This paper gives in Section 2 a description of the
state of the art in the use of EAs to optimize ANN and
DNN structures. Section 3 details the methodology
followed, in terms of the software architecture of our
algorithm. The specific medical case study on which
our approach has been tested, i.e. Obstructive Sleep
Apnoea, is described in Section 4, together with the
dataset we have worked on. Section 5 reports on the
experiments performed and on the results obtained.
Finally, Section 6 contains our conclusions and the
related future work.
2 STATE OF THE ART: EAS FOR
NNS
From a historical point of view, the attempt at finding
good configurations for ANNs by means of EAs dates
back to the nineties of last century. In those times
there was a line of research called neuro-evolution.
Basically, two different goals can be seen in this re-
search line. In the first, the aim is to find a satis-
factory structure for an ANN in terms of number of
hidden layers and other learning parameters, whereas
in the second the target is to optimize the connection
weights in an ANN. Sometimes these goals have been
faced at the same time.
The first neuro-evolution paper was probably
(Ronald and Schoenauer, 1994), in which the authors
used a Genetic Algorithm to update the weights of a
network in the simulation of the control of soft land-
ing for a toy lunar module.
In the same year two more papers, i.e. (Gruau

et al., 1994; Angeline et al., 1994) described the use of
two different EAs, respectively Genetic Programming
and Evolutionary Programming, to tackle the evolu-
tion of both network structure and learning parame-
ters.
Since then, lots of papers were written within this
line of research, too numerous to be mentioned here,
as, e.g., (Yao and Liu, 1997; De Falco et al., 1998;
Stanley and Miikkulainen, 2002; Sher, 2012; Kas-
sahun and Sommer, 2005; Siebel and Sommer, 2007;
Edlund et al., 2011).
It is worth mentioning here that in these last
months high interest in neuroevolution is growing
from the major companies and universities dealing
with Big Data, as for example Google, Sentient Tech-
nologies, MIT Media Lab, Johns Hopkins, Carnegie
Mellon, and their number continues to increase, and
so are the related efforts.
As an example, in 2017 a group of Google re-
searchers (Real et al., 2017) used a tailored EA to find
the best possible configuration for a convolutional
network. They faced both CIFAR-10 and CIFAR 100
datasets, and obtained very good results, showing that
neuro-evolution can be useful.
As a further example, in 2017 as well, researchers
from Sentient Technologies (Miikkulainen et al.,
2017) put forward an evolutionary-based methodol-
ogy for the optimization of DNN structures called
CoDeepNeat, and successfully applied it to the task
of captioning images on a magazine website in an au-
tomatic way.
Apart from applications, also research on neu-
roevolution is being carried out. As an example, in
the same year Vasconcellos and Murata (Vargas and
Murata, 2017) suggested a unified representation for
most of the ANN features, and a new method useful
to preserve diversity, called spectrum diversity. The
positive impact of their proposal was shown by the re-
sults obtained on five representative problems, among
which one related to the simulation of the motion of a
car on a hilly path.
3 METHODOLOGY
The software architecture of our approach is sketched
in Algorithm 1, that provides the pseudocode for both
the master module and a generic slave one. The mas-
ter makes here reference to a maximization problem.
The master module performs the evolution rely-
ing on a DE algorithm. Basically, an initial set,
called population, composed by NP candidate solu-
tions, called individuals, is randomly generated. Each
individual x contains the values of the parameters re-
Algorithm 1
Master Process
Begin
best_ind(0) = -1;
f(best_ind(0)) = VALMIN //very negative value
//initial generation
for ( i = 1; i <= NP; i++)
randomly initialize individual x[i](0)
for ( i= 1; i <=NP ; i++)
send solution x[i](0) to a slave
for ( i =1; i <= NP; i++)
receive from the slave the fitness
of x[i](0), f(x[i](0))
if
( f(x[i](0)) > f(best_ind(0) )
best_ind(0) = x[i](0)
f(best_ind(0) = f(x[i](0))
//perform the cycle of generations
for ( t = 1; t <= Max_Gen; t++)
for ( i = 1; i <= NP; i++)
apply DE operators and create a
trial solution ts[i](t)
for ( i = 1; i <= NP; i++)
send trial solution ts[i](t)
to a slave
//create the next population
for ( i =1; i <= NP; i++)
receive the fitness of ts[i](t)
f(ts[i](t)))
if
(f(ts[i](t) >= f(x[i](t))
x[i](t+1) = ts[i](t)
else
x[i](t+1) = x[i](t)
if
(f(ts[i](t) > f(best_ind(t))
best_ind(t) = ts[i](t)
f(nest_ind(t) = f(ts[i](t))
output the best structure found
best_ind(Max_Gen)
End
Slave process:
Begin
for (i = 0; i <= NP; i++)
begin
receive from the master the DNN
structure to evaluate
//Perform the tensorFlow actions:
train the DNN
test the DNN
send to the master the accuracy of
the DNN over the test set
end
end
End
presenting a structure for a DNN. The quality of each
individual is evaluated by means of an invocation to
the fitness function f (this computation will be exe-

cuted by a slave module). Then, starting from the in-
dividuals currently available, a new population is cre-
ated thanks to operators specific to the DE as muta-
tion, recombination, and selection. Namely, in corre-
spondence to the generic i–th individual in the current
population, by application of those operators a new
solution, referred to as trial, is created, and its qual-
ity is evaluated by means of the fitness function. The
individual with the higher fitness between the current
i-th and the trial is inserted in the i-th position of the
population being created. The creation of a new pop-
ulation from the current one is repeated for a number
of iterations, called generations, equal to Max Gen.
At the end of the evolution, the best found solution in
terms of higher fitness value is proposed to the user.
For more details on DE, interested readers can make
reference to (Price et al., 2006).
The slave module, instead, is invoked by the mas-
ter each time the fitness of a newly created individual
has to be evaluated. It receives the values of the con-
figuration parameters encoding for a specific DNN,
prepares a proper file in python language and invokes
the execution of this latter file in the TensorFlow li-
brary (Abadi et al., 2016). TensorFlow executes the
learning phase for the proposed DNN over the train-
ing set, and then yields the accuracy of the trained
network over the test set. The slave sends this value
back to the master and becomes waiting for the next
structure to train and test.
For this classification problem, we consider as the
fitness of a DNN structure the accuracy it achieves
over the test set, i.e. the ratio between the number
of the test set items correctly classified and the to-
tal number of items in the test set. With this choice,
the optimization problem becomes a maximization
problem, aiming at finding structures with as-high-as-
possible accuracy values in the range [0.0 - 100.0].
3.1 The Implementation Choices
To encode a DNN structure into a DE individual,
firstly a number of DNN parameters has to be suitably
chosen. In this preliminary work we have decided to
consider the following parameters:
• the number of hidden layers NHL
• for each of the hidden layers, the number of neu-
rons making up the layer (for the i–layer, it is
NNL i)
• the activation function AF
• the number of learning steps LS
For each of these parameters, an admissible range
has been set, as reported in Table 1.
It should be noted that all the parameters can take
on integer values, but AF, which can take values from
a set of three possible values: rectified linear unit
(relu), hyperbolic tangent (tanh), and sigmoid.
Another important choice is that related to the en-
coding for the parameter values. Actually, DE is well
suited to deal with real-valued problems, whereas the
parameters accounted here can take on integer val-
ues. To make all things consistent, each parameter has
been encoded as a real value in the range [0.0 - 1.0],
and the integer value I represented by a real value R is
given by: I = R · (MAX − MIN) + MIN, where MIN
and MAX represent the minimum and the maximum
admissible values for that parameter, respectively.
4 THE MEDICAL CASE STUDY
4.1 Obstructive Sleep Apnoea
Obstructive sleep apnea (OSA) (McNicholas and
Levy, 2000) is a breathing disorder that takes place in
the course of the sleep and is produced by a complete
or a partial obstruction of the upper airway that man-
ifests itself as frequent breathing stops and starts dur-
ing the sleep. In the medical practise, it is defined as a
cessation of airflow for at least 10 seconds, and people
with OSA disorder stop typically hundreds times per
night, during the sleeping, and each stop lasts about
10–30 seconds.
Statistics report that about 4% of the general pop-
ulation suffer from this condition to some extent, and
it is estimated that fewer than 25% of OSA sufferers
are actually aware that they have this problem (Alqas-
sim et al., 2012). These undiagnosed patients cause,
in the USA for example, a loss of 70 billion dollars,
11.1 billion in damages, and 980 deaths each year (Al-
mazaydeh et al., 2012).
Monitoring OSA, by detecting and classifying the
apnoea episodes, becomes crucial for people suffering
from this condition, especially in case of the follow-
up evaluation of some given medical therapies or cer-
tain drugs, in which it is required to check side effects,
such as sleep or breathing disturbances, like OSA
episodes. In general, the task of aiming at the evalua-
tion of the quality of sleep for a subject and at investi-
gating the presence of OSA episodes during the nights
is highly important in order to ameliorate health con-
Table 1: The ranges for all the considered parameters.
NHL NNL i AF LS
minimum 1 1 1 500
maximum 10 30 3 10,000

ditions for citizens suffering from OSA, and, at the
same time, to reduce both mortality and healthcare-
related costs. In fact, it should be remarked here that
this disease results in problems as asphyxia, hypox-
emia, and awakenings, and often has consequences as
increased heart rate or high blood pressure, and, on
the other hand, may yield long-term symptoms that
negatively influence life quality.
With respect to the literature, there are numer-
ous proposed systems to monitor and classify OSA
episodes in a less invasive and more accurate way
(Shokoueinejad et al., 2017). Among them, some ap-
proaches just use data gathered by a single-channel
ElectroCardioGram (ECG), as for example (Sannino
et al., 2014; De Falco et al., 2015) or (Al-Abed et al.,
2007) in which a three-layer Multi-Layer Perceptron
(MLP) classifier was used, or (Acharya et al., 2011)
in which a four-layered feed-forward neural network
with two hidden layers and 11 neurons was employed
to process five non-linear parameters. However, in all
these works the structures of the networks are manu-
ally configured.
4.2 The Original Database
To perform experiments on the use of an EA to find
a good parameter setting for a DNN able to clas-
sify OSA episodes by using data gathered by a ECG
signal, we have created a new dataset starting from
the apnea-ECG database (Penzel et al., 2000), freely
downloadable from www.physionet.org. The apnea-
ECG database consists of 70 recordings, one for each
patient. Only thirty-five of them contain annotations
about OSA episodes, each of them are related to 1-
minute segment of the record. So, we took into con-
sideration these 35 recordings only.
Among the 35 recordings, 20 (labelled as a01 –
a20) are related to people definitely suffering from
OSA, five (b01 – b05) are borderline, and ten (c01
- c10) are people with no OSA at all or with a very
low level of the disease.
Starting from these recordings, we have created
a new dataset and have let it undergo the classifica-
tion task by the DNN. Namely, we have taken each
recording, and for each 1-minute segment we have
computed the values of a set of twelve typical Heart
Rate Variability (HRV) parameters, related to the fre-
quency domain, the time domain, and the non-linear
domain, as better described in the next subsection.
4.3 The Obtained Dataset
Firstly, each ECG record was cleaned from power
line interference, and muscle and movement artefacts,
by using an innovative recurvise denoising scheme
(Cuomo et al., 2016). Then, the filtered signals were
processed by using Kubios (Niskanen et al., 2004), a
Matlab based software package for event-related bio-
signal analysis able to extract and analyze HRV fea-
tures. Standard linear HRV analysis was performed
according to the guidelines of the European Society of
Cardiology and the North American Society of Pacing
and Electrophysiology (of the European Society of
Cardiology et al., 1996). Additionally, non-linear fea-
tures were computed according to the literature (San-
nino et al., 2014). The computed measurements are:
• Frequency Domain:
– the power in the Ultra-low frequency band:
ULF
– the power in the Very low frequency band: VLF
– the power in the Low frequency band: LF
– the power in the High frequency band: HF
– the total Power of the signal (i.e. the sum of the
four above powers): P
– the low frequency/high frequency ratio: LF/HF
• Time Domain:
– the average value of NN intervals: ANN
– the standard deviation of the average NN inter-
vals: SDANN
– the proportion of NN50 divided by the total
number of NNs, where NN50 is the number of
pairs of successive NNs that differ by more than
50 ms: pNN50
– the square root of the mean squared difference
of successive NNs: rMSSD
• Non-linear Domain:
– the approximate entropy: AE
– the fractal dimension: FD
Each database item is constituted by those 12 val-
ues, together with the class of the instance as known
from the annotations related to that recording. These
latter will be represented by a 1 for a non-apnoea
minute and by a 2 for an apnoea minute. The resulting
dataset is composed by a total of 11,752 items, that
are then divided into a training set and a testing set
consisting of 7,051 and 4,701 items, respectively. Ta-
ble 2 shows the details of the data used for this study.
5 THE EXPERIMENTS
Both the master and the slave processes have been
implemented in C language. The hardware available
for our experiments is an iMac Pro platform endowed

with processing nodes constituted by cores running at
3,0 GHz. Ten of them have been used by us to run the
slave processes, and one to execute the master one.
Depending on the value chosen for the population size
NP, each of the cores reserved for the slaves will ex-
ecute several such processes at each generation. For
example, should NP be equal to 30, each slave would
be called upon three times at each generation.
To evaluate the fitness of each proposed DNN con-
figuration, we have made use of a TensorFlow pro-
gram based on the DNNClassifier function. Python
version is the 3.6.3, whereas for TensorFlow the 1.3.0
version has been employed. The evaluation of the
quality of each DNN requires an amount of time in-
dicatively ranging within about one and three min-
utes, depending on the structure and on the number
of steps proposed by the DE for the specific network.
The experiments have been divided into two
phases, as described in the following.
5.1 Manual Configuration
A first phase of the experiments has consisted in let-
ting a user find manually the best configuration for
the DNN by performing a wide set of trials. For each
manually-configured network the parameters values
used are within the limits shown in Table 1.
The first remark related to this phase of man-
ual settings is that the vast majority of the
manually-tested DNN configurations yields as accu-
racy 52.99%, which tells us that the problem is far
from easy. In all these situations, all the test set items
are assigned to the majority class (non-OSA), which
implies no understanding at all of the problem. As
an example of this situation, Table 3 shows the con-
fusion matrix for the DNN constituted by 5 hidden
layers with 10, 20, 20, 20, 10 neurons respectively,
rectified linear unit as the activation function, and a
number of steps equal to 3000.
As a result of this laborious manual phase, the best
configuration tailored by hand has resulted to be the
following: 3 hidden layers with 15, 15, 15 neurons re-
spectively, rectified linear unit as the activation func-
tion, and a number of steps equal to 3000. This con-
figuration yields a percentage of accuracy over the test
set equal to 57.69%.
Table 2: The details of the data used for this work.
class 1 class 2
OSA no-OSA Total
episodes episodes
Training Set 3,707 3,344 7,051
Testing Set 2,491 2,210 4,701
Total 6,198 5,554 11,752
Table 4 reports the confusion matrix obtained by
using this configuration.
As it can be seen, this DNN attempts to predict
by assigning the majority of the items related to class
1 events to the class 2. This means that actually the
problem is not well understood by the network un-
der account, and results in a large number of false
positives (top right), so that the specificity is very
low. From a medical viewpoint, a large number of
non-OSA segments is incorrectly considered as OSA
events. As a general comment, this DNN structure is
quite unsatisfactory.
5.2 DE-driven Configuration
The next phase has resided in using the parallelized
version of the DE in order to improve the above re-
sult. Before carrying out the experiments, several de-
cisions should be taken about DE. Firstly, DE can use
many different search strategies. Within this paper we
have set it as a DE/best/1/bin, the actions of which can
be found in (Price et al., 2006). Basically, to create
each new trial individual, a difference vector between
two individuals randomly chosen in the current pop-
ulation is added to the current best individual. As for
the DE search parameters, we have set them as fol-
lows: NP = 50, Max Gen = 30, F = 0.2, CR = 0.2. All
these choices have been made without any prelimi-
nary tuning phase for the values, and are based on our
experience on the use of DE to face other problems.
The value chosen for NP implies that each slave core
will be called upon five times at each generation.
The best configuration obtained at the end of the
run is a DNN composed by three hidden layers with
respectively 4, 16, and 15 units, with relu as activation
function and a number of steps equal to 5012. Its ac-
curacy over the test set is equal to 68.35%. The time
needed to find it has been of about five hours and 10
minutes.
The most important result is that a noticeable im-
provement has been obtained in terms of accuracy
over the manual configuration.
Figure 1 shows the best fitness value obtained at
each generation as a function of the generations. Also
the average of the fitness values of the individuals at
each generation is reported.
As it can be seen, already in the first generations
Table 3: The confusion matrix for the majority of hand-
made configurations.
assigned to assigned to
class 1 class 2
real class 1 2491 0
real class 2 2210 0
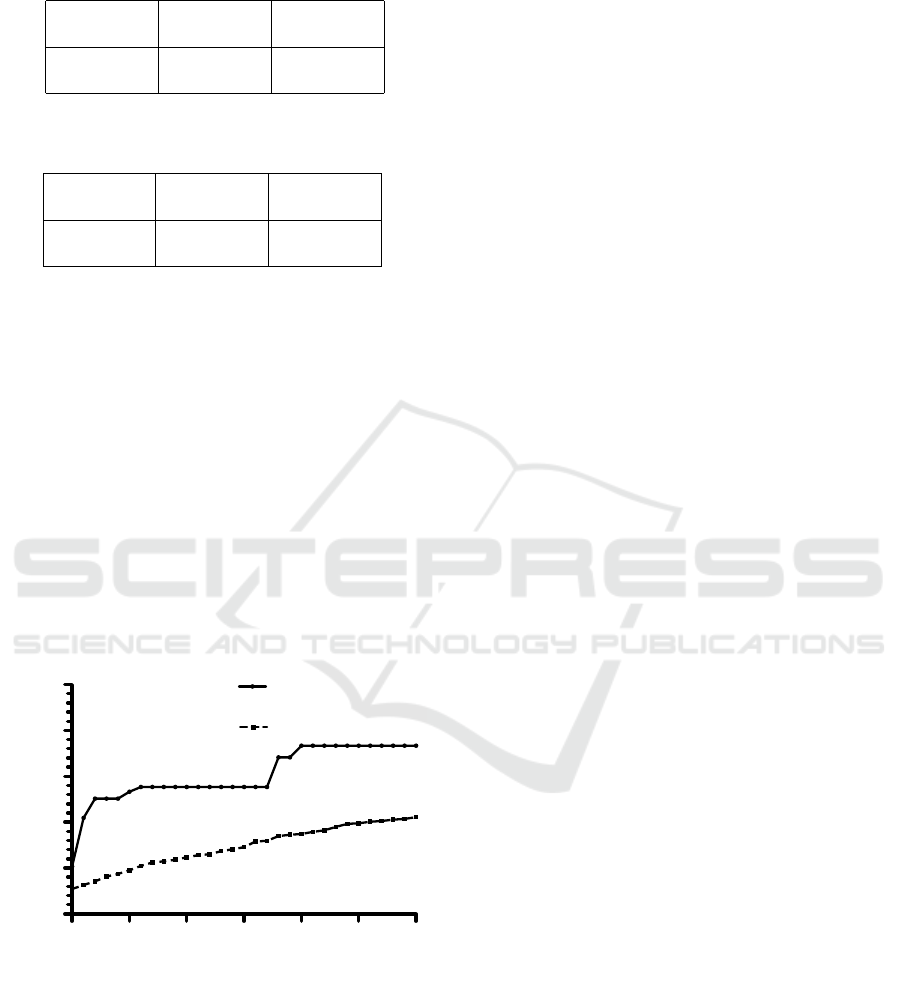
Table 4: The confusion matrix for the best hand-made con-
figuration found.
assigned to assigned to
class 1 class 2
real class 1 1018 1473
real class 2 516 1694
Table 5: The confusion matrix for the best solution provided
by DE.
assigned to assigned to
class 1 class 2
real class 1 1671 820
real class 2 668 1542
the DE allows finding improving solutions. More-
over, as the number of generations increases, so do
both the best fitness value (sometimes) and the aver-
age fitness value (very frequently). These two trends
show that the evolution is effective in finding better
and better configurations.
Moreover, Table 5 shows the confusion matrix for
the best solution evolved by DE.
In this case the results show that evolution has pro-
vided a DNN structure that does not attempt to assign
the items by relying on the majority class, so under-
standing of the problem has been obtained. The num-
ber of false positives shown in the top right corner of
the table is much lower than that for the hand-tailored
solution, about half, which is a much better situation
from the medical viewpoint.
0 5 10 15 20 25 30
50
55
60
65
70
75
best fitness
average fitness
generations
accuracy
Figure 1: The evolution of the DE run.
6 CONCLUSIONS AND FUTURE
WORK
This paper has described some preliminary steps to-
wards finding good configurations for Deep Neural
Network structures, which is a very important prob-
lem in general, and particularly in the medical do-
main, especially when highly-sized data sets are to
be faced. The first such step consists in the use of
Differential Evolution, and the second lies in using a
parallelized version based on a master-slave model in
order to reduce the turnaround time.
The preliminary results obtained here show that
this approach can be useful in easily obtaining struc-
tures allowing increases in the accuracy with respect
to those provided by humans.
There are many issues that have not been consid-
ered here, yet they are of high importance to further
improve the validity of the approach. Among them
the most important is that this approach should be
tested on more datasets, and special attention should
be paid to those with higher sizes, so as to investi-
gate its usefulness when Big Data are to be faced. In
this latter case it is very likely that the times needed to
find good Deep Neural Network structures will highly
increase, resulting in days of turnaround time. This
problem could require the use of larger, more power-
ful parallel machines, consisting of a larger number
of computing nodes so as to test many more possible
configurations at the same time.
Moreover, experiments should be conducted on
the use of distributed models for EAs (Tomassini,
2006) to find good DNN configurations. In fact, these
models have been successfully employed in recent
years for many multivariable problems, resulting in
many cases in both improvement in solution quality
and reduction in the time needed to find a good solu-
tion. Some examples of this can be found in (De Falco
et al., 2014; De Falco et al., 2017) with reference to
the design and the implementation of distributed DE
models.
REFERENCES
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z.,
Citro, C., Corrado, G. S., Davis, A., Dean, J., Devin,
M., et al. (2016). Tensorflow: Large-scale machine
learning on heterogeneous distributed systems. arXiv.
Acharya, U. R., Chua, E. C.-P., Faust, O., Lim, T.-C., and
Lim, L. F. B. (2011). Automated detection of sleep
apnea from electrocardiogram signals using nonlinear
parameters. Physiological measurement, 32(3):287.
Al-Abed, M., Manry, M., Burk, J. R., Lucas, E. A., and
Behbehani, K. (2007). A method to detect obstruc-
tive sleep apnea using neural network classification of
time-frequency plots of the heart rate variability. In
IEEE Int. Conf. of Engineering in Medicine and Biol-
ogy Society, pages 6101–6104.
Almazaydeh, L., Elleithy, K., and Faezipour, M. (2012).
Detection of obstructive sleep apnea through ecg sig-

nal features. In IEEE Int. Conf.on Electro/Information
Technology (EIT), pages 1–6.
Alqassim, S., Ganesh, M., Khoja, S., Zaidi, M., Aloul, F.,
and Sagahyroon, A. (2012). Sleep apnea monitor-
ing using mobile phones. In IEEE 14th Int. Conf.
on e-Health Networking, Applications and Services
(Healthcom), pages 443–446.
Angeline, P. J., Saunders, G. M., and Pollack, J. B. (1994).
An evolutionary algorithm that constructs recurrent
neural networks. IEEE transactions on Neural Net-
works, 5(1):54–65.
B
¨
ack, T., Fogel, D., and Michalewicz, Z. (1997). Handbook
of evolutionary computation. Release, 97(1):B1.
Cuomo, S., De Pietro, G., Farina, R., Galletti, A., and San-
nino, G. (2016). A revised scheme for real time ecg
signal denoising based on recursive filtering. Biomed-
ical Signal Processing and Control, 27:134–144.
De Falco, I., De Pietro, G., and Sannino, G. (2015). On find-
ing explicit rules for personalized forecasting of ob-
structive sleep apnea episodes. In IEEE Int. Conf. on
Information Reuse and Integration (IRI), pages 326–
333.
De Falco, I., Della Cioppa, A., Maisto, D., Scafuri, U.,
and Tarantino, E. (2014). An adaptive invasion-based
model for distributed differential evolution. Informa-
tion Sciences, 278:653–672.
De Falco, I., Della Cioppa, A., Scafuri, U., and Tarantino,
E. (2017). Exploiting diversity in an asynchronous
migration model for distributed differential evolution.
In Genetic and Evolutionary Computation Conf. Com-
panion (GECCO), pages 1880–1887.
De Falco, I., Iazzetta, A., Natale, P., and Tarantino, E.
(1998). Evolutionary neural networks for nonlinear
dynamics modeling. In Parallel Problem Solving from
Nature (PPSN), pages 593–602.
Edlund, J. A., Chaumont, N., Hintze, A., Koch, C., Tononi,
G., and Adami, C. (2011). Integrated information in-
creases with fitness in the evolution of animats. PLoS
computational biology, 7(10):e1002236.
Gruau, F. et al. (1994). Neural network synthesis using cel-
lular encoding and the genetic algorithm.
Hertz, J. A., Krogh, A. S., and Palmer, R. G. (1991). Intro-
duction to the theory of neural computation, volume 1.
Kassahun, Y. and Sommer, G. (2005). Efficient rein-
forcement learning through evolutionary acquisition
of neural topologies. In ESANN, pages 259–266.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learn-
ing. Nature, 521(7553):436–444.
McNicholas, W. and Levy, P. (2000). Sleep-related breath-
ing disorders: definitions and measurements.
Miikkulainen, R., Liang, J., Meyerson, E., Rawal, A., Fink,
D., Francon, O., Raju, B., Navruzyan, A., Duffy, N.,
and Hodjat, B. (2017). Evolving deep neural net-
works. arXiv.
Najafabadi, M. M., Villanustre, F., Khoshgoftaar, T. M.,
Seliya, N., Wald, R., and Muharemagic, E. (2015).
Deep learning applications and challenges in big data
analytics. Journal of Big Data, 2(1):1.
Niskanen, J.-P., Tarvainen, M. P., Ranta-Aho, P. O., and
Karjalainen, P. A. (2004). Software for advanced
hrv analysis. Computer methods and programs in
biomedicine, 76(1):73–81.
of the European Society of Cardiology, T. F. et al. (1996).
Heart rate variability: standards of measurement,
physiological interpretation, and clinical use. Circu-
lation, 93:1043–1065.
Penzel, T., Moody, G. B., Mark, R. G., Goldberger, A. L.,
and Peter, J. H. (2000). The apnea-ecg database. In
Computers in cardiology 2000, pages 255–258.
Price, K., Storn, R. M., and Lampinen, J. A. (2006). Differ-
ential evolution: a practical approach to global opti-
mization.
Real, E., Moore, S., Selle, A., Saxena, S., Suematsu, Y. L.,
Le, Q., and Kurakin, A. (2017). Large-scale evolution
of image classifiers. arXiv.
Ronald, E. and Schoenauer, M. (1994). Genetic lander: An
experiment in accurate neuro-genetic control. In Int.
Conf. on Parallel Problem Solving from Nature, pages
452–461.
Rumelhart, D. E., Hinton, G. E., and Williams, R. J. (1985).
Learning internal representations by error propaga-
tion. Technical report, California Univ San Diego La
Jolla Inst for Cognitive Science.
Sannino, G., De Falco, I., and De Pietro, G. (2014). An
automatic rules extraction approach to support osa
events detection in an mhealth system. IEEE jour-
nal of biomedical and health informatics, 18(5):1518–
1524.
Sher, G. I. (2012). Handbook of neuroevolution through
Erlang.
Shokoueinejad, M., Fernandez, C., Carroll, E., Wang, F.,
Levin, J., Rusk, S., Glattard, N., Mulchrone, A.,
Zhang, X., Xie, A., et al. (2017). Sleep apnea: a re-
view of diagnostic sensors, algorithms, and therapies.
Physiological measurement, 38(9):R204.
Siebel, N. T. and Sommer, G. (2007). Evolutionary re-
inforcement learning of artificial neural networks.
International Journal of Hybrid Intelligent Systems,
4(3):171–183.
Stanley, K. O. and Miikkulainen, R. (2002). Evolving neu-
ral networks through augmenting topologies. Evolu-
tionary computation, 10(2):99–127.
Tomassini, M. (2006). Spatially structured evolutionary al-
gorithms: artificial evolution in space and time.
Vargas, D. V. and Murata, J. (2017). Spectrum-diverse
neuroevolution with unified neural models. IEEE
transactions on neural networks and learning systems,
28(8):1759–1773.
Yao, X. and Liu, Y. (1997). A new evolutionary system
for evolving artificial neural networks. IEEE trans. on
neural networks, 8(3):694–713.
