
Segmentation of Cell Membrane and Nucleus
by Improving Pix2pix
Masaya Sato
1
, Kazuhiro Hotta
1
, Ayako Imanishi
2
, Michiyuki Matsuda
2
and Kenta Terai
2
1
Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
2
Kyoto University, Kyoto, Japan
Keywords: Semantic Segmentation, Generative Adversarial Network, Pix2pix, Cell Membrane and Cell Nucleus.
Abstract: We propose a semantic segmentation method of cell membrane and nucleus by improving pix2pix. We use
pix2pix which is an improved method of DCGAN. Pix2pix generates good segmentation result by the
competition of generator and discriminator but pix2pix uses generator and discriminator independently. If
generator knows the criterion for classifying real and fake images, we can improve the accuracy of
generator furthermore. Thus, we propose to use the feature maps of the discriminator into generator. In
experiments on segmentation of cell membrane and nucleus, our proposed method outperformed the
conventional pix2pix.
1 INTRODUCTION
In the field of cell biology, cell biologists segment
cell membrane and cell nucleus manually now.
Manual segmentation takes cost and time. In
addition, the segmentation results become subjective.
Therefore, an automatic segmentation method is
desired. In recent years, segmentation accuracy is
much improved by the progress of deep learning.
Thus, we can develop an automatic segmentation
method with high accuracy now.
The effectiveness of encoder-decoder
convolutional neural network (CNN) such as the
SegNet (Vijay Badrinarayanan et al., 2017) and the
U-net (Olaf Ronneberger et al., 2015) for semantic
segmentation is reported. At first, we tried to use
encoder-decoder CNN for segmentation of cell
membrane and cell nucleus. However, segmentation
accuracy is not sufficient. Therefore, we tried to use
pix2pix (Phillip Isola et al., 2017) which is the
improved version of Generative Adversarial
Networks (GAN) ((Ian Goodfellow at el., 2014),
(Emily Denton at el., 2015)). Since pix2pix can train
the transformation between input and output images,
we can use it for segmentation (Masaya Sato et al.,
2017).
Pix2pix consists of generator and discriminator.
Generator produces an output image from the input
image. Discriminator classifies whether the output
image of the generator is real or fake. Two networks
are adversarial relationship. Pix2pix generates the
good segmentation result by the competition
between generator and discriminator in comparison
with the encoder-decoder CNN. However, the
accuracy of cell membrane is still low. Further
improvement is required.
In this paper, we try to improve the generator in
pix2pix. The goodness of the generator is evaluated
by the discriminator. Thus, knowing the important
features in discriminator is important for improving
the generator. For example, cell membrane is not
broken. If discriminator judges real and fake images
by using the knowledge, the information is effective
for generator to generate good segmentation result.
Thus, we use the feature maps of discriminator in
generator. Concretely, we concatenate the feature
maps of discriminator with the encoder part of the
generator. Generator can learn how discriminator
judges real or fake and use the information to
improve the accuracy.
In experiments, we used 50 fluorescence images
of the liver of transgenic mice that expressed
fluorescent markers on the cell membrane and in the
cell nucleus. 40 images are used for training and
remaining 10 images are used for evaluating the
accuracy. Ground truth images are made by human
experts. Therefore, the number of images is small.
216
Sato, M., Hotta, K., Imanishi, A., Matsuda, M. and Terai, K.
Segmentation of Cell Membrane and Nucleus by Improving Pix2pix.
DOI: 10.5220/0006648302160220
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 4: BIOSIGNALS, pages 216-220
ISBN: 978-989-758-279-0
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
We confirmed that the segmentation accuracy is
improved in comparison with conventional pix2pix.
The effectiveness of generator using the feature
maps of discriminator is demonstrated.
This paper is organized as follows. In section 2,
we explain our proposed method. Experimental
results on segmentation of cell membrane and cell
nucleus are shown in section 3. Section 4 describes
conclusion and future works.
2 PROPOSED METHOD
As described previously, we improve pix2pix by
using the feature maps of discriminator in generator.
At first, we explain pix2pix in section 2.1. After
that, we explain the proposed method in section 2.2.
2.1 Pix2pix
Pix2pix is an improved version of DCGAN. The
difference from standard DCGAN is the input of
generator. The input of standard DCGAN is a vector
(e.g.100 dimensions) but the input of pix2pix is an
image. Pix2pix trains the transformation from input
image to output image (e.g. from gray scale to color
image).
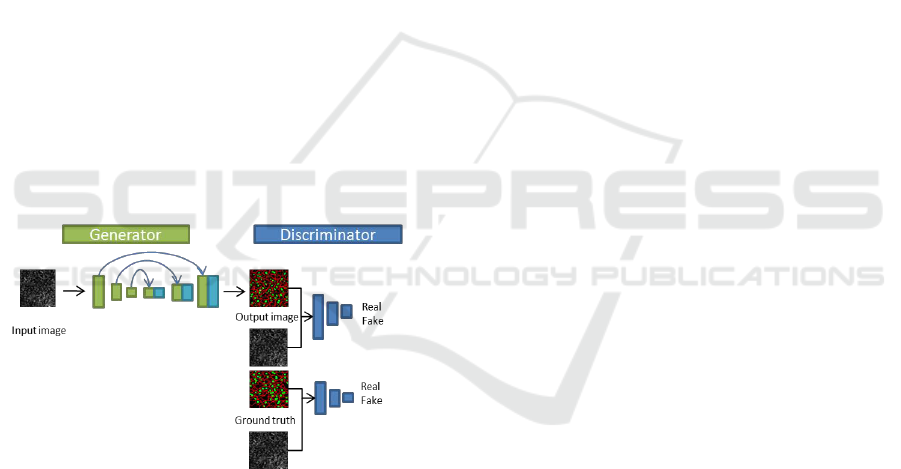
Figure 1: The structure of pix2pix.
The structure of pix2pix is shown in Figure 1.
Figure shows that generator makes the output image
from an input image and the discriminator classifies
it into real or fake class. In pix2pix, CNN is used as
the discriminator and the U-net is used as the
generator.
U-net is a kind of encoder-decoder CNN. The
main difference is the paths from encoder to decoder.
The paths prevent to lose small objects by encoder.
In other words, the multi-scale information is used to
generate an output image effectively.
Discriminator evaluates the goodness of
generator in pix2pix. However, they are independent
of each other, and the information in discriminator is
not used effectively in generator. Thus, we use
feature maps of discriminator in generator to make
good segmentation result which is similar as real
ground truth.
2.2 Our Method
In our method, the information in discriminator is
used into generator. To teach the generator how the
discriminator classifies real or fake, the feature maps
in discriminator are concatenated with encoder part
in generator. To do so, the paths are added to the
encoder part in generator from discriminator like U-
net.
Figure 2 shows the structure of our method.
Lower blue dot square shows the detail of blue dot
square in upper figure. In our method, two
generators and discriminators are used because we
want to use the feature maps of discriminator in
generator. The blue rectangles show the feature
maps of discriminator and green rectangles show the
feature maps of generator. At first, the 1st generator
which is the same as pix2pix makes an output image,
and the output image is fed into the 1st discriminator.
The feature maps of the 1st discriminator are used in
the 2nd generator. The output image is fed into the
2nd discriminator. The structure of both
discriminators is the same though that of two
generators is different.
The paths are added to the feature maps with the
same size between the 1st discriminator and the 2nd
generator. Since the size of feature map at the 1st
convolution layer in the 1st discriminator is 128 x
128 x 64, it is concatenated with the 1st convolution
layer of the encoder in the 2nd generator with 128 x
128 x 128. Thus, the size of the concatenated feature
map is 128 x 128 x (64+128). In the following
experiments, we concatenated all feature maps in the
1st discriminator to the encoder part of the 2nd
generator.
3 EXPERIMENTS
We used fluorescence images of the liver of
transgenic mice that expressed fluorescent markers
on the cell membrane and in the cell nucleus. There
are 50 images in total. The size of the image is 256 x
256 pixels. We divided those images into 40 training
images and 10 test images. We use two evaluation
measures; Global accuracy and Class average
accuracy. Global accuracy is correct classification
rate in all pixels. This is influenced by objects with
Segmentation of Cell Membrane and Nucleus by Improving Pix2pix
217
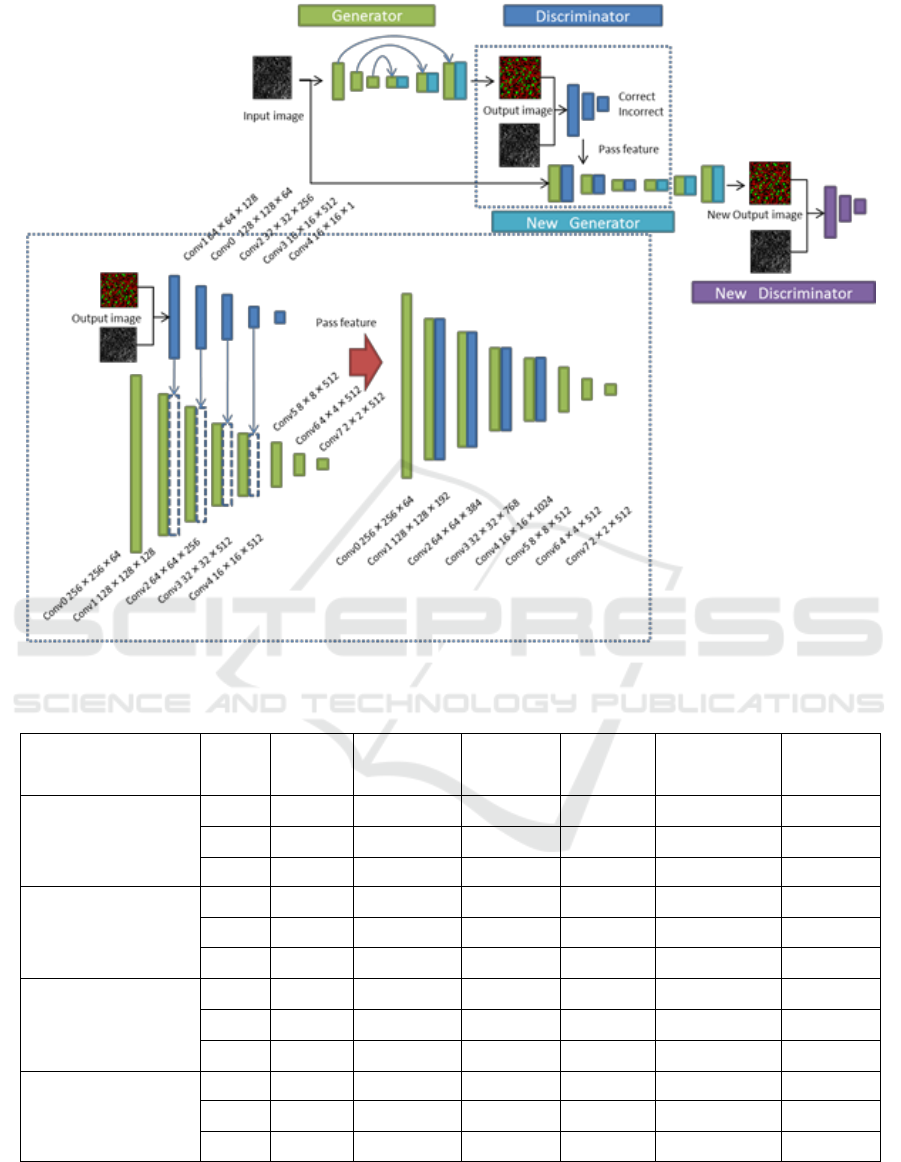
Figure 2: The structure of our method.
Table 1: The accuracy of our method.
Method
Try
time
Epoch
Global
accuracy
Class
average
Back
ground
Cell
membrane
Cell
nucleus
pix2pix
1
1438
77.15%
71.49%
86.86%
54.46%
73.14%
2
1124
77.01%
71.30%
86.77%
54.26%
72.87%
3
1102
77.11%
71.10%
87.31%
53.50%
72.48%
our
method
(N=2)
1
1490
77.24%
72.01%
85.98%
57.36%
72.69%
2
1440
77.43%
72.40%
85.94%
57.76%
73.51%
3
1442
77.34%
72.30%
85.75%
58.22%
72.92%
our
method
(N=3)
1
1402
76.99%
71.59%
85.99%
56.53%
72.24%
2
1390
76.89%
71.86%
85.33%
57.65%
72.60%
3
1446
76.95%
71.95%
85.40%
57.52%
72.94%
our
method
(N=0,1,2,3)
1
1458
78.00%
72.50%
87.08%
57.64%
72.78%
2
1468
77.96%
72.74%
86.81%
57.43%
73.98%
3
1500
78.02%
72.97%
86.50%
58.56%
73.85%
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
218

Figure 3: Partial view of the segmentation result.
large area such as background. Class average
accuracy is the mean classification rate of each class.
This is influenced by objects with small area such as
cell membrane and nucleus.
The number of epoch in training is set to 1500.
The best model is determined by the minimum loss
of generator.
We evaluate each method three times because
the accuracy changes slightly depending on the
random number. Table 1 shows the accuracy of three
times evaluations. N=2 means that only the 2nd
layer of generator uses feature maps of discriminator.
N=0,1,2,3 means that all feature maps of
discriminator are used in generator.
Table 2: The mean accuracy.
Pix2pix
Ours
(N=2)
Ours
(N=3)
Ours
(N=0,1,2,3)
Global
accuracy
77.09%
77.34%
76.95%
77.99%
Class
average
71.30%
72.24%
71.80%
72.74%
Back
ground
86.98%
85.89%
85.57%
86.80%
Cell
membrane
54.08%
57.78%
57.24%
57.88%
Cell
nucleus
72.83%
73.04%
72.59%
73.53%
Tables show that the usage of feature maps of
discriminator in generator is effective. However,
when only the 2nd or 3rd layer of the generator uses
the features maps of discriminator, the accuracy
improvement is not so large. The results show that
feature maps with multi-resolution in discriminator
are effective to improve the accuracy of generator.
In class average accuracy, our method using all
feature maps improved 1.44% in comparison with
standard pix2pix. Cell membranes are difficult
because of their complicated shapes. However, our
method improved 3.80%. This result demonstrated
the effectiveness of our method.
Figure 3 shows the segmentation results. The
segmentation accuracy of cell membrane by the
proposed method is much better than the
conventional method. Figure 4 shows partial views
of the segmentation results. The images in the upper
row are the enlarged view of a part of the image in
the upper row of Figure 3. When we focus on the
blue rectangles in Figure 4, the cell nucleus is
detected more correctly by our method. The images
in the lower row are the enlarged view of a part of
the image in the second row of Figure 3. When we
focus on the purple rectangles, the segmentation
accuracy of cell membrane by our method is better
than the conventional method. These results
demonstrated the effectiveness of our method using
feature maps of discriminator in generator.
4 CONCLUSIONS
We used feature maps of discriminator in generator
in order to improve the segmentation accuracy. By
teaching the information of discriminator to
generator, the accuracy of cell membrane is much
improved in comparison with standard pix2pix.
However, current accuracy is not sufficient for
automatic segmentation of cell membranes and
Segmentation of Cell Membrane and Nucleus by Improving Pix2pix
219

nucleus. Thus, we must improve segmentation
accuracy further. When we use the feature maps of
discriminator in generator, we just concatenate the
feature maps with the encoder part of generator. We
should use convolution before concatenation. This is
a subject for future works.
ACKNOWLEDGEMENTS
This work is partially supported by MEXT/JSPS
KAKENHI Grant Number 16H01435 “Resonance
Bio”.
REFERENCES
Badrinarayanan, V., Kendall, A. and Cipolla, R. (2017).
SegNet: A Deep Convolutional Encoder-Decoder
Architecture for Image Segmentation. IEEE
Transactions on Pattern Analysis and Machine
Intelligence, Vol.39, No.12, pp.2481-2495.
Ronneberger, O., Fischer, P. and Brox, T. (2015). U-Net:
Convolutional Networks for Biomedical Image
Segmentation. Proc. Medical Image Computing and
Computer-Assisted Intervention, pp.234-241.
Isola, P., Zhu, J., Zhou, T. and Efros, A. A. (2017). Image-
to-Image Translation with Conditional Adversarial
Networks. Proc. IEEE conference on Computer Vision
and Pattern Recognition, pp.5967-5976.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A. and Bengio,
Y. (2014). Generative adversarial nets. Advanced in
Neural Information Processing Systems, Vol.27,
pp.2672-2680.
Denton, E., Chintala, S., Szlam, A. and Fergus, R. (2015).
Deep Generative Image Models using a Laplacian
Pyramid of Adversarial Networks. Advanced in
Neural Information Processing Systems, Vol.28, pp.
1486-1494.
Sato, M., Hotta, K., Imanishi, A., matsuda, M., and Terai,
K. (2017). Segmentation of cell membrane and cell
nucleus Using pix2pix of Local Regions. International
Symposium on Imaging Frontier, p.77
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
220
