
Bayesian Network and Structured Random Forest Cooperative Deep
Learning for Automatic Multi-label Brain Tumor Segmentation
Samya Amiri
1,2
, Mohamed Ali Mahjoub
2
and Islem Rekik
3
1
Isitcom - Higher Institute of Computer Science and Communication Techniques of Hammam Sousse,
University of Sousse, Tunisia
2
LATIS Eniso - National Engineering School of Sousse, University of Sousse, Tunisia
3
BASIRA Lab, CVIP Group School of Science and Engineering, Computing, University of Dundee, U.K.
Keywords: Structured Random Forest, Bayesian Network, Deep Cooperative Network, Autocontext Model, Brain
Tumor Segmentation, MRIs.
Abstract: Brain cancer phenotyping and treatment is highly informed by radiomic analyses of medical images.
Specifically, the reliability of radiomics, which refers to extracting features from the tumor image intensity,
shape and texture, depends on the accuracy of the tumor boundary segmentation. Hence, developing fully-
automated brain tumor segmentation methods is highly desired for processing large imaging datasets. In this
work, we propose a cooperative learning framework for multi-label brain tumor segmentation, which
leverages on Structured Random Forest (SRF) and Bayesian Networks (BN). Basically, we embed both
strong SRF and BN classifiers into a multi-layer deep architecture, where they cooperate to better learn
tumor features for our multi-label classification task. The proposed SRF-BN cooperative learning integrates
two complementary merits of both classifiers. While, SRF exploits structural and contextual image
information to perform classification at the pixel-level, BN represents the statistical dependencies between
image components at the superpixel-level. To further improve this SRF-BN cooperative learning, we
‘deepen’ this cooperation through proposing a multi-layer framework, wherein each layer, BN inputs the
original multi-modal MR images along with the probability maps generated by SRF. Through transfer
learning from SRF to BN, the performance of BN improves. In turn, in the next layer, SRF will also benefit
from the learning of BN through inputting the BN segmentation maps along with the original multimodal
images. With the exception of the first layer, both classifiers use the output segmentation maps resulting
from the previous layer, in the spirit of auto-context models. We evaluated our framework on 50 subjects
with multimodal MR images (FLAIR, T1, T1-c) to segment the whole tumor, its core and enhanced tumor.
Our segmentation results outperformed those of several comparison methods, including the independent
(non-cooperative) learning of SRF and BN.
1 INTRODUCTION
The emergence of the new field of radiomics, which
addresses the conversion of medical images into
mineable data through the extraction of large
amounts of quantitative features (Aerts et al.,2014),
has led to major advances in tumor diagnosis,
phenotyping, and patient treatment planning.
Notably, the reliability of radiomics fundamentally
depends on the accuracy of the tumor boundary
segmentation, as radiomic features are generally
extracted from within and around the tumor lesion.
Hence, fully automated brain tumor segmentation
methods are highly desired. This will in part
alleviate the burden of manually segmenting tumor
lesions on brain images, as well as facilitate the task
of statistically analyzing big brain tumor image
datasets for clinical studies. The large variation in
tumor characteristics (shape, position, texture)
makes the segmentation task very challenging.
To solve this problem, several previous works
considered tumor segmentation as a classification
problem at the pixel, voxel, patch or region level
(Havaei et al., 2017; Koley et al.,2016; Lefkovits et
al.,2016; Christ at al., 2017; Folgoc et al., 2016). In
particular, Random Forest (RF) was previously used
for tumor segmentation, where basically each input
intensity image patch is mapped to a class label at
Amiri, S., Mahjoub, M. and Rekik, I.
Bayesian Network and Structured Random Forest Cooperative Deep Learning for Automatic Multi-label Brain Tumor Segmentation.
DOI: 10.5220/0006629901830190
In Proceedings of the 10th International Conference on Agents and Artificial Intelligence (ICAART 2018) - Volume 2, pages 183-190
ISBN: 978-989-758-275-2
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
183
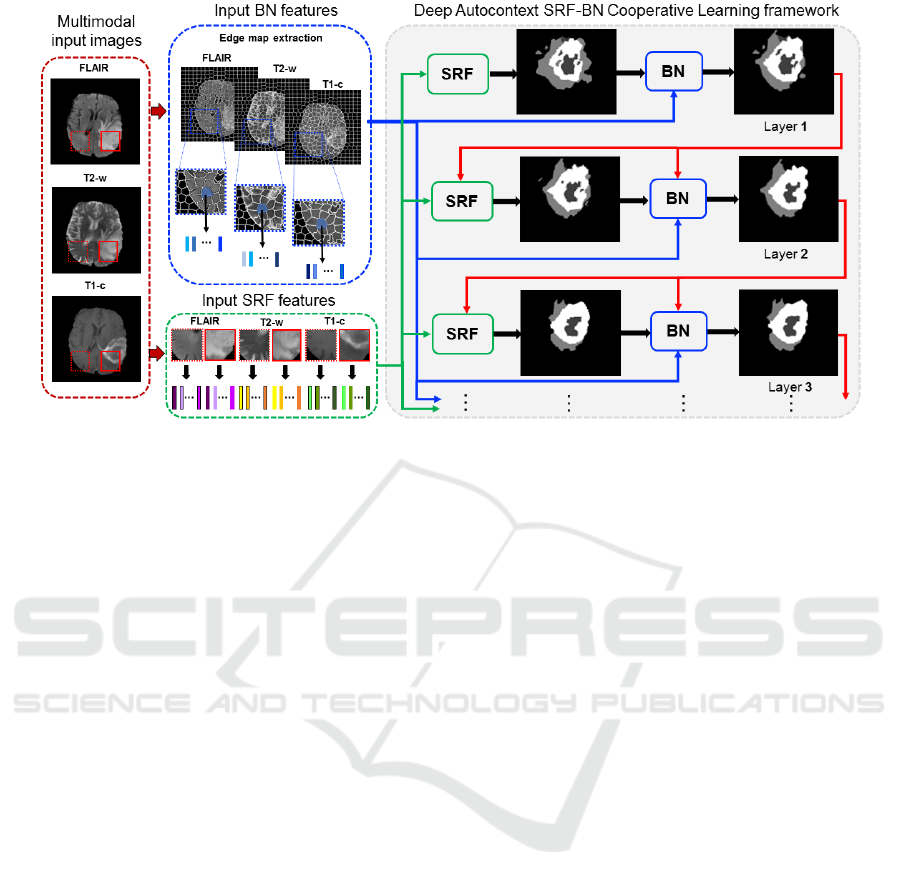
Figure 1: Proposed deep SRF-BN cooperative learning for multi-label tumor lesion segmentation. In each layer, the SRF
inputs different features extracted at the 2D patch-level and generates an intermediate segmentation result, while the BN
inputs different features extracted at the superpixel-level. In the next layer, SRF has two inputs: the original patch features
and the output segmentation result from the previous layer; while BN has three inputs: the original patch features, the output
segmentation result from the previous layer and the output SRF segmentation map in the same layer. This deep multi-layer
cooperative-learning architecture provides contextual information for both classifiers through the intermediate segmentation
maps to gradually improve their learning.
at the center pixel of the patch, thereby performing
patch-to-pixel mapping (Breiman, 2001). As a
variant of RF, Structured Random Forest (SRF) was
used to take into account the image structure when
producing the final segmentation maps, through
estimating a patch-to-patch mapping. This allows
integrating more spatial information through
averaging neighboring output label patches. SRF
demonstrated high-performance in different
challenging classification tasks such as in (Zhang et
al., 2016; Kontschieder et al., 2011; Zhang et al.,
2017).
On one hand, the increasingly popular
convolutional neural networks were used for tumor
segmentation (Havaei et al., 2017). However, fine-
tuning of an entire deep network still requires a lot
of efforts and resources, and SVM-based methods
also involve time consuming grid search and cross
validation to identify good regularization
parameters. In addition, when multiple pre-trained
deep CNN models are available, it is unclear which
pre-trained models are appropriate for target tasks
and which classifiers would maximize accuracy and
efficiency. On the other hand, among all the
graphical models as Neural Networks and decision
trees, Bayesian Networks (BNs) nicely overcome
these limitations. Indeed, they are powerful tools in
first representing probabilistic dependencies and
uncertainty between different image features (Zhang
and Ji, 2008), second modeling and fusing complex
relationships between image features of different
natures (e.g., multimodal features), and third
handling noisy as well as missing signals in images.
Together, these facts made
BNs well suited for multimodal image
classification since they are easily adaptable for
multi-label problems compared to other classifiers
such as SVM, moreover they encode dependencies
between the learned features of different class labels.
While different object segmentation and action
recognition problems in computer vision were
solved based on Bayesian graphical representations
(Panagiotakis et al., 2011; Zhang and Ji, 2011; Yang
at al., 2015), the use of BN remains absent in tumor
segmentation literature.
Although regarded as strong classifiers, both
SRF and BN might suffer from a few limitations
when used separately. For instance, SRF does not
perform well when classifying transitions between
label classes, while BN parameter learning such as
prior probabilities is challenging and
computationally expensive. Combining them
together, may help iron out the weaknesses of each
when used separately as well as leverage on their
strengths (i.e., preserving the learned structural
information for SRF and the integration of
ICAART 2018 - 10th International Conference on Agents and Artificial Intelligence
184

multimodal features for BN). Hence, we propose to
combine both SRF and BN classifiers into a multi-
layer deep architecture, where they can cooperate to
perform joint multi-label brain tumor segmentation.
Our framework incorporates different features
derived from different image components
(superpixel and patch) such as intensity features. We
further integrate contextual features, which capture
semantic relations (i.e., label relations) between
neighboring patches and enforce spatial consistency
between patches within and around the tumor lesion.
The proposed SRF-BN cooperative learning strategy
ensures the transfer of the probability maps
outputted from SRF to BN of the same layer, which
enables the integration of both patch and superpixel
level knowledge in a unified framework.
Additionally, in the spirit of auto-context model, the
output segmentation map of each layer is further
aggregated with the original input features, thereby
augmenting the inputs to subsequent layers. This
allows boosting the classification performance of
both classifiers and improving feature learning,
which progressively refines the segmentation result
from layer to layer.
2 DEEP COOPERATIVE
LEARNING FOR
MULTI-LABEL
CLASSIFICATION
PREPARATION
In the following, we present the main steps of our
multi-label cooperative-learning based segmentation
framework. Fig.1 illustrates the proposed multi-layer
architecture composed of cascaded SRF-BN blocks,
where each block ultimately outputs the BN
posterior probability map fed as semantic context to
the next SRF-BN block of the subsequent layer.
Specifically, in each block excluding the first one,
SRF inputs the intensity patch features of the
original MR scans with the segmentation result (i.e.,
semantic context) of the previous layer. In turn, the
prior probabilities required for BN learning are
statistically computed using (1) the probability
segmentation maps generated by SRF of the same
layer and (2) the BN posterior probability of the
previous layer. In the following sections, we will
present the design of the two components (SRF and
BN) making each block in our deep auto-context
multi-label segmentation architecture.
2.1 Structured Random Forest
SRF is a variant of the traditional Random Forest
classifier, which is able to handle and preserve the
structure of different labels in the image
(Kontschieder et al., 2011). While, standard RF
maps an intensity feature vector extracted from a 2D
patch centered at pixel x to the label of its center
pixel x (i.e., patch-to-pixel mapping), SRF maps the
intensity feature vector to a 2D label patch centered
at x (patch-to-patch mapping). This is achieved at
each node in the SRF tree, where the function that
splits patch features between right and left children
nodes depends on the joint distribution of two labels:
a first label at the patch center x and a second label
selected at a random position within the training
patch (Kontschieder et al.,2011). We also note that
in SRF, both feature space and label space nest
patches that may have different dimensions. Despite
its elegant and solid mathematical foundation as well
as its improved performance in image segmentation
compared with RF, SRF might perform poorly at
irregular boundaries separating different label
classes since it is trained using regularly structured
patches (Kontschieder et al., 2011). Besides, it does
not include contextual information to enforce spatial
consistency between neighboring label patches.
To address these limitations, we propose to
embed SRF into a deep autocontext framework,
where the contextual information is provided by a
Bayesian network which learns to segment the
image at the superpixel level, allowing to better
capture irregular boundaries in the image.
2.2 Bayesian Network
Various BN-based models have been proposed for
image segmentation (Zhang and Ji, 2008;
Panagiotakis et al., 2011; Zhang and Ji, 2011; Guo et
al., 2017). In our work, we adopt the BN architecture
proposed in (Zhang and Ji, 2011). As a
preprocessing step, we first generate the edge maps
from the input MR image modalities (Fig. 1). The
edge map consists of a set of superpixels {Sp
i
},i=1,..,N (or regional blobs) and edge segments
{E
j
},j=1,..,L.
We define our BN as a four-layer network,
where each node in the first layer stores a
superpixel. The second layer is composed of nodes
each storing a single edge from the edge map. The
two remaining layers store the extracted superpixel
features and edge features, respectively. During the
training stage, for BN parameters, we define the
prior probability of p(Sp
i
) as a uniform distribution
Bayesian Network and Structured Random Forest Cooperative Deep Learning for Automatic Multi-label Brain Tumor Segmentation
185

and then learn the conditional probability p(MS
pi
│Sp
i
) representing the relationship between the
superpixel features and their corresponding labels
using a mixture of Gaussians model. In addition, we
empirically define the conditional probability
modeling the relationships between each superpixel
label and each edge state (i.e., true or false edge)
p(E
j
│Pa (E
j
) ), where Pa (E
j
) denotes the parent
superpixel nodes of E
j
.
During the testing stage, we learn the BN
structure encoding the contextual relationships
between superpixels and edge segments.
Specifically, each edge node has for parent nodes the
two superpixel nodes that are separated by this edge.
In other words, each superpixel provides contextual
information to judge whether the edge is on the
object boundary or not. If two superpixels have
different labels, it is more likely that there is a true
object boundary between them, i.e. E
j
=1, otherwise
E
j
=0. Although automatic segmentation methods
based on BN have shown great results in the state-
of-the-art, they may perform poorly in segmenting
low-contrast image regions and different regions
with similar features (Zhang and Ji, 2011). To
further improve the segmentation accuracy of BN,
we propose to include additional information
through cooperative learning using SRF.
2.3 SRF-BN Cooperative Learning
(One Layer)
To take advantage of the strengths of both classifiers
and overcome their limitations, we first propose a
one-layer cooperative learning strategy, where BN
benefits from the learned patch-to-patch mapping by
SRF. First, the trained SRF generates the
segmentation result, using the feature patches
extracted from the different MRI modalities of the
testing subject. Then, BN uses the SRF segmentation
result to define the prior probabilities p(Sp
i
) for each
superpixel region Sp
i
. Hence, with this cooperative
learning, the BN prior probabilities are estimated
based on the input SRF segmentation probability
maps. Such one-layer cooperative learning strategy
only boosts the BN performance since it is
performed in one way (from SRF to BN), while SRF
does not benefit from BN learning.
2.4 Deep SRF-BN Cooperative
Learning (Two Layers)
To address the aforementioned limitation of the one-
layer SRF-BN architecture, we further propose to
deepen the cooperative learning between SRF and
BN in the spirit of auto-context model (Tu and Bai,
2010). Basically, in the proposed deep auto-context
SRF-BN cooperative learning architecture, each
layer inputs the segmentation result of the previous
layer to boost the per formance of both SRF and BN
classifiers. In each layer, excluding the first one,
SRF classifier inputs the segmentation result of the
previous layer along with the original input
multimodal feature patches (Fig. 1). This allows the
integration of contextual features learned at both the
patch level (from SRF in the previous layer) and
superpixel level (from BN in the previous layer).
Similarly, BN inputs the segmentation result of the
previous layer along with the original input
multimodal superpixel features, while adding the
probability segmentation map output of the SRF in
the same layer. In this way, BN prior probabilities
are updated in each layer based on the posterior
probability of the previous layer and the SRF
probability map in the same layer.
2.5 Preprocessing and Features
To improve the performance of our segmentation
framework, we perform a few preprocessing steps.
Hence, we apply the N4 filter for inhomogeneity
correction, and use the histogram linear
transformation for intensity normalization. To train
the previous models, we use conventional features
(e.g., patch intensity) and we also propose a rich
feature set as follows:
▪ Statistical Features: First order operators
(mean, standard deviation, max, min, median,
Sobel, gradient); higher order operators
(laplacian, difference of gaussian, entropy,
curvatures, kurtosis, skewness); texture features
(Gabor filter); spatial context features
(symmetry, projection, neighborhoods)
(Prastawa et al., 2004).
▪ Symmetric Features: This is originally used to
describe and to exploit the symmetrical
properties of the brain structure. Thus, we define
the symmetry descriptor characterizing the
differences between symmetric pixels with
respect to the mid-sagittal plane. The adopted
symmetry measure is the intensity variance.
3 RESULTS AND DISCUSSION
In this section, we display the evaluation results of
our proposed brain tumor segmentation framework
on the Brain Tumor Image Segmentation Challenge
(BRATS, 2015) dataset. It contains brain MRI scans
ICAART 2018 - 10th International Conference on Agents and Artificial Intelligence
186
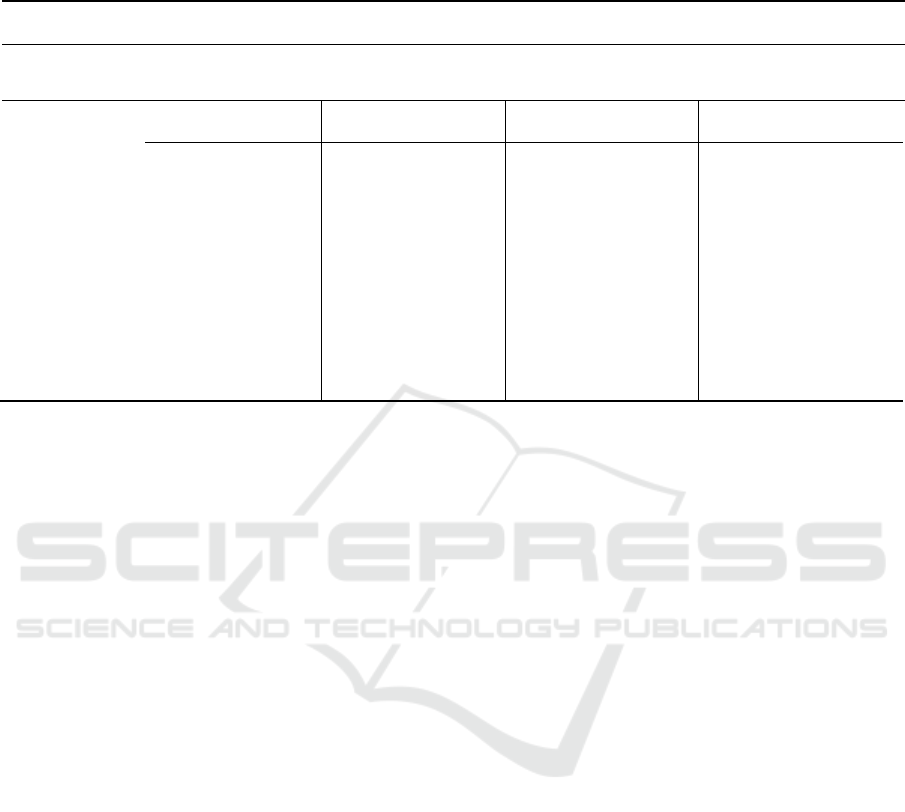
Table 1: Segmentation results of the proposed framework and comparison methods averaged across 50 patients.(HT: whole
Tumor; CT: CoreTumor; ET: Enhanced Tumor; L=i, i=1,_,3 denotes the number of layers; * indicates outperformed
methods with p-values<0.05.
Results
Dice score
Features
Intensity
Intensity+symmetry
descriptor
Intensity + statistical
features
Intensity + statisical
features+symmetry
descriptor
Methods
HT
CT
ET
HT
CT
ET
HT
CT
ET
HT
CT
ET
Deep-AC
SRF-BN(L=3)
86,8
75,9
73,8
87,2
76,3
75
89,09
78,4
78,9
89,1
80,92
79,2
Deep-AC
SRF-BN (L=2)*
85
73,65
70,6
85,4
74,87
73,3
88,79
77,8
78,1
88,9
78,2
78,9
SRF-BN (L=1)*
79,2
70,15
69
80
70,85
69
82,5
72,6
70
83,6
72,88
70,05
AC-SRF*
75
58
32
75,29
58,69
32,5
80
70,05
37,12
80,23
73
37,5
SRF*
72
56
31
72,9
57
31
75
60
35
75,8
61
35,2
BN*
62,96
42
30
65
43
30,8
70,8
45
32
71,3
45
33
for more than 200 patients with high-grade gliomas.
For each patient, the four MRI modalities along with
the corresponding manually labeled glioma's
segmentation are available; they are rigidly
coregistered and resampled to a common resolution.
To generate the oversegmented MR image
modalities we extract the edge-map from the FLAIR
MRI using SLIC oversegmentation algorithm
(Achanta et al., 2010) then we apply it for the
corresponding T1.c and T2 MRIs. We fix the
number of superpixels to 1000 and the compactness
to 10.
For the BN model, the conditional probabilities
modeling the relationships between the superpixel
labeling and the edge state are fixed as follows:
p(E
j
│Pa (E
j
) ) = 0.8 if the parent region nodes have
different labels and 0.2 otherwise. For the SRF, we
use a 10x10 feature patches and a 7x7 label patches
to train 15 trees using 500 iterations for the node
tests.
In our experiments we show a comparison to
several baseline methods: SRF and BN used solely,
the auto-context SRF (AC-SRF) and SRF-BN
cooperative learning approach (SRF-BN (L=1)).
Besides, we test the influence of the layer's number
on our deep auto-context SRF-BN framework. The
classifiers were trained using leave-one-patient
cross-validation experiments. The quality of the
obtained segmentation was evaluated on the basis of
the manually annotated ground truth using the well-
known Dice index. In Fig. 2, we show some
qualitative segmentation results and in Table. 1 and
Fig. 3 we provide the mean Dice index over 50
testing subject randomly taken from the Brats
dataset.
According to the qualitative and quantitative
results, the proposed segmentation approach clearly
outperforms the baseline methods, independently of
the number of layers, with highly statistical
significance (p_value <0.05). This proves that (1)
leveraging the two classifiers does alleviate their
limitations (2) the integration of the superpixel
features and patch features in a deep manner boosts
the performance of the segmentation framework (3)
the information fusion of multiple image modalities.
The feature set highly influences the
classification results of our framework as well as the
baseline methods. In this respect, the weak effect of
the symmetry descriptor can be explained by the
miss-detection of the mid-sagittal plane for some
subjects.
In addition, we have performed 5-cross
validation on the whole dataset (220 subjects) to
benchmark our results against (Zhao et al., 2016)
which integrated a Fully Convolutional Neural
Network and Conditional Random Fields for
BRATS 2015 segmentation. Our deep SRF-BN
cooperative learning still outperformed all
comparison methods (p-value <0.05) and in
particular the proposed method in (Zhao et al.,
2016): 0.88 vs 0.8 for whole, 0.78 vs 0.68 for core,
and 0.68 vs 0.65 for enhanced tumor. Although our
results slightly dropped using using 5-fold cross-
validation, they still achieve the best performance.
Bayesian Network and Structured Random Forest Cooperative Deep Learning for Automatic Multi-label Brain Tumor Segmentation
187
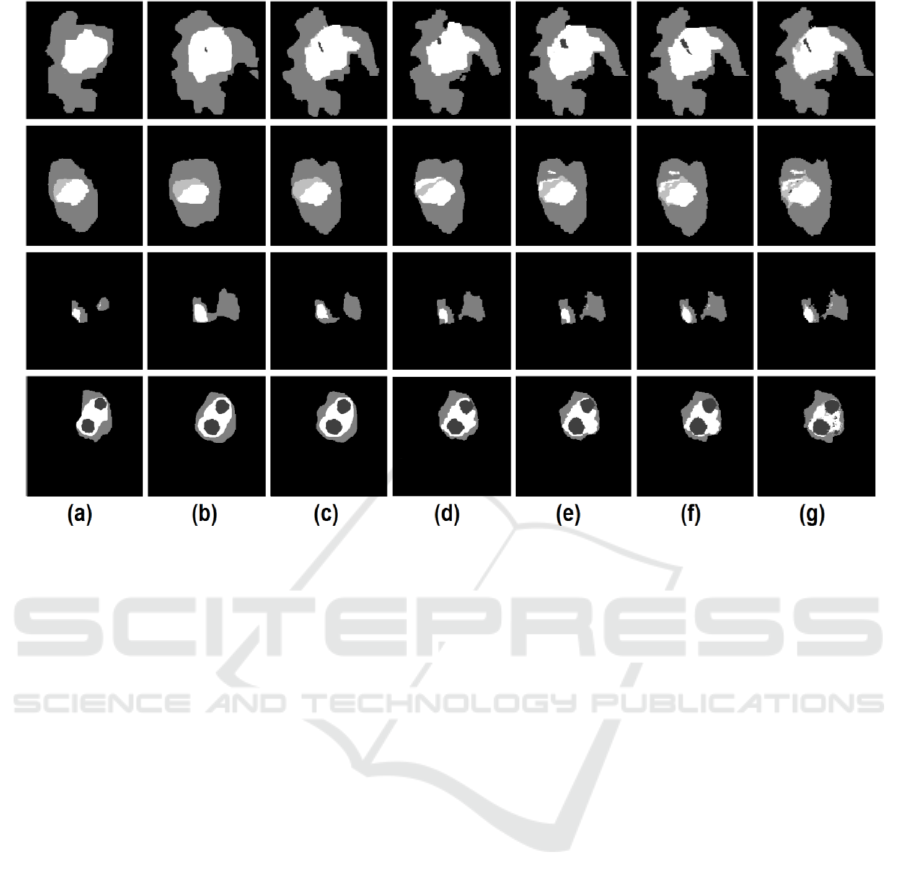
Figure 2: Qualitative segmentation results for four representative subjects using different segmentation methods: (a) the BN
segmentation result; (b) the SRF segmentation result, (c) the auto-context SRF; (d) SRF+BN segmentation result; (e) our
method (2 layers); (f) our method (3 layers); (g) the ground truth segmentation map.
Also, all proposed and comparison methods
significantly improved when using 3 modalities
compared with 1 by 8-9%. The training time took
about 5 hours and testing on one image took about 4
minutes.
We would like to note that the use of the term
learning ‘transfer’ and ‘deep’ learning was meant in
the broad sense of both words: (1) mutual
autocontext information (semantic map) transfer
between BN and SRF classifiers for progressively
improving their performances, and (2) ‘deepening’
our SRF-BN architecture to gradually improve their
cooperative learning differs from deep neural
networks, although both architectures can go deeper.
However, unlike deep one-step CNN architectures,
our method is able to consider appearance and
spatial consistency between neighboring superpixels
and patches via the gradual autocontext feed
between SRF and BN. We also stopped at layer
(L=3) in depth since the improvement became
negligible at L>3.
Although the proposed framework showed good
segmentation results thanks to the deep cooperation
between the two classifiers (BN and SRF); a few
limitations can be pointed out for further
improvements:
(1) Edge-map estimation. The considered edge-map
is estimated from a single modality (i.e.,
FLAIR), which limits the learned Bayesian
mapping to one type of imaging data.
Estimating edge-maps from different modalities
(e.g., T1.c and T2 MRIs) will help capture
different radiomic properties of the tumor lesion
especially around its boundary.
(2) Unidirectional flow between classifiers. The
learning transfer between the classifiers of the
same block as well as through the pipeline is
unidirectional, which means it can only go from
one classifier to the next one. Hence there is no
mutual benefit between the classifiers of the
same block.
(3) Hemispheric brain symmetry detection. The
brain symmetry method that we used (Loy and
Eklundh, 2006) fails in detecting the mid-sagittal
plane in a few cases. This might produce
unreliable symmetric features.
We anticipate that addressing these limitations will
further boost up the performance of our proposed
framework. We plan to investigate these in our
future work on a larger dataset.
ICAART 2018 - 10th International Conference on Agents and Artificial Intelligence
188
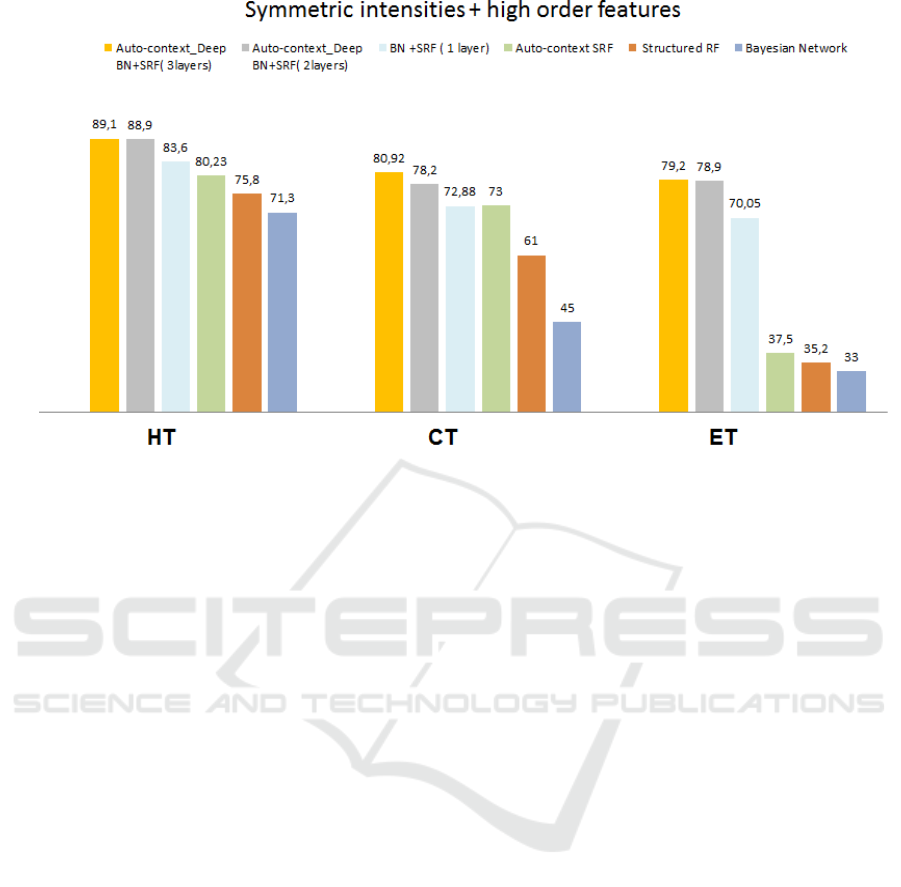
Figure 3: Average Dice index across 50 subjects using our proposed framework and all comparison methods for the three
tumor tissue classes (whole:HT, core:CT, and enhanced:ET).
4 CONCLUSIONS
In this paper, we proposed an automatic brain tumor
segmentation method based on a cooperative
learning between two classifiers, Structured RF and
BN, embedded within a deep auto-context
architecture. The experimental results prove the
efficiency of our proposed concept. Thus, SRF-BN
cooperative learning method outperforms the two
classifiers used solely, which proves that their
combination alleviates their limitations. Moreover,
the application of the deep auto-context architecture
has shown better performances for both the
quantitative and the qualitative results demonstrating
its effectiveness to boost the two classifiers and to
improve the feature learning.
Since the obtained results showed the
effectiveness of stacking SRF and BN within a
multi-label segmentation framework, we intend to
explore other architectures composed of these two
classifiers while addressing the limitations of our
proposed framework. Besides we will compare our
method with deep learning methods using multiple
BRATS testing datasets, including BRATS 2013.
REFERENCES
Aerts, H. J., Velazquez, E. R., Leijenaar, R. T., Parmar,
C., Grossmann, P., Cavalho, S., ... & Hoebers, F.
(2014). Decoding tumour phenotype by noninvasive
imaging using a quantitative radiomics approach.
Nature communications, 5.
Havaei, M., Davy, A., Warde-Farley, D., Biard, A.,
Courville, A., Bengio, Y., ... & Larochelle, H. (2017).
Brain tumor segmentation with deep neural
networks. Medical image analysis, 35, 18-31.
Koley, S., Sadhu, A. K., Mitra, P., Chakraborty, B., &
Chakraborty, C. (2016). Delineation and diagnosis of
brain tumors from post contrast T1-weighted MR
images using rough granular computing and random
forest. Applied Soft Computing, 41, 453-465.
Lefkovits, L., Lefkovits, S., & Szilágyi, L. (2016,
October). Brain Tumor Segmentation with Optimized
Random Forest. In International Workshop on
Brainlesion: Glioma, Multiple Sclerosis, Stroke and
Traumatic Brain Injuries (pp. 88-99). Springer, Cham.
Zhang, J., Gao, Y., Park, S. H., Zong, X., Lin, W., &
Shen, D. (2016, October). Segmentation of
perivascular spaces using vascular features and
structured random forest from 7T MR image.
In International Workshop on Machine Learning in
Medical Imaging (pp. 61-68). Springer International
Publishing.
Kontschieder, P., Bulo, S. R., Bischof, H., & Pelillo, M.
(2011, November). Structured class-labels in random
forests for semantic image labelling. In Computer
Vision (ICCV), 2011 IEEE International Conference
on (pp. 2190-2197). IEEE.
Zhang, L., & Ji, Q. (2008, June). Integration of multiple
contextual information for image segmentation using a
bayesian network. In Computer Vision and Pattern
Recognition Workshops, 2008. CVPRW'08. IEEE
Computer Society Conference on (pp. 1-6). IEEE.
Bayesian Network and Structured Random Forest Cooperative Deep Learning for Automatic Multi-label Brain Tumor Segmentation
189

Panagiotakis, C., Grinias, I., & Tziritas, G. (2011). Natural
image segmentation based on tree equipartition,
bayesian flooding and region merging. IEEE
Transactions on Image Processing, 20(8), 2276-2287.
Zhang, L., & Ji, Q. (2011). A Bayesian network model for
automatic and interactive image segmentation. IEEE
Transactions on Image Processing, 20(9), 2582-2593.
Yang, S., Yuan, C., Wu, B., Hu, W., & Wang, F. (2015).
Multi-feature max-margin hierarchical Bayesian
model for action recognition. In Proceedings of the
IEEE Conference on Computer Vision and Pattern
Recognition (pp. 1610-1618).
Tu, Z., & Bai, X. (2010). Auto-context and its application
to high-level vision tasks and 3d brain image
segmentation. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 32(10), 1744-1757.
Prastawa, M., Bullitt, E., Ho, S., & Gerig, G. (2004). A
brain tumor segmentation framework based on outlier
detection. Medical image analysis, 8(3), 275-283.
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., &
Süsstrunk, S. (2010). Slic superpixels (No. EPFL-
REPORT-149300).
Zhao, X., Wu, Y., Song, G., Li, Z., Fan, Y., & Zhang, Y.
(2016, October). Brain Tumor Segmentation Using a
Fully Convolutional Neural Network with Conditional
Random Fields. In International Workshop on
Brainlesion: Glioma, Multiple Sclerosis, Stroke and
Traumatic Brain Injuries (pp. 75-87). Springer, Cham.
Loy, G., & Eklundh, J. O. (2006, May). Detecting
symmetry and symmetric constellations of features.
In European Conference on Computer Vision (pp.
508-521). Springer, Berlin, Heidelberg.
Breiman, L. (2001). Random forests. Machine learning,
45(1), 5-32.
Christ, P. F., Ettlinger, F., Grün, F., Elshaera, M. E. A.,
Lipkova, J., Schlecht, S., ... & Rempfler, M. (2017).
Automatic Liver and Tumor Segmentation of CT and
MRI Volumes using Cascaded Fully Convolutional
Neural Networks. arXiv preprint arXiv:1702.05970.
Folgoc, L. L., Nori, A. V., Alvarez-Valle, J., Lowe, R., &
Criminisi, A. (2016). Segmentation of brain tumors
via cascades of lifted decision forests. In Proceedings
MICCAI-BRATS Workshop (pp. 26-30).
Zhang, L., Wang, Q., Gao, Y., Li, H., Wu, G., & Shen, D.
(2017). Concatenated spatially-localized random
forests for hippocampus labeling in adult and infant
MR brain images. Neurocomputing, 229, 3-12.
Guo, H., Hu, H., & Fan, G. (2017). A novel segmentation
method of medical CBCT image in liver organ based
on Bayesian network. Biomedical Research, 1-1.
ICAART 2018 - 10th International Conference on Agents and Artificial Intelligence
190
