
Clone Detection for Ecore Metamodels using N-grams
¨
Onder Babur
Eindhoven University of Technology, 5600 MB, Eindhoven, The Netherlands
Keywords:
Model-driven Engineering, Model Clone Detection, R, Vector Space Model, Clustering.
Abstract:
Increasing model-driven engineering use leads to an abundance of models and metamodels in academic and
industrial practice. A key technique for the management and maintenance of those artefacts is model clone
detection, where highly similar (meta-)models and (meta-)model fragments are mined from a possibly large
amount of data. In this paper we extend the SAMOS framework (Statistical Analysis of MOdelS) to clone
detection on Ecore metamodels, using the framework’s n-gram feature extraction, vector space model and
clustering capabilities. We perform a case analysis on Ecore metamodels obtained by applying an exhaustive
set of single mutations to assess the precision/sensitivity of our technique with respect to various types of
mutations. Using mutation analysis, we also briefly evaluate MACH, a comparable UML clone detection tool.
1 INTRODUCTION
Model-driven engineering (MDE) promotes the use of
models (and metamodels to which they conform) as
central artefacts in the software development process.
While this is recommended for the sake of ease of de-
velopment and maintenance of software artefacts (no-
tably source code), increasing MDE adoption leads to
an abundance of models in use
1
. Some examples of
this include the academic efforts to gather models in
repositories, or simply large-scale MDE practices in
the industry (Babur et al., 2017). This leads to a chal-
lenge in the management and maintenance of those
artefacts. One of those challenges is the identification
of model clones, which can be defined in the most
general sense as duplicate or highly similar models
and model fragments. To draw a parallel, code clones
have attracted the attention of the source code analysis
community, who had to deal with the maintenance of
large amounts of artefacts (source code) for a longer
time than the MDE community. There is a significant
volume of research on code clones, elaborating the
drawbacks of having clones (e.g. major source of de-
fects, higher maintenance cost, less reusability, etc.),
while providing techniques and public tools for their
detection (Roy et al., 2009).
Model clone detection, on the other hand, is a rela-
tively newer topic. Many researchers have drawn par-
allels from code clones, and claimed that a lot of the
1
In this context, we refer to metamodels and models
shortly as models, as metamodels are models too.
issues there can be directly translated into the world
of models. While the problem domain is indeed eas-
ily relatable, the solution proves to be a challenge.
To mention two of the arguments there, source code
clone detection usually works on linear text or ab-
stract syntax tree of the code while models are general
graphs (Deissenboeck et al., 2010); and many other
aspects are inherently different for models, such as
tool-specific representations, internal identifiers and
abstract vs. concrete syntaxes (St
¨
orrle, 2013).
There are several approaches for model clone de-
tection in the literature (Deissenboeck et al., 2010),
yet we are particularly interested in ones with a pub-
licly available tool to be reused in our studies. A
good portion of such tools are either limited to, tai-
lored for, or evaluated on MATLAB/Simulink mod-
els, and built on top of a backend originally intended
for code clone detection. Notable examples are Con-
QAT/CloneDetective (Deissenboeck et al., 2008); and
Simone/NiCaD (Alalfi et al., 2012). Another interest-
ing approach for Simulink models is ModelCD based
on graph comparison (Pham et al., 2009), but the tool
is not publicly available. Last but not least, St
¨
orrle
presents an approach and an accompanying avail-
able tool, MQ
lone
, for UML models (St
¨
orrle, 2015).
There, the author elaborately describes and classifies
UML model clones; noting the differences compared
to code clones and Simulink model clones. Further-
more, the author reports for MQ
lone
a much higher
performance and scalability compared to ConQAT
and ModelCD. Another recent account of relatively
Babur, Ö.
Clone Detection for Ecore Metamodels using N-grams.
DOI: 10.5220/0006604604110419
In Proceedings of the 6th International Conference on Model-Driven Engineering and Software Development (MODELSWARD 2018), pages 411-419
ISBN: 978-989-758-283-7
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
411

low scalability of those tools can be found in the liter-
ature (Str
¨
uber et al., 2016). MQ
lone
is integrated into
the publicly available tool suite MACH
2
, though in
a very limited manner - with almost no control over
the rich set of algorithms and settings developed by
St
¨
orrle. Other approaches, though of relatively less
importance for the scope of this paper, involve clone
detection and/or model matching for UML sequence
diagrams (Liu et al., 2006) and business process mod-
els (Dijkman et al., 2011).
In pursuit of detecting clones in large repositories
of Ecore metamodels and large evolving industrial
domain-specific language (DSL) ecosystems based
on Eclipse Modelling Framework (EMF), we have
investigated the feasibility of existing tools for our
purpose with three major requirements: (1) concep-
tual and technological applicability to Ecore meta-
model clones; (2) sensitivity to all possible meta-
model changes, in other words accuracy; and (3) scal-
ability for large datasets. As a starting point, we have
turned to MACH as a promising candidate. While
scoring very high with respect to the first and third
requirement (as metamodels are similar to UML class
diagrams), it has underperformed in terms of the sec-
ond one as we will show in Section 5.
We have eventually taken an orthogonal approach
by extending and applying the SAMOS framework
(Statistical Analysis of MOdelS), a novel tool de-
veloped for large-scale analysis of models (Babur,
2016). We wish to exploit the underlying capabil-
ities of the framework - incorporating information
retrieval-based fragmentation, natural language pro-
cessing and statistical algorithms - for model clone
detection. In this paper, we describe how we have
extended and tailored SAMOS for metamodel clone
detection. To mine metamodel clones, we have used
the n-gram feature extraction facility of the frame-
work, integrating additional scoping capability and
extended distance measures and a density based clus-
tering algorithm. We have evaluated our technique
using the mutation analysis approach (Stephan et al.,
2013) based on single mutations, where we performed
a case analysis to assess the sensitivity of our tech-
nique for each case. We finally draw conclusions on
the applicability of SAMOS to metamodel clone de-
tection and suggest potential future work in terms of
our technique and its further evaluation.
2
http://www2.compute.dtu.dk/ hsto/tools/mach.html
2 BACKGROUND: STATISTICAL
ANALYSIS OF MODELS
We outline here the underlying concepts of
SAMOS (Babur, 2016) and (Babur et al., 2016)
inspired by information retrieval (IR) and machine
learning (ML) domains. IR deals with effectively
indexing, analyzing and searching various forms
of content including natural language text docu-
ments (Manning et al., 2008). As a first step for
document retrieval in general, documents are col-
lected and indexed via some unit of representation.
Index construction can be implemented using vector
space model (VSM) with the following major com-
ponents: (1) a vector representation of occurrence of
the vocabulary in a document, named term frequency,
(2) zones (e.g. ’author’ or ’title’), (3) weighting
schemes such as inverse document frequency (idf),
and zone weights, (4) Natural Language Processing
(NLP) techniques for handling compound terms,
detecting synonyms and semantically related words.
The VSM allows transforming each document into
an n-dimensional vector, thus resulting in an m × n
matrix for m documents. Over the VSM, document
similarity can be defined as the distance (e.g. eu-
clidean or cosine) between vectors. These can be
used for identifying similar groups of documents in
the vector space via an unsupervised ML technique
called clustering (Manning et al., 2008).
SAMOS applies this workflow to models: starting
with a metamodel-driven extraction of features. Fea-
tures can be, for instance, singleton names of model
elements (very similar to the vocabulary of docu-
ments) or n-gram fragments (Manning and Sch
¨
utze,
1999) of the underlying graph structure. N-grams
originate from computational linguistics and repre-
sent linear encoding of (text) structure. An exam-
ple in our context, an example n-gram for a UML
class diagram would be for n = 2 a Class containing
a Property (Babur and Cleophas, 2017). Via compar-
ison schemes (e.g. whether to check types), weight-
ing schemes (e.g. Class weight higher than Property)
and NLP (stemming/lemmatisation, typo and syn-
onym checking, etc. ), it computes a VSM. Applying
various distance measures suitable to the problem at
hand, SAMOS applies different clustering algorithms
(via the R statistical software) and can output auto-
matically derived cluster labels or diagrams for visu-
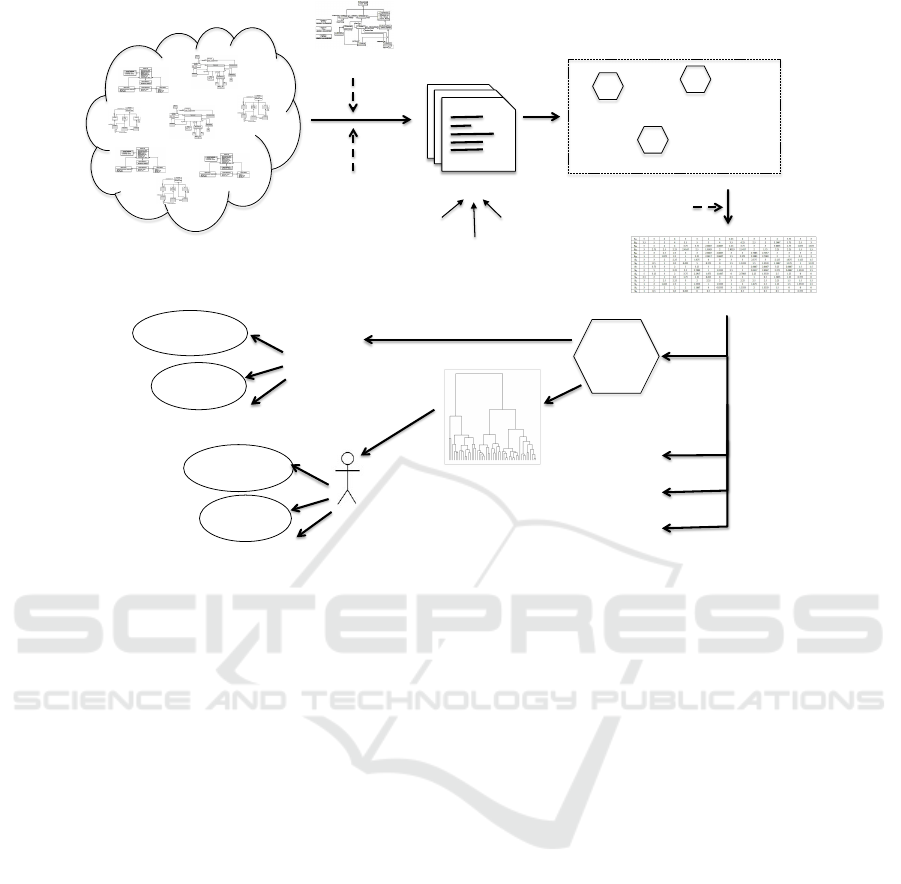
alisation and manual inspection/exploration. Figure 1
roughly illustrates the workflow, with key steps of the
workflow and several application areas including do-
main analysis and clone detection.
MODELSWARD 2018 - 6th International Conference on Model-Driven Engineering and Software Development
412
Set$of$models$
Metamodel$
Features$
NLP$
Tokeniza8on$
$Matching$scheme$
Weigh8ng$scheme$
VSM$
Distance$
calcula8on$
Clustering$
Dendrogram$
Automated$extrac8on$
Inferred$
clusters$
Extrac8on$$
scheme$
Filtering$
Synonym$
detec8on$
…$
Data$selec8on,$
filtering$
Clone$
detec8on$
…$
Classifica8on$
Analysis$
…$
Repository$
management$
Domain$
analysis$
…$
NJgrams$
Metrics$
…$
Manual$
inspec8on$
Figure 1: Overview of SAMOS workflow.
3 METAMODEL CLONES
Metamodel clones might exist due to a wide range
of reasons including copy-paste or clone-and-own
approaches for development (Deissenboeck et al.,
2010), or lack of abstraction mechanisms in meta-
models (Sutii et al., 2017). In our research, we are
interested in finding similar (fragments of) metamod-
els, with several problems at hand. A non-exhaustive
list is as follows:
• Clone detection in DSL ecosystems for quality as-
surance/refactoring,
• Empirical studies on DSL evolution and compari-
son of DSLs from related domains,
• Clone detection in metamodel reposito-
ries/datasets for repository management and
data preprocessing,
• Plagiarism detection in the assignments of our
metamodelling course.
While metamodel clones have not been specifi-
cally studied in the literature, they possess a certain
parallelism to UML domain models, class diagrams
in particular. Hence our conceptualisation of meta-
model clones is mostly adopted from (St
¨
orrle, 2015).
St
¨
orrle presents a series of arguments involving sev-
eral characteristics of models such as secondary no-
tation, internal identifiers, and most importantly em-
phasises that names of model elements are essential
parts of models. We agree with all of the arguments
in the context of EMF metamodel clones, and the
contrast with the common model clone classification
scheme for Simulink models. For Simulink mod-
els, the classification includes Type-I clones for ex-
act clones, Type-II for renamed clones with consis-
tently changed identifiers, Type-III near-miss clones
with small changes in model elements (Alalfi et al.,
2012). A notable change is that St
¨
orrle’s classification
rules out Type-II clones due to the indispensability of
element names. Our clone classification is the fol-
lowing, adding a few NLP-related items to St
¨
orrle’s
classification and omitting Type-D (for the scope of
this paper):
• Type-A. duplicate model fragments except sec-
ondary notation (layout, formatting), internal
identifiers.
– Plus any cosmetic change in the names (lower-
/uppercase, snake-/camelcase, etc. )
• Type-B. duplicate model fragments with small
percentage of changes to names, types, attributes,
few additions/removals of parts.
– Plus potentially many syntactic/semantic
changes in the names such as typos, synonyms.
• Type-C. duplicate with substantial percentage of
changes/additions/removals of names, types, at-
tributes and parts.
Clone Detection for Ecore Metamodels using N-grams
413

4 EXTENDING SAMOS FOR
CLONE DETECTION
We have extended various features of SAMOS, in or-
der to apply it to metamodel clone detection. In this
section, we elaborate the extensions, compartmen-
talised as key steps of the workflow.
Scoping. While originally the framework handles
whole models and extracts all the model elements
contained, we introduce the notion of scoping to de-
fine the granularity of independent data elements. For
Ecore metamodels, we define three scopes: whole
model, EPackage and EClass. The scope guides the
extraction by mapping a model into one (e.g. whole
model) or more data points (e.g. per EPackage con-
tained). Given a fixed scope, we adopt the approach
in (St
¨
orrle, 2015) and cover all the model elements
under transitive containment closure of that starting
model element. Note that scoping does not affect the
features (i.e. columns) in the vector space, but only
the data points (i.e. rows).
Extracting Model Element Information. The
main unit of information extracted by SAMOS has
previously been the so called type-name pair, which
essentially maps to a vertex in the underlying graph
of the model. Such a pair encodes the domain
type (eType) information (e.g. EClass) and the name
(e.g. Book) of a model element. For proper clone
detection, we need to cover all the information in
a model element, including attributes (e.g. whether
an EClass is abstract), cardinalities (e.g. of ERefer-
ences), and so on. We also need to explicitly capture
the edges (e.g. containment) to include for compari-
son. Therefore, we have extended the original feature
hierarchy in SAMOS with (1) AttributedNode, which
holds all the information of a vertex including domain
type, name, type, attributes, etc. as key-value pairs;
and (2) SimpleType, which holds only type informa-
tion - e.g. the edge type of containment in this work.
There are several implementation details to men-
tion further about the extraction:
• Our current implementation covers the full Ecore
meta-metamodel for extraction, except EAnnota-
tions, EFactories and generic types.
• Observing that most attributes in Ecore metamod-
els are used with default values, we only encode
non-default values in our AttributedNodes to re-
duce the data size and speed up the comparison.
• Another change from (Babur et al., 2016) is
that we push the type information (e.g. what
EDataType is assigned to an EAttribute) into the
AttributedNode, rather than representing it as a
separate vertex connected to the original vertex.
This is done so that the framework avoids match-
ing irrelevant features just because of matching
types, e.g. all EAttributes with the type String.
EPackage(
EPackage(
BIBTEX(
EPackage(
BIBTEX(
EClass(
LocatedElement(
EA7ribute(
loca<on(
EClass(
Bibtex(
EReference(
entries(
EClass(
Entry(
EA7ribute(
key(
EPackage(
Types(
EDataType(
Integer(
contains(
contains(
contains(
contains(
contains(
contains(
contains(
typeof(
supertype(
typeof(
EDataType(
EString(
typeof(
EDataType(
EInteger(
Figure 2: A simplified graph representation for a model.
To exemplify, see the graph in Figure 2, where
the vertices are simplified to domain type and name
information with the rest of the attributes being hid-
den. Several example AttributedNode features to be
extracted in our approach would be:
• v
1
= {name : LocatedElement, type :
EClass, abstract : true},
• v
2
= {name : location, type : EAttribute, eType :
EString, lowerBound : 1},
• v
3
= {name : Bibtex, type : EClass},
• v
4
= {name : entries, type : ERe f erence, eType :
Entry, unique : true, . . .},
• v
5
= {name : Entry, type : EClass, abstract :
true}.
Encoding Structure in N-grams. Referring
to (Babur and Cleophas, 2017) we have basically two
options for representing structure as features. We
can either (1) ignore the model structure completely,
using vertices as is, i.e. the unigram setting; or (2)
encode structure in linear chunks, i.e. the n-gram
(with n > 1) setting. In terms of the conceptual
feature hierarchy, one can think of unigrams as
corresponding to SimpleFeatures, while n-grams
are aggregate features with multiple SimpleFeatures
contained. Based on the underlying graph for a
model, we can describe n-grams as n consecutively
connected vertices. In contrast with (Babur and
Cleophas, 2017), we also incorporate the edges in the
n-grams as SimpleTypes. Some bigrams (n = 2) from
Figure 2 would be:
MODELSWARD 2018 - 6th International Conference on Model-Driven Engineering and Software Development
414

• b
1
= (v
1
, contains, v
2
),
• b
2
= (v
5
, hasSupertype, v
1
).
N-gram Comparison. As n-grams consist of Sim-
pleFeature vertices, we first define the extended ver-
tex comparison to account for attributes with the fol-
lowing multiplicative vertex similarity formula:
vSim(n
1
, n
2
) = nameSim(n
1
, n
2
)∗typeSim(n
1
, n
2
)∗
eTypeSim(n
1
, n
2
) ∗ attrSim(n
1
, n
2
) (1)
where nameSim is the NLP-based similarity be-
tween the names, typeSim and eTypeSim between the
domain types and eTypes of the AttributedNodes, and
for attributes other than name, type and eType, at-
tribute similarity being:
attrSim(n
1
, n
2
) = 1 −
# unmatched attributes
total # attr. for types
(2)
The framework allows relaxing those categories
(e.g. inserting a reducing multiplier of 0.5 for
non-matching types). N-gram comparison is the
semi-relaxed formula (Babur and Cleophas, 2017)),
i.e. given n-grams p
1
and p
2
with 2n − 1 elements (n
vertices and n −1 edges), the n-gram similarity is:
nSim(p
1
, p
2
) = ctxMult(p
1
, p
2
) ∗
2n−1
∑
i=1
vSim(v
i
1
, v
i
2
)
(3)
ctxMult(P
1
, P
2
) =
1 + |nonzero vSim matches|
1 + (2n − 1)
(4)
Weighting. SAMOS supports type-based and idf
weighting for features. While the latter is to be inves-
tigated further in the future (e.g. for ordering of clones
in a clone report), we exploit type-based weight-
ing to reduce the representative importance of cer-
tain features. Features with more important types are
given higher weights; for instance, matching EClasses
should be favored against e.g. matching EParameters.
Distance Measurement. Originally SAMOS
adopted a regular VSM approach: a choice of a
distance measure (e.g. cosine and Manhattan) and
calculation of the distance over the whole VSM.
We have identified several shortcomings in the
context of clone detection, and extended the distance
measurement.
• Manhattan distance is an absolute measure and
cosine is normalised but size-agnostic. Therefore
we cannot use them directly for clone detection.
There are several measures in the literature (im-
plemented in R) that fulfil the requirements of
normalisation and size-sensitivity, such as Bray-
Curtis and canberra (Deza and Deza, 2009).
• VSM assumes orthogonality of the features, and
takes all columns into account for distance calcu-
lation. This is violated in our case where our fea-
tures are often (partly) similar to many other fea-
tures and hence not orthogonal. For clone detec-
tion, we limit the distance calculation to the union
of originally contained features by the two enti-
ties, rather than all the features in the dataset.
For the reasons above, we have integrated a
masked variant of Bray-Curtis distance (Equation 5)
into SAMOS, extending the default distance func-
tion in the R package vegan. Given an N dimen-
sional vector space, data points (e.g. model, EClass,
etc. depending on the scope) of P consisting of fea-
tures P
1
, . .. ,P
m
and Q consisting of Q
1
, . .. ,Q
n
, p and
q the corresponding vectors on the full vector space
for P and Q, the masked bray-curtis distance is over
the vector subspace P ∪Q (size ≤ m + n) as:
bray(P, Q) =
∑
P∪Q
i
|p
i
− q
i
|
∑
P∪Q
i
(p
i
+ q
i
)
, (5)
Clustering. As the final step of the SAMOS work-
flow, we wish to apply clustering on top of the cal-
culated distances to obtain the clone clusters. This in
turn can be boiled down to finding non-singleton (size
≥ 2) and sizeable (size ≥ n) groups of data points
that are similar (distance ≤ t); with n and t thresholds
depending on the application case. While SAMOS
originally supports k-means and hierarchical cluster-
ing, we have opted for Density-Based Spatial Cluster-
ing of Applications with Noise (dbscan) (Ester et al.,
1996). The algorithm (implemented in R package db-
scan) uses two parameters, minimum Points (i.e. n)
and ε distance (i.e. t) to compute density-reachable
regions as clusters, with non-reachable regions being
labelled as noise. While technical details are beyond
the scope of this paper, dbscan possesses some prop-
erties that we desire for clone detection. dbscan:
• can detect clusters in various (non-convex, non-
spherical) shapes,
• can natively detect noise, i.e. non-clones,
• is suitable and efficient for large datasets.
5 MUTATION ANALYSIS
We have adopted a conceptual framework for validat-
ing our approach (Stephan and Cordy, 2014). The
Clone Detection for Ecore Metamodels using N-grams
415

framework suggests using mutation analysis to evalu-
ate model clone detection techniques. In this section,
we detail our assumptions, case design and goals. We
first introduce a brief evaluation of MACH using the
mutations (for UML class diagrams) and report its
shortcomings. Finally we present the results of our
techniques on the mutated dataset and discuss the po-
tential strengths and weaknesses.
Case Design. First we make the simplifying as-
sumption (for the scope of this paper) that we are in-
terested in EClass clones in Ecore metamodels. In-
specting the Ecore meta-metamodel, we identified 36
mutations that we believe to represent possible and
noteworthy atomic/small changes in an EClass. They
contain addition, removal and changing of model ele-
ments, renaming and finally reorder, move and swap
operations. Using a medium-sized base EClass con-
sisting of 1 super type, 5 EAttributes, 2 ERefer-
ences and 1 EOperation in turn with 1 EParameter
and 1 EException, we used the mutation framework,
WODEL (G
´
omez-Abajo et al., 2016), where possible
to automatically inject those mutations into the base
metamodel. WODEL contains a DSL for specifying
the mutations and the mutation workflow in a very
convenient and maintainable way.
The mutations are mostly trivial operations of
adding, removing, etc. We added three specific mu-
tations involving element names: cosmetic renaming
(such as lower vs. upper case, camel vs. snake case),
adding a typo, and replacing with a synonym. We
further added two regular and two corner cases of
move and swap operations (last rows of Table 1) in
order to demonstrate the approximate nature of infor-
mation representation in our technique. The regular
case for move involves simply moving a model ele-
ment elsewhere. In contrast, the corner case for move
involve moving a model element A contained in B
into a distinct container B’, with the condition that
vsim(B, B
0
) = 1 (same vertex similarities) - a change
easily detectable using graph isomorphism but possi-
bly not by approximate techniques such as ours. Swap
mutations are designed in a similar manner.
Last but not the least, we manually replicated all
the mutations on a comparable UML class diagram in
order to evaluate MACH.
Brief Evaluation of MACH. St
¨
orrle has developed
a variety of algorithms with different settings in the
MQ
lone
clone detector. However they are not entirely
available, being integrated in a rather limited way
into the MACH toolset. So we proceeded to analyse
the output of MACH. We fed MACH all the mutated
cases and observed what it calculated as their pairwise
difference with respect to the base model. We noted
down the (changes in the) absolute similarity values
and identified several shortcomings of the tool. In our
tests MACH seemed to:
• ignore types (e.g. of properties, parameters), mod-
ifiers, modifiers (e.g. final Class), cardinalities,
exceptions,
• not handle Class renaming properly; i.e. not
recognise the two classes as clones,
• not recognise changing of Class super type,
• not recognise all move/swaps,
• not consistently handle typos and cosmetic
changes in the element names; i.e. treat them as
complete renaming,
• not support semantic relatedness of words,
e.g. synonymy.
Goals. Ideally, we wish to obtain zero distance for
mutations leading to Type A clones (G1), and positive
distance for mutations leading to higher level clones
(G2). For the latter, we also wish to have the distance
reasonably small (e.g < 0.05 as a breaking point to-
wards Type C) given that all the mutations in this sec-
tion are atomic/small (G3), and match an intuitive as-
sessment of distance, e.g. changing just a EParameter
is less significant than changing an EAttribute (G4).
A final desirable property is that bigger changes lead
to higher distances than their smaller counterparts,
e.g. distance for introducing a typo in the name is
smaller than complete renaming; or changing the type
smaller than removing the element altogether (G5).
Evaluation and Discussion of SAMOS. We have
in turn applied our clone detection technique and re-
port here the distance measure between each case and
the base metamodel. We have used different feature
schemes (unigrams and bigrams) using Bray-Curtis
distance the optional scheme (raw/weighted). Table 1
gives a representative subset of the results.
In general, the results look promising, though not
without certain errors and weaknesses for particular
settings - most prominent errors are bolded in the ta-
ble. G1 is not violated by any technique in the two
cases with cosmetic changes and reordering. G2 is
violated in a number of cases. Unigrams (not unex-
pectedly, as they ignore structural context) evaluate
quite some mutations with zero distance. Furthermore
move/swap mutations are mostly undetected by our
technique. G3 is not uniformly satisfied but the re-
sults are generally acceptable given a relaxed thresh-
old range of 0.05−0.10. While relatively large values
for mutations involving EOperations don’t matter too
much, considering that removing it is in fact removing
all its content as well (e.g. EParameters). Arguably
MODELSWARD 2018 - 6th International Conference on Model-Driven Engineering and Software Development
416
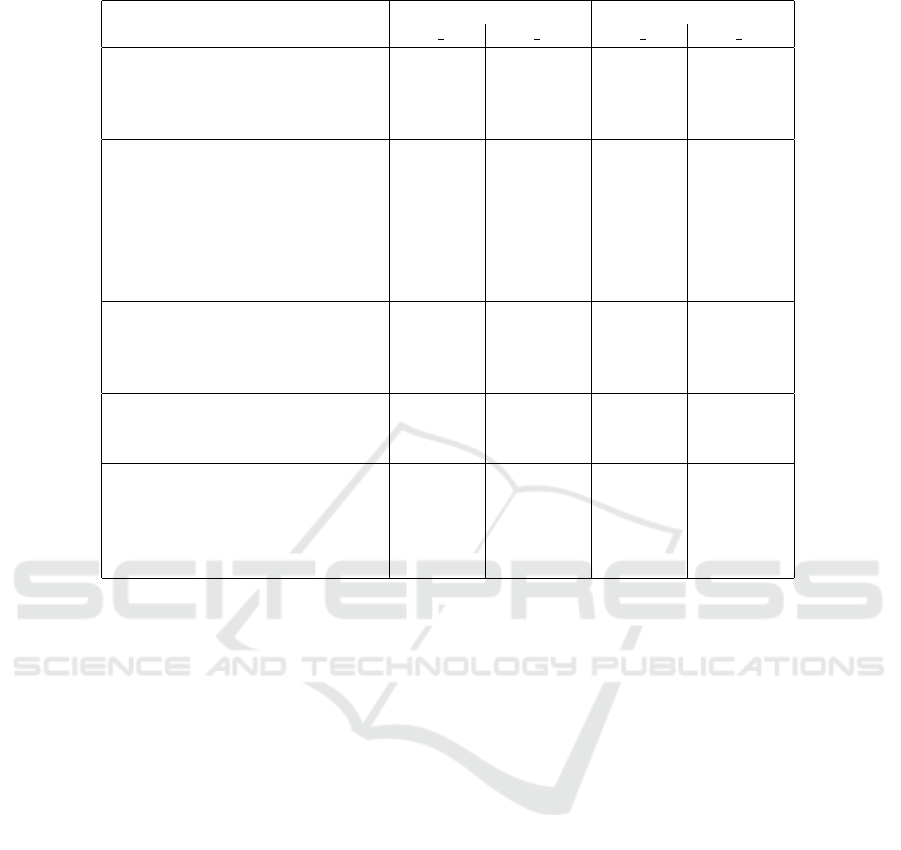
Table 1: Distances to the base model for each mutant (rows) and different setting (columns).
Mutation applied Unigram Bigram
bray raw bray wght bray raw bray wght
addEClassSupertype 0.000 0.000 0.043 0.045
addEClassEAttribute 0.047 0.048 0.050 0.052
addEClassEOperation 0.089 0.039 0.062 0.056
. . .
changeEClassNameRandom 0.098 0.201 0.743 0.778
changeEClassSupertype 0.000 0.000 0.014 0.012
changeEAttributeNameRandom 0.098 0.100 0.010 0.010
changeEAttributeType 0.072 0.073 0.005 0.005
changeEAttributeAttribute 0.013 0.013 0.001 0.001
changeEOperationNameRandom 0.107 0.066 0.045 0.022
. . .
removeEClassSuperType 0.000 0.000 0.050 0.049
removeEClassEAttribute 0.051 0.053 0.055 0.058
removeEClassEOperation 0.119 0.047 0.097 0.075
. . .
changeEAttributeNameCosmetic 0.000 0.000 0.000 0.000
changeEAttributeNameTypo 0.004 0.005 0.001 0.001
changeEAttributeNameSynonym 0.002 0.002 0.001 0.001
reorder element 0.000 0.000 0.000 0.000
move element 0.000 0.000 0.032 0.033
swap element 0.000 0.000 0.000 0.000
move into similar container 0.000 0.000 0.000 0.000
swap with similar container 0.000 0.000 0.000 0.000
the biggest issue, as bolded in the table, is with bi-
grams for renaming an EClass: due to the nature of
feature extraction (i.e. bigrams), vertices with high
number of outgoing edges (e.g. EClasses typically
having many types of elements) are over-represented
in the vector space. They are present in many fea-
tures, hence any change leads to a larger distance in
the vector space. G4 in turn is partly improved by ap-
plying weighted distances for each technique. Indeed,
fine-tuning of the weighting (possibly with more ad-
vanced schemes) would further improve G3 (partly)
and G4; nevertheless we find the results satisfactory
given the scope of this work. Finally, G5 is relatively
well achieved overall.
6 DISCUSSION
In this section we discuss several aspects of our ap-
proach and the techniques we developed.
Underlying Framework. We have built our clone
detection technique on top of SAMOS, exploiting its
capabilities such as NLP and statistical algorithms for
free. The framework has allowed us easily extend it,
e.g. in terms of extraction of new features, addition of
new distance measures. This is one of the strengths of
our approach, also considering recent developments
within SAMOS such as support for distributed com-
puting and more sophisticated NLP. Using R as the
back-end enables us to further experiment with ad-
vanced statistical and data mining techniques. More-
over, SAMOS is in principle generic, as it can be ap-
plied to any graph-based model with a corresponding
feature extraction implementation.
Accuracy. We have evaluated our technique with
mutation analysis in order to assess the accuracy in
terms of correctly yielding the expected distance be-
tween the mutated and base model. As given in Ta-
ble 1, our technique with the different settings leads
to varying degrees of accuracy; we believe to have
achieved a good overall accuracy using bigrams. Note
that some of the issues with the sensitivity can be
remedied with more optimised or advanced weight-
ing schemes for each setting. Qualitative case anal-
ysis based on mutations further allows us to pinpoint
the weaknesses of our approach and improve it where
applicable.
Other Practical Aspects. Several aspects other
than accuracy are reported in the literature as im-
Clone Detection for Ecore Metamodels using N-grams
417

portant for applying model clone detection in prac-
tice (Deissenboeck et al., 2010; Stephan and Cordy,
2014). In this work, we have fixed scoping of EClass,
thus do not run into the nested clones problem.
This would be somewhat important for e.g. EPackage
scope, but even more so for other types of models.
Other aspects including clone ranking, reporting and
inspection, visualisation are also left as future work.
Threats to Validity. A threat to validity due to the
preliminary nature of this study is the limited evalu-
ation on a synthetic dataset using a single mutation
analysis approach. Further evaluation using chains
of mutations which lead to Type C clones, and even-
tually using a real dataset is needed. It is at this
phase not clear to us what the frequency of the various
changes (i.e. mutations) is in reality, which directly
contributes to the overall accuracy of our approach
(cf. the weakness of bigrams for certain mutations).
An evaluation on a real dataset, combined with addi-
tional comparative evaluation of existing model clone
detectors would be necessary to properly assess the
precision and more importantly our relative recall.
7 CONCLUSION AND FUTURE
WORK
In this paper we present a novel model clone detec-
tion approach based on SAMOS using information
retrieval and machine learning techniques. We have
extended SAMOS with additional scoping, compari-
son schemes, customised distance measures and clus-
tering algorithms in the context of metamodel clone
detection. We have evaluated our approach using mu-
tation analysis and identified the strengths and weak-
nesses of our approach in a case-based manner.
As future work, we plan to further extend SAMOS
with additional features (e.g. n-grams with n > 2 and
subtrees), customised, improved weighting schemes,
distance measures and statistical algorithms. Another
next step is to extend state-of-the-art model clone
detectors such as Simone, ConQAT and MQ
lone
for
metamodel clone detection for evaluating those tools
separately and comparatively with our approach for
precision and relative recall.
REFERENCES
Alalfi, M. H., Cordy, J. R., Dean, T. R., Stephan, M., and
Stevenson, A. (2012). Models are code too: Near-
miss clone detection for simulink models. In Software
Maintenance (ICSM), 2012 28th IEEE Int. Conf. on,
pages 295–304. IEEE.
Babur,
¨
O. (2016). Statistical analysis of large sets of mod-
els. In 31th IEEE/ACM Int. Conf. on Automated Soft-
ware Engineering, pages 888–891.
Babur,
¨
O. and Cleophas, L. (2017). Using n-grams for the
automated clustering of structural models. In 43rd
Int. Conf. on Current Trends in Theory and Practice
of Computer Science, pages 510–524.
Babur,
¨
O., Cleophas, L., and van den Brand, M. (2016). Hi-
erarchical clustering of metamodels for comparative
analysis and visualization. In Proc. of the 12th Eu-
ropean Conf. on Modelling Foundations and Applica-
tions, 2016, pages 3–18.
Babur,
¨
O., Cleophas, L., van den Brand, M., Tekinerdogan,
B., and Aksit, M. (2017). Models, more models and
then a lot more. In Grand Challenges in Modeling, to
appear.
Deissenboeck, F., Hummel, B., Juergens, E., Pfaehler, M.,
and Schaetz, B. (2010). Model clone detection in prac-
tice. In Proc. of the 4th Int. Workshop on Software
Clones, pages 57–64. ACM.
Deissenboeck, F., Hummel, B., J
¨
urgens, E., Sch
¨
atz, B.,
Wagner, S., Girard, J.-F., and Teuchert, S. (2008).
Clone detection in automotive model-based devel-
opment. In Software Engineering, 2008. ICSE’08.
ACM/IEEE 30th Int. Conf. on, pages 603–612. IEEE.
Deza, M. M. and Deza, E. (2009). Encyclopedia of Dis-
tances. Springer.
Dijkman, R., Dumas, M., van Dongen, B., K
¨
a
¨
arik, R.,
and Mendling, J. (2011). Similarity of business pro-
cess models: Metrics and evaluation. Inf. Systems,
36(2):498–516.
Ester, M., Kriegel, H.-P., Sander, J., and Xu, X. (1996).
A density-based algorithm for discovering clusters a
density-based algorithm for discovering clusters in
large spatial databases with noise. In Proc. of the
Second Int. Conf. on Knowledge Discovery and Data
Mining, KDD’96, pages 226–231. AAAI Press.
G
´
omez-Abajo, P., Guerra, E., and de Lara, J. (2016). Wodel:
a domain-specific language for model mutation. In
Proceedings of the 31st Annual ACM Symposium on
Applied Computing, pages 1968–1973. ACM.
Liu, H., Ma, Z., Zhang, L., and Shao, W. (2006). Detect-
ing duplications in sequence diagrams based on suffix
trees. In Software Engineering Conf., 2006. APSEC
2006. 13th Asia Pacific, pages 269–276. IEEE.
Manning, C. D., Raghavan, P., Sch
¨
utze, H., et al. (2008).
Introduction to information retrieval, volume 1. Cam-
bridge University Press.
Manning, C. D. and Sch
¨
utze, H. (1999). Foundations of
Statistical Natural Language Processing. MIT Press.
Pham, N. H., Nguyen, H. A., Nguyen, T. T., Al-Kofahi,
J. M., and Nguyen, T. N. (2009). Complete and ac-
curate clone detection in graph-based models. In Pro-
ceedings of the 31st Int. Conf. on Software Engineer-
ing, pages 276–286. IEEE Computer Society.
Roy, C. K., Cordy, J. R., and Koschke, R. (2009). Compari-
son and evaluation of code clone detection techniques
MODELSWARD 2018 - 6th International Conference on Model-Driven Engineering and Software Development
418

and tools: A qualitative approach. Science of Com-
puter Programming, 74(7):470 – 495.
Stephan, M., Alafi, M. H., Stevenson, A., and Cordy, J. R.
(2013). Using mutation analysis for a model-clone
detector comparison framework. In Int. Conf. on Soft-
ware Engineering, pages 1261–1264. IEEE.
Stephan, M. and Cordy, J. R. (2014). Model clone detector
evaluation using mutation analysis. In ICSME, pages
633–638.
St
¨
orrle, H. (2013). Towards clone detection in uml domain
models. Software & Systems Modeling, 12(2):307–
329.
St
¨
orrle, H. (2015). Effective and efficient model clone de-
tection. In Software, Services, and Systems, pages
440–457. Springer.
Str
¨
uber, D., Pl
¨
oger, J., and Acret¸oaie, V. (2016). Clone
detection for graph-based model transformation lan-
guages. In Int. Conf. on Theory and Practice of Model
Transformations, pages 191–206. Springer.
Sutii, A. M., van den Brand, M., and Verhoeff, T. (2017).
Exploration of modularity and reusability of domain-
specific languages: an expression DSL in metamod.
Computer Languages, Systems & Structures.
Clone Detection for Ecore Metamodels using N-grams
419
