
Data Format for Storing ANT+ Sensors Data
Petr Je
ˇ
zek and Roman Mou
ˇ
cek
Department of Computer Science and Engineering, New Technologies for the Information Society,
Faculty of Applied Sciences, University of West Bohemia, Plzen, Czech Republic
Keywords:
ANT+, NIX, Sensor, Data/metadata Storage, EEGBase, Data Format, eHealth.
Abstract:
Medical treatment of sudden and especially chronic diseases has become more expensive. People suffering
from a variety of diseases had been traditionally treated in hospitals for a long time. Fortunately, the current
situation has been changing also thanks to relatively cheap body sensors and development of systems for
home treatment. It brings inconsiderable cost savings and improves patients’ comfort. On the other hand,
it puts demands on the used technical infrastructure and home treatment system developers who must solve
integration of different systems. A crucial point is a definition of unified data formats facilitating transfer and
storage of data to/in remote databases. There are standards and APIs such as Zigbee, Bluetooth low energy or
ANT+ that define a protocol for data transfer. However, they do not define a suitable format for long term data
storing. In this paper, data coming from ANT+ sensors have been studied and metadata related to all kinds of
body sensors and raw data and metadata specific to individual sensors have been defined. Then a framework
organizing data and metadata obtained from ANT+ sensors into an open and general data format suitable for
long term storage of sensor data is introduced. Finally, a sample use-case showing the transfer of data from
a sensor into a data storage is presented.
1 INTRODUCTION
Medical treatment of sudden and especially chronic
diseases has become more expensive, especially with
aging population. For instance, there were around 23
million people in the world affected with heart failure
in 2011 (Bui et al., 2011). These people had been usu-
ally treated in hospitals for a long time. The situation
has been changing at present because relatively cheap
solutions for home treatment have appeared in the
market (Surie et al., 2008), (Kyriacou et al., 2009) and
patients can be moved from hospitals to their homes
sooner. It brings advantages of a better comfort for
patients and makes treatment generally cheaper.
Home treatment systems use a set of wearable sen-
sors, usually powered from batteries, for monitoring
of health or fitness level. They have to operate for
a long time period without possibility to change bat-
teries frequently. That is why new protocols with low
energy consumption such as ZigBee (Farahani, 2008),
Bluetooth Low Energy (Heydon, 2012) or ANT (Za-
loker, 2014) have been developed. Data from these
sensors are transferred to remote servers where they
are processed and visualized. Body Area Network
(BAN) is an integration of sensors providing a large
data collection of body parameters. When the number
of sensors connected to BAN increases, requirements
for the management, long term storage and sustain-
ability of acquired data also increases.
Although there are some low energy consumption
standards for data transfer, these are too fragmented
to allow easy manipulation with obtained data. Of
course, these standards also do not provide means
for long term storage and management of transferred
data. As a solution this paper presents how to use
a general data format called NIX (Stoewer et al.,
2014) for encapsulating and storing ANT+ sensor
data.
The paper is organized as follows. Section 2 deals
with sensor infrastructure and description of data ob-
tained from sensors. Section 3 describes existing
ANT+ profiles; the most suitable profiles for eHealth
domain are selected. Section 4 introduces a frame-
work that facilitates conversion of sensor data to the
NIX format. Section 5 presents the usage of proposed
transformation, a simple use-case is provided. Sec-
tion 6 summarizes the work and provides an outlook
to the future.
396
JeÅ¿ek P. and MouÄ ek R.
Data Format for Storing ANT+ Sensors Data.
DOI: 10.5220/0006229103960400
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 396-400
ISBN: 978-989-758-213-4
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 STATE OF THE ART
There are several approaches aiming to manage Body
Area Networks data. These solutions use semantic
web technologies or cloud infrastructures. Three lay-
ers ontology describing data from different sensors is
presented in (Mehmood et al., 2014). This ontology
facilitates development of tools used for processing of
sensor data. A Sensor-Cloud infrastructure (Yuriyama
and Kushida, 2010) represents physical sensors as vir-
tual sensors stored in a cloud infrastructure. Semantic
Sensor Web (Sheth et al., 2008) is based on annota-
tion of sensors data by means of Semantic Web. Such
annotated data can be distributed on the Internet.
3 ANT+ PROFILES DISCUSSION
The ANT protocol is popular because of its low en-
ergy consumption and existence of suitable means
for description of body parameters. ’Device profiles’
(called ANT+) define data over the network in a con-
sistent way (Innovations, 2013) and facilitate devel-
opment of sensors and management of sensor data in
application programs.
ANT+ profiles support a large scale of activities
such as cycling, walking, or measurement of body pa-
rameters such as heart rate, blood pressure, weight, or
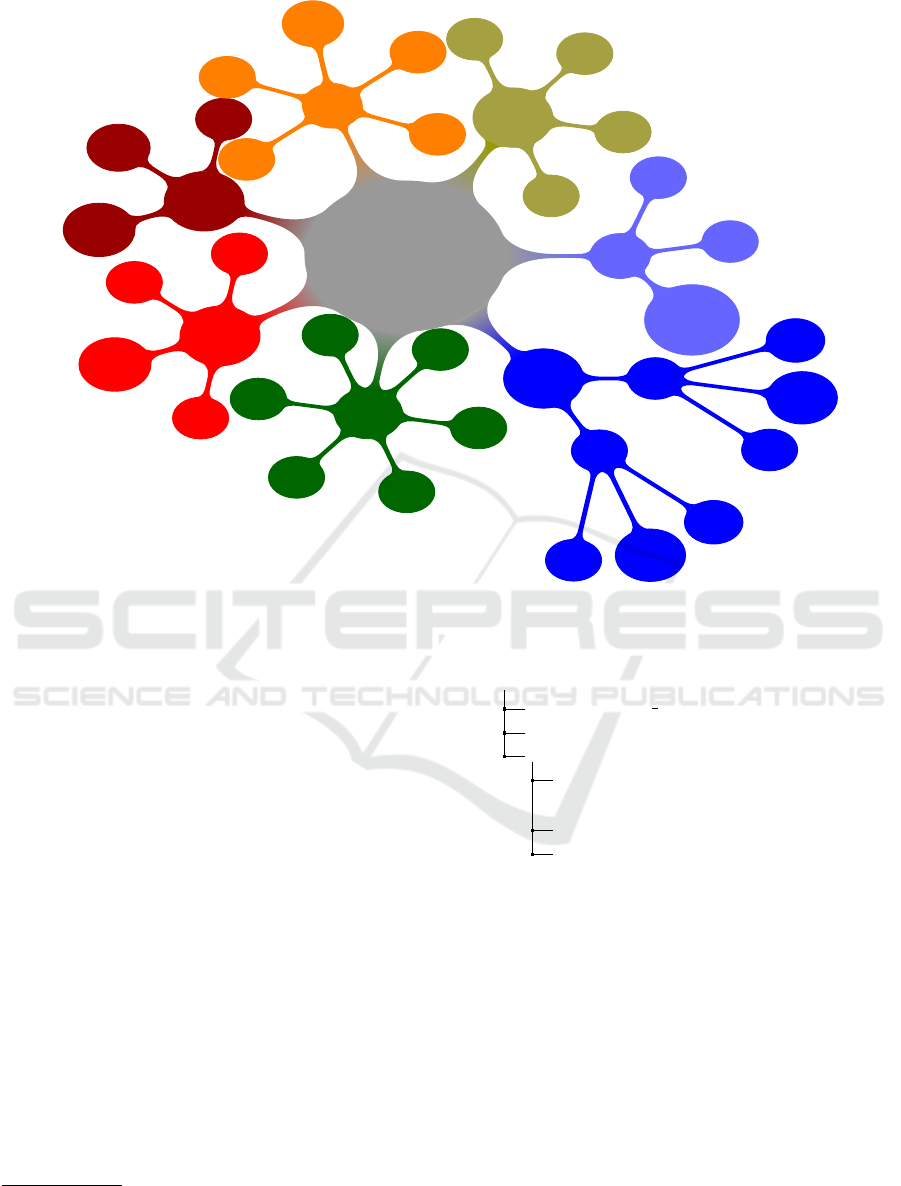
muscle oxygen. When browsing individual profiles
in a detail we find the attributes that are common for
all profiles, for example a device name, device status,
manufacturer, signal strength or battery status. Then,
there are attributes varying for individual profiles. In-
dividual profile attributes represent raw sensor data
or domain specific metadata while common attributes
describe general metadata (see Figure 1).
4 PROPOSED FRAMEWORK
4.1 Prerequisites
Due to the absence of a suitable format/data struc-
ture for sensor data representation we have designed
a framework for collection and storage of sensor raw
data and metadata in a defined structure. The used
data structure/format has to be robust, flexible and
widely accepted by scientific community to cover
a heterogeneous nature of sensor data, provide a long
term data sustainability, and ensure its re-usability in
third-party systems.
4.2 Format Discussion
Within a working group of International Neuroin-
formatics Coordinating Facility (INCF) (Bjaalie and
Grillner, 2007) and its Task Force on Electrophys-
iology
1
there were introduced two approaches to-
wards defining a standard on electrophysiology data.
The first one uses the Hierarchical Data Format
(HDF5) (HDF5 Group, 2013). HDF5 is portable
and extensible format supporting an unlimited vari-
ety of datatypes that is designed for flexible and ef-
ficient I/O operations with high volume and com-
plex data. The second approach uses odML (Grewe
et al., 2011) as a free form tree-like structure of
sections, properties and values suitable for meta-
data description. This simple, platform-independent
and human-readable format also ensures compatibil-
ity with other systems developed within the commu-
nity such as (Zehl et al., 2014), (Le Franc et al.,
2014), and (Davison et al., 2013). The next step of the
task force, merging of these two approaches (Teeters
et al., 2013), resulted in the proposal of the NIX for-
mat (Stoewer et al., 2014) that provides a data model
for storing experimental data in HDF5 together with
its metadata internally organized in the odML format.
The NIX format is currently used in Helmholtz (Davi-
son et al., 2013) and EEGBase (Jezek and Moucek,
2012) projects.
Although the NIX format was intended to be used
in electrophysiology, its general definition makes it
suitable for any time series data.
4.3 Proposed Mapping
We selected ANT+ profiles relating to person health
and/or fitness level. Figure 1 shows common meta-
data (see the central circle) and domain specific raw
data and metadata (see other circles). The NIX model
consists of several main elements: Block, DataAr-
ray, Tag, MultiTag, Source, Group, and Dimension.
Each element includes a set of attributes (such as id,
name, specific attributes) and link to metadata orga-
nized in the odML structure. We used a simplified
NIX model for mapping ANT+ elements. The Source
element represents an ANT+ device, DataArray rep-
resents raw data, Dimension represents description of
graph axes, and Block wraps a complete record.
1
http://www.incf.org/programs/datasharing/
electrophysiology-task-force
Data Format for Storing ANT+ Sensors Data
397
ANT+ device
name, ANT+
device status,
ANT+ device
number,
Battery status,
Manufacturer
identification,
Manufacturer
specific data,
ANT+ signal
strength,
Product
information
Bicykle
power
Torque
Speed
Torque
Cadence
Torque
Effective-
ness
and
Pedal
Smooth-
ness
Bicycle
Speed
and
Cadence
Speed
Sensor
Latest
Speed
Event
Time
Cumu-
lative
Wheel
Revolu-
tions
Stopped
flag
Cadence
Sensor
Latest
Cadence
Event
Time
Cumu-
lative
Pedal
Revolu-
tions
Stopped
flag
Blood
Pres-
sure
Systolic
Diastolic
Time-
stamp
Mean
Ar-
terial
Morning
/
evening
Heart
Rate
Heart
Rate
Mon-
itor
Last
Heart
Beat
Time
of Pre-
vious
Heart
Beat
Heart
Beat
Count
Heart
Rate
Muscle
Oxygen
Mon-
itor
Hemo-
globin
Con-
cen-
tration
Satu-
rated
Hemo-
globin
Capa-
bilities
Weight
Scale
Hydra-
tion
Body
fat
Active,
Basal
Metabolic
Rate
Muscle
Mass
Bone
Mass
Stride,
Speed,
Dis-
tance
Distance
Speed
Calories
Stride
Count
Figure 1: ANT+ Profiles Network.
5 USE CASE
The presented framework serves mainly to designers
and programmers of the systems for home monitor-
ing. Let’s assume the following use-case. A program-
mer wants to implement a system for heart rate moni-
toring of elderly people. There are the following sys-
tem requirements: sensors have to be easy to use and
sensors data must be easily transferred to a computer
where they are stored and evaluated. Moreover, dur-
ing regular medical checks a physician uses long term
records to check health condition of the patient and
eventually starts a treatment. It means that both the
patient and physician have access to the infrastructure
that ensures data storing and management as well as
data security, consistency and sustainability.
In this use-case we used the Garmin Premium
Strap Heart Rate Monitor as a representative of ANT+
supporting devices. The Android SDK
2
was used to
read ANT+ data into Android smart phones. We in-
tegrated this SDK into a custom mobile application
MoBio
3
that reads data from ANT+ sensors and store
2
http://developer.android.com/sdk/
3
https://github.com/NEUROINFORMATICS-GROUP-
FAV-KIV-ZCU/MoBio
Section
name = heart rate
type = metadata
Properties
Device name = Strap heart rate
monitor
Device number = 1
Product Information = Garmin
Premium Strap Heart Rate Monitor
Figure 2: Metadata from the heart rate sensor in odML
structure.
them on a SD card. The user of MoBio can pair avail-
able sensors, record data and visualize them. This
solution is available to a large number of users due
to the existence of cheap Android smart phones and
heart rate monitor straps on the market.
Since our framework was also integrated into Mo-
Bio, recorded data can be stored in the NIX format.
MoBio parses the record, metadata are transferred
into a structure with one section and several proper-
ties (see an example in Figure 2) and continuously
read heart beats data are stored into the DataArray el-
ement (see an example in Figure 3). The Source ele-
ment has an attribute metadata that contains a link to
HEALTHINF 2017 - 10th International Conference on Health Informatics
398
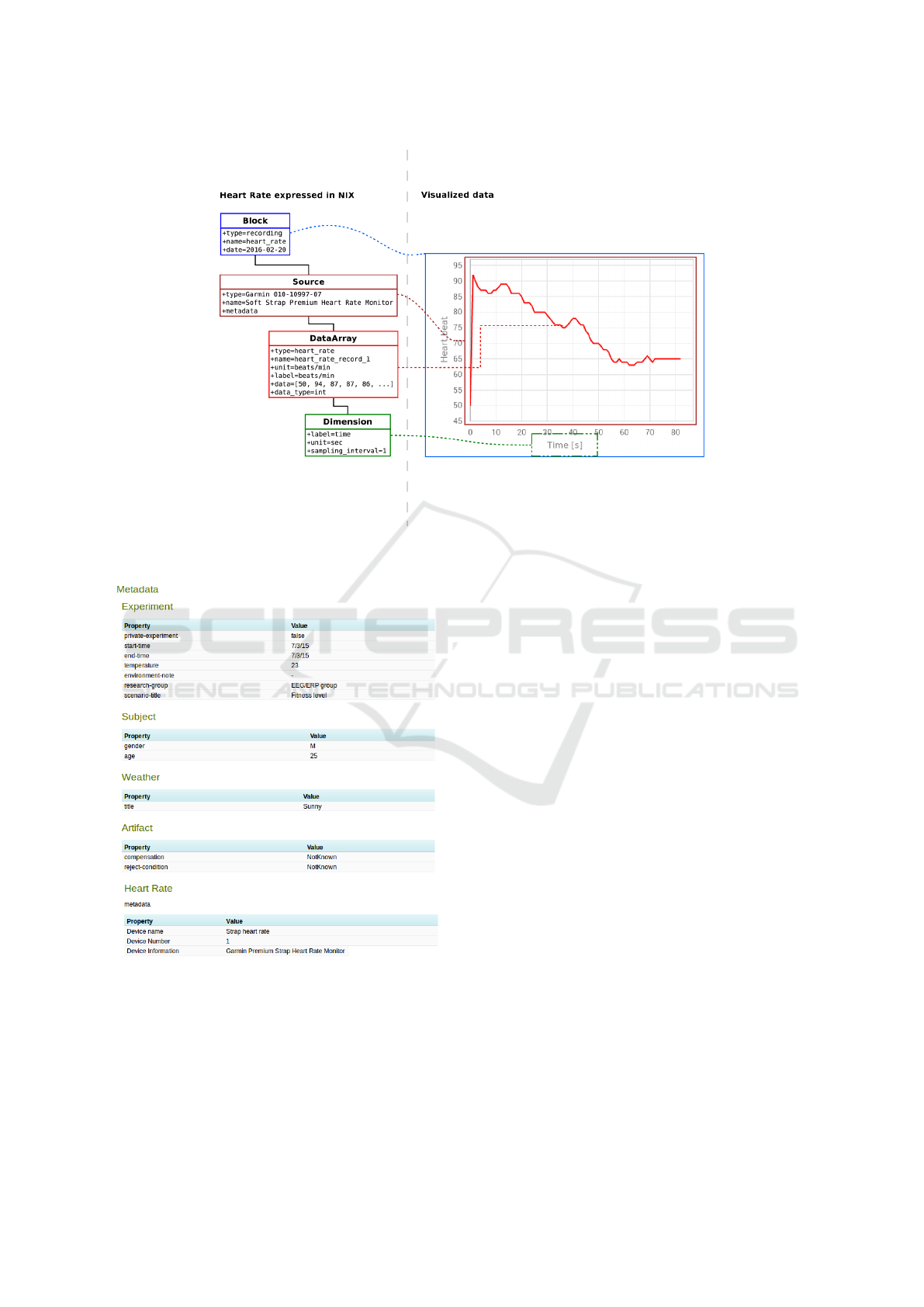
Figure 3: Heart rate record in the NIX format.
Figure 4: Metadata stored in EEGBase.
the odML structure.
Once the data and metadata are stored they can be
transferred to a suitable database. Figure 4 shows the
metadata stored and visualized in EEGBase. A com-
plete description of the experiment contains metadata
from the Heart Rate strap. The raw data are stored as
well.
6 CONCLUSIONS AND FUTURE
WORK
Together with raising popularity of sensors for home
treatment several low energy standards have been de-
fined. These standards enable data to be transferred
from body sensors into common computers where
they are processed and visualized. ANT+ supported
by significant sensors producers is one of the most
used standards. Although transfer protocols and sev-
eral APIs for working with sensors are defined, an
open standard for storing sensors data are not substan-
tially provided. Since home treatment systems use
proprietary data formats, they cannot be easily inte-
grated with variety of sensors.
In this paper we overcome these difficulties by
designing a framework that maps data from ANT+
sensors into the open and generally applicable NIX
format. The format brings advantages of two layers
structure, metadata are structured using the flexible
odML format and data are organized using the HDF5
format. Two layered organization of ANT+ sensors
data is also a significant contribution of this work.
The functionality of the framework is shown on
a simple use case. Within our future work the test-
ing of a large collection of sensors followed by data
transfer to a few databases is supposed. We also plan
to invite developers of home treatment systems to in-
tegrate the framework into their solutions. When the
framework is fully tested, we start to work on the
Data Format for Storing ANT+ Sensors Data
399

transformation of data using the Bluetooth low energy
standard into the NIX format as well.
ACKNOWLEDGEMENTS
This publication was supported by the project
LO1506 of the Czech Ministry of Education, Youth
and Sports under the program NPU I.
REFERENCES
Bjaalie, J. G. and Grillner, S. (2007). Global neuroinformat-
ics: the International Neuroinformatics Coordinating
Facility. J Neurosci, 27(14):3613–5.
Bui, A. L., Horwich, T. B., and Fonarow, G. C. (2011). Epi-
demiology and risk profile of heart failure. Nature
Reviews Cardiology, 8(1):30–41.
Davison, A. P., Brizzi, T., Guarino, D., Manette, O. F.,
Monier, C., Sadoc, G., and Fr
´
egnac, Y. (2013).
Helmholtz: a customizable framework for neurophys-
iology data management. Frontiers in Neuroinformat-
ics, (25).
Farahani, S. (2008). ZigBee Wireless Networks and
Transceivers. Newnes, Newton, MA, USA.
Grewe, J., Wachtler, T., and Benda, J. (2011). A bottom-up
approach to data annotation in neurophysiology. Fron-
tiers in Neuroinformatics, 5(16).
HDF5 Group (2013). Hierarchical data format.
http://www.hdfgroup.org/HDF5/.
Heydon, R. (2012). Bluetooth low energy: the developer’s
handbook. Prentice Hall.
Innovations, D. (2013). Ant message protocol and usage.
Jezek, P. and Moucek, R. (2012). SYSTEM FOR
EEG/ERP DATA AND METADATA STORAGE
AND MANAGEMENT. NEURAL NETWORK
WORLD, 22(3):277–290.
Kyriacou, E., Pattichis, C., and Pattichis, M. (2009). An
overview of recent health care support systems for
eemergency and mhealth applications. In Engineering
in Medicine and Biology Society, 2009. EMBC 2009.
Annual International Conference of the IEEE, pages
1246–1249.
Le Franc, Y., Gonzalez, D., Mylyanyk, I., Grewe, J., Jezek,
P., Moucek, R., and Wachtler, T. (2014). Mobile meta-
data: bringing neuroinformatics tools to the bench.
Frontiers in Neuroinformatics, (53).
Mehmood, N. Q., Culmone, R., and Mostarda, L. (2014).
An ontology driven software framework for the
healthcare applications based on ant+ protocol. In
Advanced Information Networking and Applications
Workshops (WAINA), 2014 28th International Confer-
ence on, pages 245–250. IEEE.
Sheth, A., Henson, C., and Sahoo, S. (2008). Semantic sen-
sor web. Internet Computing, IEEE, 12(4):78–83.
Stoewer, A., Kellner, C., Benda, J., Wachtler, T., and Grewe,
J. (2014). File format and library for neuroscience
data and metadata. Front. Neuroinform. Conference
Abstract: Neuroinformatics 2014.
Surie, D., Laguionie, O., and Pederson, T. (2008). Wire-
less sensor networking of everyday objects in a smart
home environment. In Intelligent Sensors, Sensor
Networks and Information Processing, 2008. ISSNIP
2008. International Conference on, pages 189–194.
Teeters, J. L., Benda, J., Davison, A. P., Eglen, S., Ger-
hard, S., Gerkin, R. C., Grewe, J., Harris, K., Jackson,
T., Moucek, R., Pr
¨
opper, R., Sessions, H. L., Smith,
L. S., Sobolev, A., Sommer, F. T., Stoewer, A., and
Wachtler, T. (2013). Considerations for developing
a standard for storing electrophysiology data in hdf5.
Frontiers in Neuroinformatics, (69).
Yuriyama, M. and Kushida, T. (2010). Sensor-cloud infras-
tructure - physical sensor management with virtual-
ized sensors on cloud computing. In Network-Based
Information Systems (NBiS), 2010 13th International
Conference on, pages 1–8.
Zaloker, J. (2014). Ant/ant+. Arrow M2M representative.
Zehl, L., Denker, M., Stoewer, A., Jaillet, F., Brochier, T.,
Riehle, A., Wachtler, T., and Gr
¨
un, S. (2014). Han-
dling complex metadata in neurophysiological exper-
iments. Frontiers in Neuroinformatics, (29).
HEALTHINF 2017 - 10th International Conference on Health Informatics
400
