
Selection of Aflatoxin B1 Mimic Epitope Peptides by Phage Display
Ke Wu
#a
, Xiaoyan Qiu
#b
and Renrong Liu
*c
School of Life Science, Jiangxi Science & Technology Normal University, Jiangxi, Nanchang, China
#
These authors contributed equally to this work
Keywords:
Aflatoxin B1, Phage Display Peptide Library, Mimic Epitope, ELISA.
Abstract:
Aflatoxin B1 (AFB1) is a natural pollutant with strong toxicity and carcinogenicity. It not only causes huge
economic losses, but also poses a serious threat to human, livestock and poultry health. To acquire the
mimic epitope peptide of aflatoxin B1 and establish a non-toxic detection system of aflatoxin, an anti-
afb13c7 monoclonal antibody was used as the target molecule, and the mimic epitopes of AFB1 were
screened from the phage random 7 peptide library. A total of 39 phage clones were selected for verification.
36 of them could specifically bind to the antibody, and 30 of them could be inhibited by AFB1, whose DNA
was extracted and sequenced. The results showed that 30 positive phage particles were actually 17 phage
clones. The common sequence was histidine (H) - proline (P) - tryptophan (W), abbreviated as xxxxhpw,
xxxhpwx, xxhpwxx, xhpwxxx (X was any amino acid). The linear range, detection limit and half inhibitory
concentration (IC50) of the 17 positive phage particles were similar. The linear range was 1-2000 pg / ml.
1 INTRODUCTION
a
Aflatoxin B1 (AFB1) is a secondary metabolite
produced by Aspergillus flavus, Aspergillus
parasiticus and Aspergillus wasabi, which has been
proved to be carcinogenic, teratogenic and
mutagenic (Riikka, 2017, Rushing, 2018). AFB1 is
easy to pollute wheat, rice and other agricultural
products, which poses a serious threat to human,
livestock and poultry health (Anja, 2016, Zhao,
2015). The method of monitoring and detecting
AFB1 is important to prevent its harm, which has
great research significance.
Domestic and foreign scholars have done a lot of
research on the detection methods of AFB1.
Since the immunoassay for AFB1 have to use the
toxin both in free and conjugated forms, it may pose
a toxicity risk to kit manufacturers and users, so it is
an urgent problem to find the substitute of AFB1
standard (Liu, 2016). The epitope peptide of AFB1
McAb was successfully screened by phage display
technology in this study.
a
https://orcid.org/0000-0002-6663-5928
b
https://orcid.org/0000-0003-2425-8316
c
https://orcid.org/0000-0001-7415-5169
2 MATERIALS AND METHODS
2.1 Reagents and Instruments
The anti-AFB1 monoclonal antibody was made in
Laboratory of Jiangxi Normal University of science
and technology. Aflatoxin B1 was purchased from
Solarbio. IPTG and X-gal were purchased from
Golden clone Biotechnology Co., Ltd. Random
phage 7 peptide library and E.coli ER2738 were
purchased from NEW ENGLAND BioLabs. Agar
Powder was purchased from Chembase.
Polyethylene glycol. PEG-8000 was purchased from
Shanghai Shenggong biology Co., Ltd. Mulitiskan
MK3 microplate reader (Thermo, USA).
Centrifuge5804r freezing centrifuge (Eppendorf,
USA). Washing machine (Thermo, USA). Pipette
gun (Thermo, USA). Polystyrene 96-well microtiter
plates (Costar).
2.2 Panning and Identification of
Positive Mimetic Peptides
The specific operation process is as follows (Wang,
2018, Shi, 2019)
The solid phase carrier (Polystyrene 96-well
microtiter plates) was coated with monoclonal
antibody and incubated with phage library for 60
Wu, K., Qiu, X. and Liu, R.
Selection of Aflatoxin B1 Mimic Epitope Peptides by Phage Display.
DOI: 10.5220/0011508600003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 1279-1283
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
1279

minutes. The unconjugated phage was washed out,
and the specific binding phage was washed down to
amplify the phage. The above steps were repeated
for 3 times and the phage titer was determined. The
final product was confirmed by positive clone
sequencing, and the binding of the selected peptide
to the monoclonal antibody was detected by ELISA.
The specific steps are as follows.
A. The anti-AFB1 monoclonal antibody was
dissolved in the PBS of 0.1M pH 7.4 (100, 75,
50μg/mL), and the 100μL/pore 4°C package was
spent overnight.
B. Discard the liquid in the hole, wash it 3 times
with TBST (including 0.1% Tween20), put the board
upside down on a clean tissue and pat it hard to
remove the residual solution. Add 3% BSA-PBS
300μL/hole 4°C to seal for 2 hours.
C. Wash it 3 times with TBST (including 0.1%-
0.05% Tween20), add the phage peptide bank
(Ph.D.-7 library) 100μL per hole (dillute it with
TBST at 1:10, containing phage about 2×1011pfu),
and shake gently at 16-22°C. It should be 1h.
D. Discard the liquid in the hole, wash it 10
times with TBST, and wash away the unbined
bacteriophages.
E. Add the lotion (Gly-HCL pH2.2) 100μL per
hole, shake it gently at 16-22°C for 8 minutes, suck
out the lotion, and add 15μL neutralizing buffer.
F. Except for 10μL for titer determination after
neutralization, the rest are added to 20mL
inoculation with the LB culture medium of E.coli
ER2738, which is in the pre- logarithmic growth
stage for amplification culture.
G. After 4.5 hours of oscillation culture of 37°C,
4°C 10000rpm centrifugal for 10 minutes, 1/6
volume of PEG/NaCl is added to the supernatant,
and 4°C precipitates overnight.
H.4°C centrifugal 15min (10000rpm), remove
the upper clearing, then use 1 mL TBST suspended
phages, and add 1/6 volume of PEG / NaCl ice to
incubate 60 minutes. 4°C centrifugal 15min
(10000rpm), de-clearing, precipitation with 200μL
TBST suspension and 10μL to measure titer.
A. Inoculated with E.coli ER2738
monobacterium colony in 5-10mL LB medium,
rocker cultured to logarithmic mid-term (OD600 at
about 0.5).
B. When cells grow, melt the upper agar and
divide it into 3mL equal parts in the sterilization test
tube, and dilute one tube for each phage. Store at
45°C for later use.
C.37°C warm up the LB/IPTG/Xgal plate, and
take one tablet for each phage dilution.
D. The bacteriophages to be tested are diluted
with a 10x series of LB medium.
E. When the bacterial culture reaches the middle
logarithmic, it is divided into 200μL equal parts in
the trace centrifugal tube, and each phage dilutes one
tube.
F. Add 10μL bacteriophages with different
dilution degrees to each tube, quickly oscillate and
mix well, and warm up 16-22oC for 1-5 min. Add
infected cells to the upper agar culture tube pre-
temperatured at 45°C, mix one tube quickly at a
time, and immediately pour them on the
LB/IPTG/Xgal plate pre-temperatured at 37°C. Tilt
the plate appropriately to spread the upper agar
evenly.
G. After the plate is cooled for 5 minutes, put it
upside down at 37°C for the night. Check the plate
and count the number of spots on the plate with
~102 phage plaques. Then multiply this number by
dilution factor to obtain the empty spot formation
unit (pfu) titer per 10μL phage. DNA of positive
phage clones was extracted and sequenced
A. Inoculate overnight cultures of E.coli ER2738
at 1: 100 dilution on LB medium and 1 mL into
culture tubes. One tube per clone to be identified.
B. Using a sterilized toothpick or tip, pick a blue
plaque into the 1 mL culture tube above. Note: The
plaque should be selected from a total of less than
100 plaque to ensure that each plaque selected
contains only one DNA sequence.
C. 37 °C shaker culture 4.5-5 h.
D. The cultures were transferred into
microcentrifuge tubes and centrifuged at 4C 30 sec
(10000 rpm). The supernatant is transferred into a
fresh tube and centrifuged again. 80% supernatant
was transferred to fresh centrifuge tube with pipette.
This was the amplified phage reservoir, stored at
4 °C or diluted with sterile glycerin 1: 1 and stored
at -20 °C.
E. coli ER2738 was inoculated in 20 mL LB
medium and cultured at 37 °C to prologarithmic
stage. Alternatively, the overnight culture of E.coli
ER2738 was diluted in a 1: 100 ratio in 20 mL LB
medium.
F. Adding 5L phage reservoir to each tube of
Ecoli ER 2738 culture medium, aerated at 37 °C for
4.5 h.
G. The above cultures were transferred into
centrifuge tubes and centrifuged at 10,000 rpm for
10 min. The supernatant is moved into a fresh
centrifuge tube and centrifuged again.
H. Take 80% supernatant in fresh centrifuge tube,
add 1 / 6 volume of PEG / NaCl. Precipitation at
4 °C overnight.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
1280

I.4℃ 10000rpm 15 min Centrifugally precipitate
and discard the supernatant.
J. Precipitate was resuspended in 1 mL TBS. The
suspension was transferred to a microcentrifuge tube
and centrifuged at 4 °C for 5 min (10000 rpm) to
remove the residual cells.
K. supernatant was transferred into a fresh
microcentrifuge tube and added 1 / 6 volume of PEG
/ NaCl to precipitate. Ice action 15-60 min.
Centrifuge 10 min (10000 rpm) at 4 °C, discard the
supernatant, and centrifuge briefly to remove the
residual supernatants. The precipitate was
resuspended in 50 L TBS and the titer of phage was
determined. 4 °C storage.
A. Monoclonal antibody against AFB1 was
dissolved in 0.1 M pH 7.4PBS (10 μg / mL) and
coated overnight with 100 μL / well 4 °C.
B. PBST washed plate three times, 3% skim milk
300μL / hole 4 °C closed 2h.
C. Addition of 1 μg / mL of AFB1 (soluble in 20%
methanol PBS) 50 μL and PBST diluted 50 μL.
(Dilution ratio was twice the maximum dilution for
identification of specifically bound positive phage
clones with absorbance values of 1.0-1.5), 37 °C 1 h.
D. PBST washed 6 times and added horseradish
peroxidase labeled anti-M13 monoclonal antibody
(100 μL / well); 37 °C acts for 1h. Wash 6 times.
Benzidine (OPD) color, 2M H2SO4 terminated
reaction, OD value was measured. The original
peptide library coated as positive control, McAb
coated with original peptide as negative control,
monoclonal antibody coated with PBST as blank
control. According to the ratio of OD value (S) of
test sample to OD (N) of negative control, the S / N
was more than 2.1. The phage which can be blocked
by AFB1 molecule is the mimotope of AFB1.
500μl of the phage-containing supernatant was
transferred to a fresh microfuge tube and added with
200μl PEG/NaCl. The mix was let stand at room
temperature for 10 minutes, then it was centrifuged
for 10 minutes, supernatant was discarded. Pellet
was suspended thoroughly in 100μl Iodide Buffer
and added with 250μl ethanol. The resuspended
solution was incubated for 10 minutes at room
temperature and spun for 10 minutes, supernatant
was discarded, the pellet was washed in 70% ethanol
and dried briefly under vacuum. The pellet was
suspended in 30μl TE buffer. Take 5ul of the above
solution for nucleic acid electrophoresis, and send
the rest to the Shanghai Shenggong biology Co., Ltd
for sequencing.
2.3 Establishment of Competitive
ELISA Standard Curve with AFB1
Mimic Epitope
Dilute AFB1 standard stock solution into a series of
concentration standard solutions. According to the
competitive steps under the optimized conditions,
the luminescence value was determined through
experiments, and the standard curve was established
(the logarithm of the toxin standard solution
concentration was the abscissa and the binding rate
was the ordinate). Specific steps are as follows
A. After amplification and purification of
positive phage particles, the optimal antibody
coating concentration and phage concentration were
determined by matrix titration.
B. anti-AFB1 monoclonal antibody soluble in 0.1
M pH7.4PBS, 100 μ L / pore 4°C was coated
overnight. The original peptide library was coated as
positive control, the monoclonal antibody coated
with primitive peptide library as negative control
and the mAb coated with PBST as blank control.
C. PBST washed plate three times, 3% skim milk
300μL / hole 4°C closed 2h.
D. Adding concentrations of 200, 100, 50, 25,
12.5, 6.25, 4, 2, 1, 0.5, 0.25, 0.125, 0.10, 0.05, 0 ng /
mL AFB1 standard (dissolved in 20% methanol PBS)
50 μ L and positive phage 50 μL, shake and mix,
keep moisture at 37°C for 1 hour.
E. PBST washed 6 times, added horseradish
peroxidase labeled anti-M13 monoclonal antibody
(100 μL / pore), 37°C for 1 h.
F. PBST washed 6 times. The reaction was
terminated by 2M H2SO4. The optical density (OD
value) at 450 nm was determined. The binding rate
(%) = B / B0 × 100% (B0 is the OD value without
AFB1 and B is the value of OD with AFB1).
Drawing Competition Inhibition Curve.
3 RESULTS
3.1 Analysis of DNA Sequence and
Polypeptide Core Series
After DNA sequencing, the amino acid sequence
was translated by the DNAMAN software. The 30
phage particles are actually 17 phage clones., No.
A1-17 The common sequence of aflatoxin mimic
epitope peptide is histidine (H) - proline (P) -
tryptophan (W), abbreviated as xxxhpwx, xhpwxxx,
xxxxhpw, xxhpwxx. X is any amino acid. The DNA
and amino acid sequences are shown in Table 1.
Selection of Aflatoxin B1 Mimic Epitope Peptides by Phage Display
1281
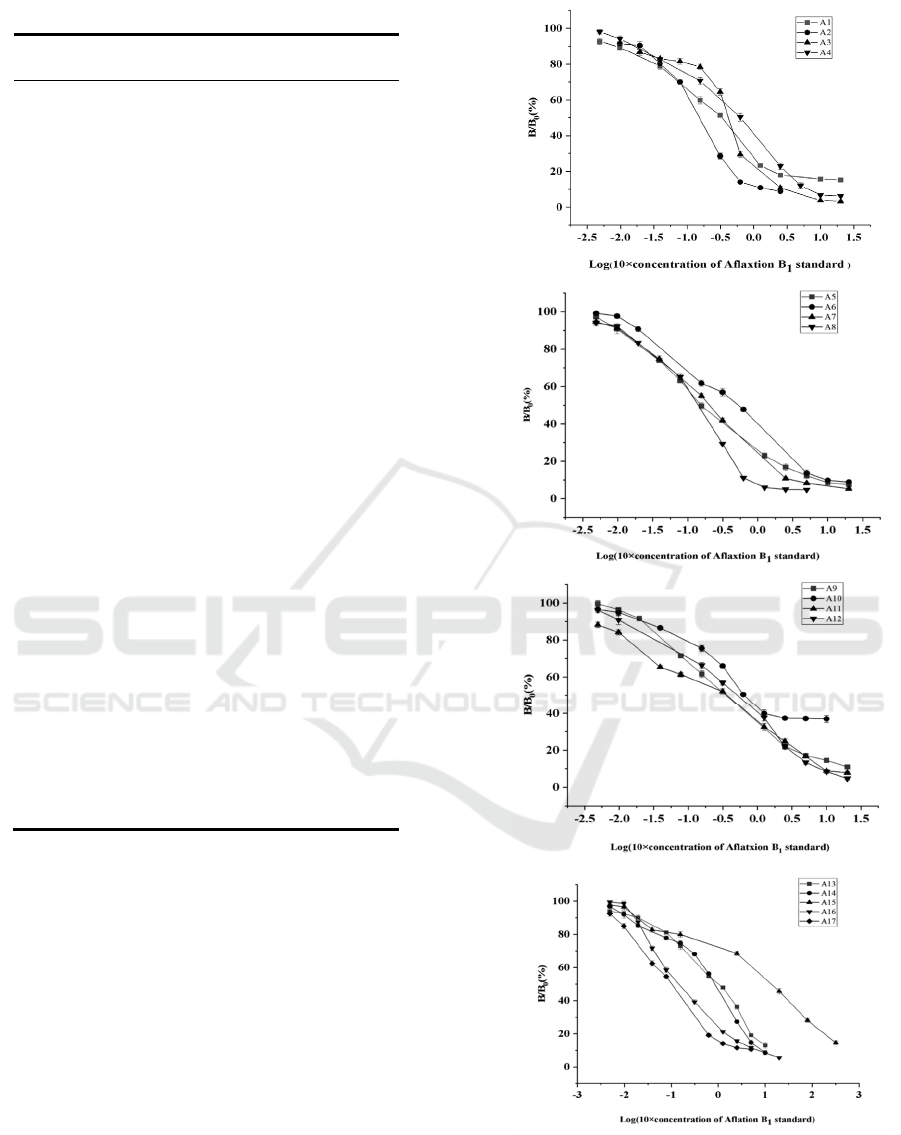
Table. 1 The inserted DNA sequences of positive phages
Ph
age
Nu
mber
DNA sequence
Petide
sequence
A1 1
AGATACAAAGTAG
GAACCAGC
SMFHPW
S
A2 2
CCACGCACCGTAG
GAACCAGA
GAWHP
WS
A3 1
ATAGTAGGAACCT
CAACCACC
YHPWSW
W
A4 1
GTCGTAAACACCG
TAGGCACC
QHLWHP
W
A5 6
ACCGGACACAAAG
TAGGAACC
WPVFHP
W
A6 1
CTCCAAAAAGTAG
GAACCAGC
EVFHPW
S
A7 2
ATAGGCCGAACCG
TAGGAACC
YPAWHP
W
A8 1
CTAAAAGGCACCG
TAGGCACC
DFPWHP
W
A9 2
CTAGTAGGCACCC
ACATAAGC
DHPWVY
S
A1
0
3
CGCGTAGGCACCC
CAGGATAA
AHPWGP
I
A1
1
2
AGCCAAACCGTAG
GAACCAGA
SVWHPW
S
A1
2
2
ATAGTAGGAACCA
GACACTGC
YHPWSV
T
A1
3
1
AAACACGTAGGCA
CCTAAACC
FVHPWI
W
A1
4
2
TTATGCGTAGGAA
CCAAAACC
NTHPWF
W
A1
5
1
CAATTACAAATAG
TAGGAACC
VNVYHP
W
A1
6
1
TGAAACGTAGGCA
CCGTAACC
TLHPWH
W
A1
7
1
CACGTCGGAATAG
TAGGCACC
VQPYHP
W
3.2 Establishment of Standard Curve
of Non-toxic ELISA with AFB1
Mimic Epitope
The standard curve of ELISA was established by
coating 96 well plate with anti AFB1 monoclonal
antibody (3c7) and incubating with phage particles
a1-a17 with mimic epitope of AFB1. The results
showed that the standard curve of ELISA based on
these phage particles showed a good linear
relationship. The linear range, detection limit and
half inhibitory concentration (IC50) were basically
similar, and the linear range was 1-2000 pg / ml (as
shown in Fig1).
Figure 1 The standard curves of competitive ELISA of
A1-A17
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
1282

4 CONCLUSION
Appropriate concentration of target protein is
beneficial to the screening of mimic peptides.
Reducing target protein concentration may improve
the specificity of washing. In order to screen antigen
mimic epitopes with high affinity, the detergent
concentration of the first round of elutriation was
0.1%, and that of the second round of elutriation was
0.5%. The number of ligands that have affinity with
the target protein at the beginning of panning is very
small. If the detergent concentration is too high, the
ligands with weak binding force may be lost. In the
second round, due to the amplification of ligands, the
concentration of detergent can be increased, and high
affinity ligands are more easily elutriated.
ACKNOWLEDGMENTS
This work was financially supported by grants from
Natural Science Foundation of China (No.31960501)
REFERENCES
Anja, S.T, Kerstin, B, Jorg,K,etal.(2016) Neutralisation of
factor VIII inhibitors by anti-idiotypes isolated from
phage-displayed libraries [J]. Thrombosis &
Haemostasis Journal of the International, 116:32-41.
Liu,J, Tan, L.M, Wang, J,etal.(2016) Complete
biodegradation of chlorpyrifos by engineered
Pseudomonas putida cells expressing surface-
immobilized laccases [J]. 157: 200-207.
Riikka,P, Elena, B. P, Rodrigo, B, etal.(2017)Microarray-
Based Immunoassay with Synthetic Mimotopes for the
Detection of Fumonisin B [J]. Analytical chemistry,
89: 6216-6223.
Rushing, B.R.& Selim, M.I. Aflatoxin B1: A review on
metabolism, toxicity, occurrence in food, occupational
exposure, and detoxification methods [J]. Food
Chemical Toxicology, 2018, 124: 81-100.
Shi, L.Y, Zhou, H, Zhao, M.(2019)Construction of
atherosclerotic antibody library using mammalian cell
surface antibody display technology[J].Chinese
Journal of Pathophysiology
Wang, Q, Fang, J, Pan, Q.H, etal.(2018) Efficient and
Stable Delivery of Multiple Genes to Fish Cells by a
Modified Recombinant Baculovirus System [J].
International Journal of Molecular Sciences, 19:3767.
Zhao, L, Ning, B, Bai, J, etal. (2015) Selection of
bisphenol A - Single-chain antibodies from a non-
immunized mouse library by ribosome display [J].
Analytical Biochemistry, 488: 59-64.
Selection of Aflatoxin B1 Mimic Epitope Peptides by Phage Display
1283
