
Enhancing Biomedical Scientific Reviews Summarization with
Graph-based Factual Evidence Extracted from Papers
Giacomo Frisoni
a
, Paolo Italiani
b
, Francesco Boschi
c
and Gianluca Moro
d
Department of Computer Science and Engineering (DISI),
University of Bologna, Via dell’Universit
`
a 50, I-47522 Cesena, Italy
Keywords:
Abstractive Document Summarization, Event Extraction, Semantic Parsing, Biomedical Text Mining, Natural
Language Processing, Natural Language Understanding.
Abstract:
Combining structured knowledge and neural language models to tackle natural language processing tasks is
a recent research trend that catalyzes community attention. This integration holds a lot of potential in doc-
ument summarization, especially in the biomedical domain, where the jargon and the complex facts make
the overarching information truly hard to interpret. In this context, graph construction via semantic parsing
plays a crucial role in unambiguously capturing the most relevant parts of a document. However, current
works are limited to extracting open-domain triples, failing to model real-world n-ary and nested biomedi-
cal interactions accurately. To alleviate this issue, we present EASumm, the first framework for biomedical
abstractive summarization enhanced by event graph extraction (i.e., graphical representations of medical evi-
dence learned from scientific text), relying on dual text-graph encoders. Extensive evaluations on the CDSR
dataset corroborate the importance of explicit event structures, with better or comparable performance than
previous state-of-the-art systems. Finally, we offer some hints to guide future research in the field.
1 INTRODUCTION
The main difficulty when dealing with text-related
tasks is taming the ambiguity of the language, where
a plethora of linguistic phenomena and writing styles
can express the same fact, often not explicitly re-
porting background knowledge for the mentioned en-
tities. Despite the unprecedented progress enabled
by deep learning in the natural language processing
(NLP) field, facts and events are still not sacred to
large transformer-based language models, which—
even with hundreds of billions of parameters (Brown
et al., 2020)—difficulty separate discrete semantic
relations from surface language structures (Bender
et al., 2021). Such superficiality mainly translates
into hallucinations (production of fabricated content)
(Zhou et al., 2021) and fragility (vulnerability to ad-
versary attacks) (Zhang et al., 2020a), creating dis-
cussions about the proper use of the term “artificial
understanding”.
Working at a semantic level is crucial in summa-
a
https://orcid.org/0000-0002-9845-0231
b
https://orcid.org/0000-0002-9710-3748
c
https://orcid.org/0000-0002-4394-3768
d
https://orcid.org/0000-0002-3663-7877
rization tasks, where models need to rephrase and
summarize long and often labyrinthine portions of
text. The biomedical literature further emphasizes
this problem, with (i) scientific documents convey-
ing precise domain-specific information, (ii) a nar-
row margin for interpretation and rephrasing, and (iii)
the non-tolerance of factual mistakes. At the same
time, given the fast-growing volume of biomedical
literature (Landhuis, 2016), providing clinicians and
researchers with tools aimed to automatically grasp
the key points of a certain topic is becoming a pre-
rogative for efficient knowledge discovery (Moradi
and Ghadiri, 2019; Frisoni et al., 2020a; Frisoni and
Moro, 2020; Frisoni et al., 2020b).
To solve these issues, the community has recently
highlighted the need for integrating multi-relational
knowledge (Colon-Hernandez et al., 2021), like ex-
ternal knowledge graphs (Yasunaga et al., 2021) or
structured representations obtained via semantic pars-
ing (Zhang et al., 2020b) or latent semantic corre-
lations (Domeniconi et al., 2016b,a; Frisoni et al.,
2020c). If the combination of language models and
knowledge graphs constitutes a research path already
explored, the same cannot be said for the second
case, where most contributions are limited to flat
168
Frisoni, G., Italiani, P., Boschi, F. and Moro, G.
Enhancing Biomedical Scientific Reviews Summarization with Graph-based Factual Evidence Extracted from Papers.
DOI: 10.5220/0011354900003269
In Proceedings of the 11th International Conference on Data Science, Technology and Applications (DATA 2022), pages 168-179
ISBN: 978-989-758-583-8; ISSN: 2184-285X
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
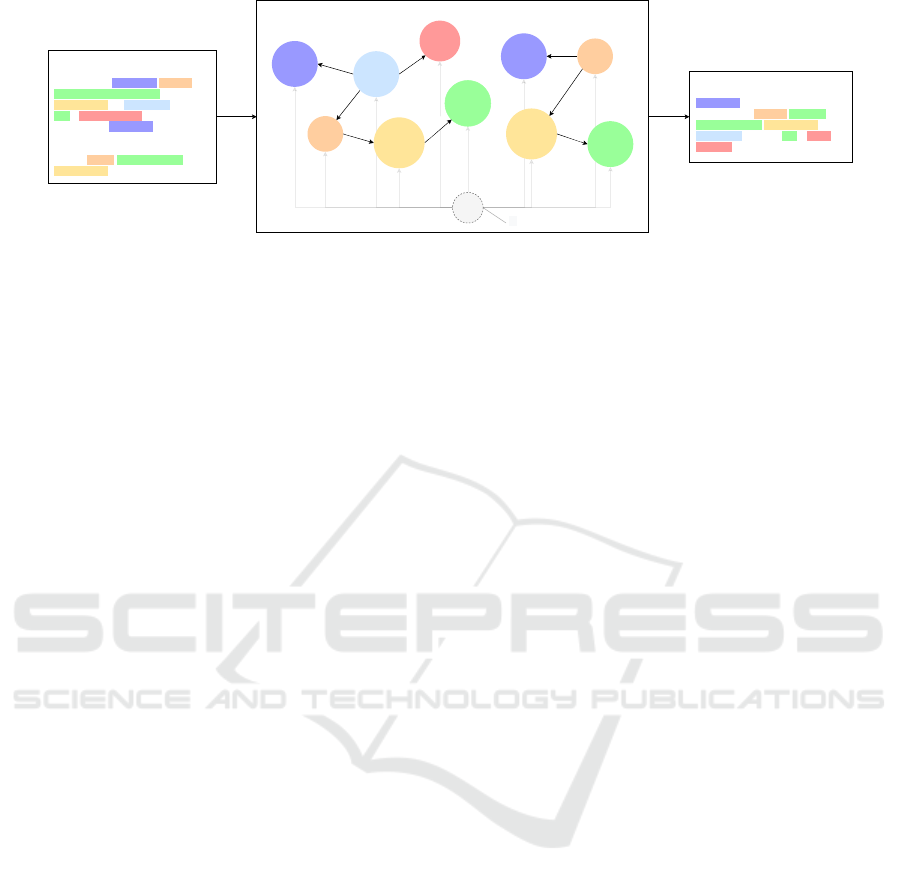
Input Article
We show that Troponin I inhibits
capillary endothelial cell
proliferation by interaction with the
cell's bFGF receptor.
In a nutshell, Troponin I (TnI) is a
novel cartilage-derived angiogenesis
inhibitor, first demonstrated by Moses
et al. to inhibit endothelial cell
proliferation ...
Troponin I
inhibits
capillary
endothelial
cell
Theme
proliferation
Theme
Cause
interaction
Theme
bFGF
receptor
Theme
Event Graph
GGP
-Reg
Cell
Cell
proliferation
Binding
GGP
Summary
Troponin I (TnI) is an angiogenesis
inhibitor that inhibits capillary
endothelial cell proliferation by
interaction with the cell's bFGF
receptor. ...
Troponin I
GGP
inhibits
-Reg
endothelial
cell
Cell
proliferation
Cell
proliferation
...
Cause
Theme
Theme
Figure 1: Sample biomedical abstractive summarization guided by events. The event graph localizes relevant information for
entities and triggers, providing a global context.
open-domain triplet-based extractions (i.e., subject-
predicate-object tuples), risking deriving incomplete
or incorrect facts non-useful for specific domains like
biomedicine (Bui et al., 2012; Frisoni et al., 2021). In
this context, event extraction (Frisoni et al., 2021)—
an advanced semantic parsing technique for deriving
n-ary and potentially nested interactions between par-
ticipants having a specific semantic role—appears as
a promising direction. We point out to the reader
that there is a well-known terminological discrepancy
between “event” and “evidence” in the biomedical
NLP community. Although the concept of “event”
is by definition associated with temporality, the field
of biomedical event extraction has evolved over the
years and today refers to a structured prediction task
concerning more generally complex relationships be-
tween entities playing arbitrary semantic roles, like
“cure” and “cause”. As such, biomedical events are
released from the presence of a temporal element
(e.g., “vitamin D modulates the immune system”). In
this paper, the keyword “event” therefore stays for
medical evidence mentioned in the scientific litera-
ture, in accordance with previous works. Still, we are
aware that this term may be misleading and requires
revision (Frisoni et al., 2021).
We propose EASUMM, the first model employing
event extraction for abstractive single-document sum-
marization, using a tandem architecture to integrate
traditional text encoders with graph representations
learned by a graph neural network (GNN). By exper-
imenting on the CDSR dataset (Guo et al., 2021), we
demonstrate biomedical event extraction graphs can
indeed help the model to preserve the essential global
context and keep the connection between the most rel-
evant entities, thus generating a higher quality sum-
mary (Figure 1).
The rest of the paper is organized as follows. First,
in Section 2, we examine related work. Then, Section
3 describes our event-based strategy for deriving se-
mantic graphs from text. Next, Section 4 details our
model, from the architecture to the training process.
Section 5 presents our experimental setup, while Sec-
tion 6 exhibits the results obtained. Finally, Section 7
closes the discussion and points out future directions.
2 RELATED WORK
Abstractive Document Summarization. Summa-
rizing text implies compressing the input document
into a shorter version, retaining salient informa-
tion, and discarding redundant or unnecessary at-
tributes. An abstractive summarizer is asked to gen-
erate new sentences, rather than simply selecting
the core ones, thus imitating a paraphrasing process
closer to human-like interpretation.
Neural models have achieved unprecedented re-
sults in recent years, mainly thanks to encoder-
decoder frameworks. In a nutshell, an encoder maps
the source tokens into a sequence of continuous rep-
resentations, while a decoder generates the summary
step-by-step. Remarkably, transformer-based archi-
tectures and self-supervised pre-training techniques
have been responsible for a profound impetus in ab-
stractive summarization (Liu and Lapata, 2019; Dong
et al., 2019; Rothe et al., 2020; Zhang et al., 2019; Qi
et al., 2020; Lewis et al., 2020)—even in low-resource
(Moro and Ragazzi, 2022) and multi-document set-
tings (Moro et al., 2022), promoting the creation of
large unlabeled corpora.
However, according to large-scale human evalua-
tions (Maynez et al., 2020), nowadays text generators
are highly prone to hallucinate content that is unfaith-
ful to the input document. For this reason, latest con-
tributions (Pasunuru and Bansal, 2018; Arumae and
Liu, 2019; Huang et al., 2020a) tend to include rein-
forcement learning modules to improve informative-
ness and consistency.
Graph-enhanced Summarization. Graphs are one
of the most effective forms for introducing external
Enhancing Biomedical Scientific Reviews Summarization with Graph-based Factual Evidence Extracted from Papers
169

knowledge into summarization models, allowing dif-
ferent quality improvements (e.g., coherence, factu-
ality, low redundancy, long-range dependencies, in-
formativeness, semantic coverage) depending on how
they are constructed.
Particularly, graph structures have long been used
for extractive summarization. In this sense, early
approaches, such as TextRank (Mihalcea and Tarau,
2004), propose to build a connectivity network with
inter-sentence cosine similarity and document-level
relations (Wan, 2008). Alternative neural systems de-
sign graph-based attention to identify important sen-
tences (Tan et al., 2017).
As for abstractive summaries, results are based on
the cross-cutting success of GNNs, which allow ap-
plying deep learning to highly structured data with-
out imposing linearization or hierarchical constraints.
Fernandes et al. (2019) extend standard sequence en-
coders with GNNs to leverage named entities and en-
tity coreferences inferred by existing NLP tools, sur-
passing models that use only the sequential structure
or graph structure. This also relates to the recent
graph verbalization trend (Song et al., 2018; Koncel-
Kedziorski et al., 2019; Agarwal et al., 2021), where
inputs may originate from both knowledge graphs
and information extraction or semantic parsing tech-
niques (e.g., abstract meaning representation, AMR).
Instead of directly generating text from a graph in
a data-to-text scenario, An et al. (2021) redefine the
task of scientific papers summarization by utilizing
a graph-enhanced encoder on top of a citation net-
work. Following a similar text-graph complementary
view—where graphs are used in addition to docu-
ment encoder—several researchers have tried to au-
tomatically build and incorporate a straightforward
and machine-readable knowledge representation of
the underlying text (Fan et al., 2019; Huang et al.,
2020b; Zhu et al., 2021), also considering different
level of granularities (Ji and Zhao, 2021). To this end,
OpenIE (Angeli et al., 2015) and Stanford CoreNLP
(Manning et al., 2014) are by far the two most popular
libraries, focusing on triplets and coreference resolu-
tion, respectively.
Notably, numerous newly introduced graph-
guided summarizers adopt LSTM models to effect
information propagation (Koncel-Kedziorski et al.,
2019; Fernandes et al., 2019; Huang et al., 2020a;
Zhu et al., 2021; An et al., 2021; Ji and Zhao, 2021),
achieving competitive performance compared to pre-
trained language models at a lower computational and
environmental cost.
3 GRAPH CONSTRUCTION
The vast majority of Relation Extraction (RE) sys-
tems focus primarily on directed or undirected extrac-
tive binary relations, which results in a list of triples
connecting only entity pairs. However, in biomedi-
cal science, flat triples are notoriously not adequate
to represent the complete biological meaning of the
original document, potentially leading to the extrac-
tion of incomplete, uninformative, or erroneous facts
(Bui et al., 2012; Frisoni et al., 2021). On the con-
trary, Event Extraction (EE) systems can handle n-
ary complex relations with nested and overlapping
definitions. According to the BioNLP-ST competi-
tions (Kim et al., 2009, 2011; N
´
edellec et al., 2013),
events are composed of a trigger (a textual mention
which clearly testifies their occurrence, e.g., “inter-
acts”, “regulates”), a type (e.g., “binding”, “regular-
ization”), and a set of arguments with a specific role,
which can be entities or events themselves. Figure 2
showcases some crucial differences in the expressive-
ness between traditional RE and EE outputs.
We construct graphs from raw documents apply-
ing DeepEventMine (shortened as DEM) (Trieu et al.,
2020), an EE system with state-of-the-art results on
seven biomedical tasks. Even when gold entities are
unavailable, DEM can detect events from raw text
with promising performance, which means that it is
able to perform named entity recognition and we do
not need to provide annotations for triggers and enti-
ties. Built on top of SciBERT (Beltagy et al., 2019),
DEM starts from enumerating all possible text spans
of a sentence (up to a certain length), then performs
a flow of entity and trigger detection, role detection,
event and modification detection in an end-to-end-
manner through custom layers.
Like other relational data, events can be shaped
as multi-relational graphs (Frisoni et al., 2021). We
model graphs taking inspiration from the definition
of Event Graphs proposed in (Frisoni et al., 2022).
The graph G = (V, E) consists of a finite set of nodes
V = v
1
, ...v
|V |
and a set of edges E ⊆ V ×V , where
edge e
i, j
connects node v
i
to node v
j
. Edges are di-
rected, labeled, and unweighted, with no cycles. A
node represents a trigger or an entity, while an edge
models an entity-trigger or a trigger–trigger relation,
with the second applying for nested events. Enti-
ties that don’t belong to any event are ignored during
graph construction. Node connections are encoded
in an adjacency matrix A ∈ R
|V |×|V|
, where a
i j
= 1
if there is a directed link from v
i
to v
j
, and 0 other-
wise. Nodes and edges in G are associated with type
information. We operate graph rewiring by adding a
master node connecting all event nodes to enhance the
DATA 2022 - 11th International Conference on Data Science, Technology and Applications
170
Input sentence
Troponin I inhibits
capillary
endothelial cell
proliferation
by interaction with the
cell's bFGF receptor.
Troponin I
inhibits
capillary
endothelial
cell
Theme
proliferation
Theme
Cause
interaction
Theme
bFGF
receptor
interaction
Theme
cell's bFGF
receptor
inhibits
inhibits cell
proliferation by
I
is with
cell
proliferation
inhibits
inhibits cell
proliferation by
inhibits
Troponin
interaction with
cell's bFGF
receptor
inhibits cell
proliferation by
inhibits cell
proliferation by
capillary
endothelial cell
proliferation
inhibits
capillary cell
proliferation
inhibits
inhibits
endothelial cell
proliferation
inhibits
cell
inhibits
bFGF
receptor
with
Event Extraction
OpenIE Extraction
GGP
-Reg
Cell
Cell
proliferation
Binding
GGP
Figure 2: Difference between semantic graphs obtained
with closed-domain Event Extraction and traditional open-
domain Relation Extraction on a real-world biomedical
sentence. The first prediction is made with DeepEvent-
Mine MLEE, while the second comes from OpenIE 5.1
(https://github.com/dair-iitd/OpenIE-standalone). An event
graph maps complex interactions mentioned in the text
to a linkage between the trigger (dark gray) and entity
(light gray) nodes, labeling edges and participants with pre-
defined roles and types specified in an ontology. On the
other hand, a graph extracted with OpenIE collects a pos-
sible set of subject-predicate-object triplets; since OpenIE
is not aligned with an ontology, nodes and entities are text
phrases. Comparing the two graphs, the latter is merely
extractive, error-prune, and devoid of additional metadata;
worse, it does not capture semantic and structured intercon-
nections between n-ary participants, often ignoring crucial
conditions for the correctness of a triplet or extracting in-
complete facts difficult to merge with post-processing.
information flow and ensure we end up with a single
graph rather than a set of small disjoint graphs.
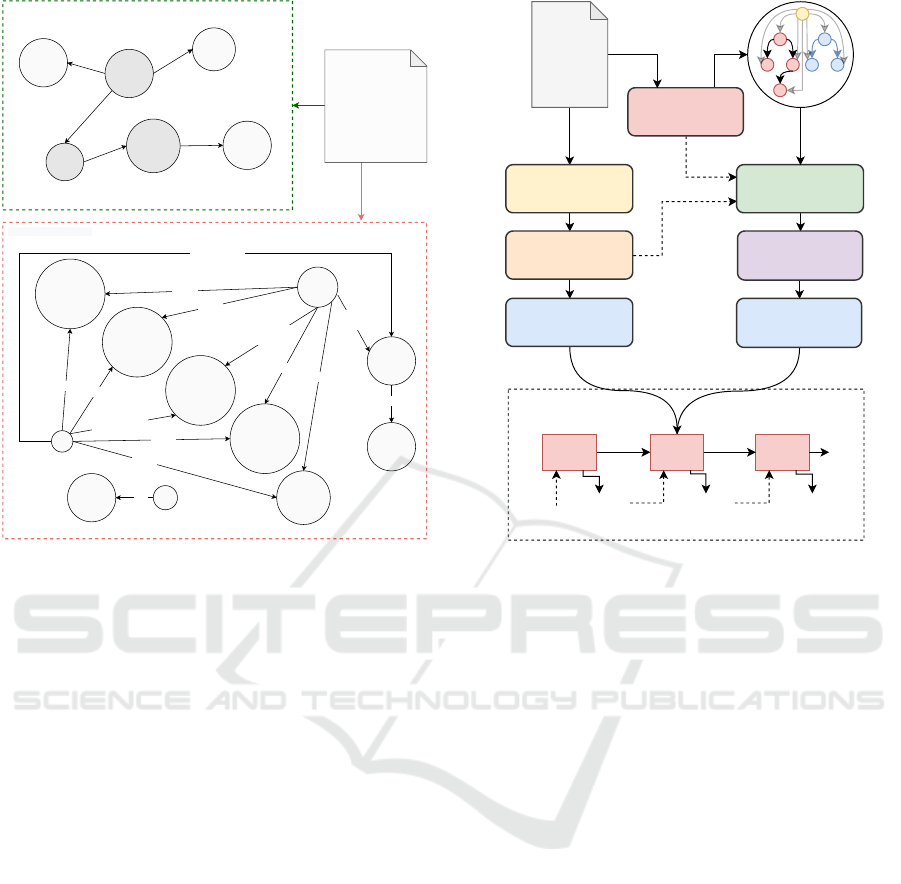
4 MODEL
Our model follows a biencoder-decoder architecture
(depicted in Figure 3), taking inspiration from (Huang
et al., 2020a). It takes two inputs, the sequence of all
tokens present in the document x = x
k
and the multi-
relational heterogeneous event-graph G (constructed
as explained in Section 3).
DeepEventMine
Input
Article
SciBERT
Bi-LSTM Layer
GAT Layers
C
t
Attention Layer
Attention Layer
Generated Summary
Node Init
C
v
t
...
<s>
Word1 Word2 Word3
Figure 3: Our event-augmented summarization framework.
The summary is generated by attending both the event graph
and the input document.
4.1 Encoders
4.1.1 Document Encoder
The sequence of tokens x is fed to SciBERT, also used
in the first layer of DEM. We take token embeddings
from the output of the last layer and we pass them
to a multi-layer bidirectional LSTM (BiLSTM), thus
obtaining the sequence of encoder hidden states h
k
.
4.1.2 Graph Encoder
Node Initialization. We initialize each node feature
v
i
by considering both its text span and entity/trigger
type. First, we average the per-token hidden states h
k
corresponding to the matched text. Then, we concate-
nate the obtained representation to the argument type
embedding s
a
(or trigger type embedding s
t
) learned
by DEM. In this regard, we believe that type metadata
can play a crucial role in augmenting the understand-
ing capacity of the model and resolving ambiguities.
The master node is represented by a vector of zeros.
Contextualized Node Encoding. The graph G is
passed to a Graph Attention Network (GAT) variant
introduced in (Koncel-Kedziorski et al., 2019), work-
ing with a self-attention setup where N independent
heads are calculated and concatenated before a resid-
ual connection is applied. Basically, each node em-
Enhancing Biomedical Scientific Reviews Summarization with Graph-based Factual Evidence Extracted from Papers
171

bedding ˆv
i
is obtained from a weighted average of its
neighboring nodes N (v
i
):
ˆ
v
i
= v
i
+ k
N
n=1
∑
j∈N (v
i
)
α
n
i j
W
0,n
v
j
, (1)
α
n
i, j
=
exp
(W
1,n
v
i
)
>
W
2,n
v
j
∑
z∈N (v
i
)
exp
(W
1,n
v
i
)
>
W
2,n
v
z
, (2)
where α
n
i j
is the attention mechanism correspond-
ing to the n-th attention head, applied to node v
i
and
node v
j
. W
∗
are trainable parameters.
4.2 Decoder
The decoder uses a multi-layer unidirectional LSTM
that generates summary tokens recurrently, exploiting
at each time step t the graph and document context
vectors c
t
, c
v
t
.
4.2.1 Attending to the Graph
The graph context vector is computed considering the
decoder hidden state s
t
:
c
v
t
=
∑
i
a
v
i,t
ˆ
v
i
, (3)
where a
v
i,t
denotes the attention mechanism (com-
puted using (Bahdanau et al., 2015)) corresponding to
the i-th node at time step t:
a
v
i,t
= softmax
u
T
0
tanh(W
3
s
t
+ W
4
ˆ
v
i
)
. (4)
u
∗
are also trainable parameters.
4.2.2 Attending to the Document
Similarly, the document context vector is computed
over input tokens by considering c
v
t
and encoder hid-
den states h
k
:
c
t
=
∑
k
a
k,t
h
k
, (5)
where a
k,t
denotes the attention corresponding to
the k-th input document token at time step t:
a
k,t
= softmax(
u
T
1
tanh(W
5
s
t
+ W
6
h
k
+ W
7
c
v
t
)
.
(6)
4.2.3 Token Prediction
The decoder hidden state s
t
is concatenated to the
document and graph context vectors, expressing the
salient content coming from both sources. This final
representation is used to compute the probability dis-
tribution of the vocabulary vocab at time step t:
P
vocab,t
= softmax (W
out
[s
t
|
c
t
|
c
v
t
]). (7)
We also include a copy mechanism as in (Huang
et al., 2020a) to check out the embedding of the token
generated at previous time step y
t−1
:
P
copy,t
= σ (W
copy
[s
t
|
c
t
|
c
v
t
| y
t−1
]). (8)
P
copy,t
∈ [0, 1] is used as a soft switch to choose
between generating a token from the vocabulary by
sampling from P
vocab,t
, or copying a token from the
input sequence by sampling from the attention distri-
bution a
k,t
. The probability of generating the token w
at time t is given by:
P
t
(w) = P
copy,t
P
vocab,t
(w)+
(1 − P
copy,t
)
∑
k:w
k
=w
a
k,t
.
(9)
4.2.4 Training Objective
We consider a negative log-likelihood loss function
function between the generated summary
ˆ
y and the
ground-truth y:
L = −
1
|D|
∑
(y,x)∈D
log p
θ
(y | x, G), (10)
where x are the source documents and y and are
the target summaries from training set D, G is the
graph constructed from x, and θ = {W
∗
, u
∗
} is the
set of the model trainable parameters.
5 EXPERIMENTAL SETUP
5.1 Dataset
We evaluate our model on the CDSR dataset (Guo
et al., 2021), designed for assessing the automated
generation of lay language summaries from biomed-
ical scientific reviews. Besides creating accurate and
factual summaries, this task also requires a joint style
transition from the original language of healthcare
professionals to that of the general public. These
properties make CDSR a perfect testbed for our so-
lution. The training, validation, and test sets contain
5178, 500, and 999 samples. As for EE, each source
document was split into a set of sentences and passed
to DEM; the results were saved in standoff .a∗ files.
The total numbers of events, entities, and triggers ex-
tracted by DEM are shown in Table 5.
DATA 2022 - 11th International Conference on Data Science, Technology and Applications
172

5.2 Training Details and Parameters
All experiments were run using a single NVIDIA
GeForce RTX 3090. We used the cased version of
SciBERT to extract token embeddings. The LSTM
models consist of 2 layers with 256-dimensional hid-
den states (128 for each direction in the encoder one).
The number N of self-attention heads of the graph
encoder GAT is set to 4. We used the version of
DEM pre-trained on the MLEE task
1
(Pyysalo et al.,
2012)—the EE benchmark linked to the biomedical
domain most aligned to CDSR based on empirical
tests (see Appendix A.2).
5.3 Baseline Methods and Comparisons
We perform extensive ablation studies by testing dif-
ferent EASUMM variants (hereinafter shortened as
EAS), which we denote through the suffix, with “−”
symbolizing a module exclusion and “+” an addi-
tion/substitution:
• −G stands for the graph encoder exclusion;
• +RB indicates the use of RoBERTa (Liu et al.,
2019) instead SciBERT to generate source docu-
ment tokens embeddings;
• −TYPE refers to the node type exclusion during
the initialization.
For a comparative analysis, we also experiment with
two extractive methods:
• Oracle extractive: it creates an oracle summary
by selecting the set of sentences in the document
that generates the highest ROUGE-2 score with
respect to the gold standard summary (i.e. extrac-
tive upper bound);
• BERT (Devlin et al., 2019): inter-sentence trans-
former layers and sigmoid classifier on top of
BERT outputs, with Oracle extractive used as su-
pervision for training;
and two abstractive methods:
• Pointer generator (See et al., 2017): standard
seq2seq model with a pointer network that allows
both copying words from the source and generat-
ing words from a fixed vocabulary;
• BART (Lewis et al., 2020): full-transformer pre-
trained on large corpora by reconstructing text af-
ter a corruption phase with an arbitrary noising
function. Besides the CDSR fine-tuning on the
vanilla version, we also take into account a vari-
ant with additional pre-training steps on PubMed
to compensate the limited training data. Specifi-
1
http://nactem.ac.uk/MLEE
cally, we use the PMC articles dataset
2
, contain-
ing 300K PubMed abstracts.
5.4 Evaluation
Quantitative Analysis. Following (Guo et al.,
2021), we use ROUGE (Lin, 2004) to evaluate the
summarization performance. ROUGE-n measures
overlap of n-grams between the model-generated
summary and the human-generated reference sum-
mary, and ROUGE-L measures the longest matching
sequence of words using the longest common sub-
sequence. We report the ROUGE-1, ROUGE-2 and
ROUGE-L scores computed using pyrouge
3
.
Given the additional scientific → public language
translation objective of the CDSR task, other than in-
formativeness, we are interested in assessing the ease
with which a reader can understand a passage, de-
fined as readability. We use three standard metrics
for this goal: Flesch-Kincaid grade level (Kincaid
et al., 1975), Gunning fog index (Gunning, 1952),
and Coleman-Liau index (Coleman and Liau, 1975).
Their formulae are as follows:
• Flesch-Kincaid grade level
0.39
total words
total sentences
+
11.8
total syllables
total words
− 15.59,
(11)
• Gunning fog index
0.4
words
sentences
+ 100
complex words
words
, (12)
where complex words are those words with three
or more syllables.
• Coleman-Liau index
0.0588L − 0.296S − 15.8, (13)
where L is the average number of letters per 100
words and S is the average number of sentences
per 100 words.
All these evaluation metrics—which we compute
using textstat
4
—estimate the years of education
generally required to understand the text. Lower
scores indicate that the text is easier to read; scores
of 13-16 correspond to college-level reading ability
in the United States education system.
2
https://www.kaggle.com/cvltmao/pmc-articles
3
https://github.com/bheinzerling/pyrouge
4
https://pypi.org/project/textstat/, version 0.72
Enhancing Biomedical Scientific Reviews Summarization with Graph-based Factual Evidence Extracted from Papers
173
Qualitative Analysis. Automatic evaluation met-
rics for judging summarization and simplification per-
formance are not able to capture all the quality aspects
of the inferenced text. To further assess the properties
of our generated summaries, we conduct an in-depth
human evaluation study to analyze desired quality di-
mensions and identify primary error sources. We ran-
domly sample 50 CDSR test set instances and hire
three native or fluent English speakers with biomedi-
cal competencies (average age: 24.6 years old; aver-
age time for completion: 2 hours; education level: 1
PhD and 2 master students; no compensation). Selec-
tion criteria ensure that our evaluators are representa-
tive of the college-educated lay public. Specifically,
we presented each human rater with the source doc-
ument, the generated summary, and the ground-truth,
asking to judge the prediction along three quality cri-
teria with a Likert scale from 1 (worst) to 5 (best).
Detailed guidelines are in Appendix A.3.
• Informativeness. Does the summary provide
enough and necessary content coverage from the
input article?
• Fluency. Does the text progress naturally? Is
it grammaticaly correct (e.g., no fragments and
missing components) and coherent whole?
• Understandability, CDSR-related (Guo et al.,
2021). Is the summary easier to understand than
the source?
We also ask evaluators to binary label whether sum-
maries contain any of the following types of unfaith-
ful errors: (i) Hallucination, fabricated content not
present in the input; (ii) Deletion or substitution, in-
correctly missing or edited elements (e.g., entities
with altered semantic role); (iii) Repetitiveness, re-
peated fragments.
6 RESULTS
6.1 Automated Summary Evaluation
6.1.1 Evaluation on Full Dataset
Table 1 shows the results of our proposed mod-
els and baseline methods. EASUMM gives better
ROUGE scores than all its variants. In particular,
we can appreciate the improvement with respect to
EASUMM−G, demonstrating the positive effect of
event graphs. We can also see how a more domain
coherent language model like SciBERT contributes to
better results than RoBERTa. Additionally, the graph
encoder in the RoBERTa implementation does not
seem to provide any progress over the solution with-
out it. The contribution of a type-augmented node
initialization technique is also clear and shows once
again how useful the semantic information extracted
by DEM is. EASUMM significantly outperforms
BERT, pointer generator, and plain Bi-LSTM archi-
tectures but struggles to beat large generative trans-
former models like BART (quality gap of ≈6 ROUGE
points), despite greater readability on average. This
behavior suggests a future direction of building our
model on top of a large pre-trained encoder-decoder
model. We also note the importance of extending
training data with other biomedical corpora.
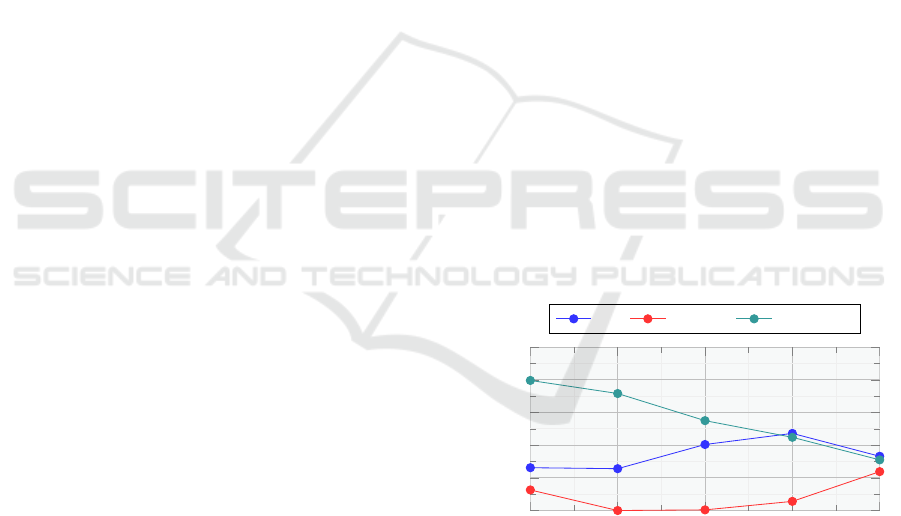
6.1.2 Evaluation on Subsets
As reported in Appendix A.2, the amount of events
extracted in each document is contained, resulting in
sparse graphs with few nodes. We hypothesize that
the graph encoder contribution could be limited by
this fact, expecting a more noticeable performance
gap concerning EASUMM−G for those documents
containing a larger number of events extracted per
sentence (shortened as EEpS). Following this line, we
build four subsets where source documents have an
EEpS greater than 0.1, 0.2, 0.3, and 0.4. Table 2
reports the ROUGE scores on each of the four sub-
sets for the different model variants and BART
BASE
.
As EEpS increases, the performance gap between the
solutions with graph encoder and solutions without
graph encoder widens, proving our supposition. We
can also notice how the EASUMM performance gets
closer to BART
BASE
.
0 0.1 0.2 0.3 0.4
0
1
2
3
4
5
EEpS
˜
R gap
EAS EAS+RB BART:EAS
Figure 4: Performance gap—measured as
˜
R (average of
ROUGE-1, ROUGE-2, and ROUGE-L)—between event-
augmented models and the number of extracted events per
sentence (EEpS). With EAS and EAS+RB, the gap is mea-
sured w.r.t. variants without the graph encoder. BART:EAS
tracks the gap between the fine-tuned BART
BASE
and EAS.
6.2 Inference Time and CO2 Impact
In line with recent graph-enhanced summarizers (Zhu
et al., 2021; An et al., 2021; Ji and Zhao, 2021), we do
not utilize an encoder-decoder architecture based on
DATA 2022 - 11th International Conference on Data Science, Technology and Applications
174
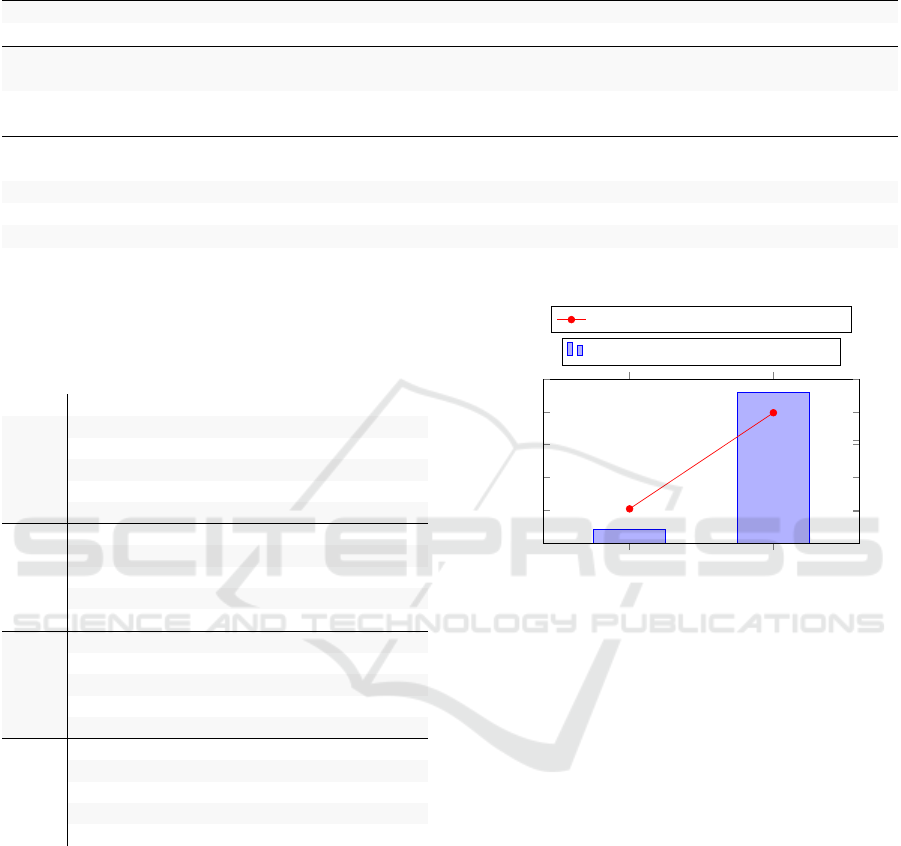
Table 1: Automated evaluation on the full testset of CDSR with ROUGE and readability metrics. Top: extractive models.
Middle: abstractive models. Bottom: our event-augmented abstractive models. Best scores for each model type are in bold.
Model ROUGE-1 ROUGE-2 ROUGE-L Flesch-Kincaid Gunning Coleman-Liau
ORACLE EXTRACTIVE 53.56 25.54 49.56 14.85 13.45 16.13
BERT 26.60 11.11 24.59 13.44 13.26 14.40
POINTER GENERATOR 38.33 14.11 35.81 16.36 15.86 15.90
BART
BASE
51.39 20.81 48.56 14.31 18.13 14.00
BART
LARGE
52.53 21.83 49.75 13.59 14.16 14.45
BART
LARGE
+PUBMED 52.66 21.73 49.97 13.30 13.80 14.28
Ours
EAS−G+RB 44.23 18.03 41.68 14.05 17.86 14.05
EAS+RB 44.12 17.82 41.60 13.57 17.29 13.77
EAS−G 44.68 17.95 42.25 12.41 16.76 12.82
EAS−TYPE 45.41 18.36 42.99 12.14 16.40 12.91
EAS 46.30 18.73 43.78 12.42 16.68 13.06
Table 2: ROUGE performance on four testset subsets, de-
pending on the minimum number of extracted events per
sentence (EEpS). ↑ and ↓ symbols denote the score increase
and decrease w.r.t. the previous subset, respectively.
EEpS Model R-1 R-2 R-L
BART
BASE
49.55 18.89 46.60
EAS−G+RB 44.45 17.65 41.13
EAS+RB 45.97 17.88 42.95
EAS−G 45.41 17.60 42.38
> 0.4
EAS 47.29 ↑ 18.50 ↑ 44.60 ↑
> 0.3
BART
BASE
49.75 19.12 46.70
EAS−G+RB 44.53 17.09 41.54
EAS+RB 44.97 16.96 42.09
EAS−G 43.87 16.77 41.14
EAS 46.77 ↑ 17.95 ↓ 44.14 ↑
BART
BASE
49.81 19.31 46.84
EAS−G+RB 44.15 16.92 41.34
EAS+RB 44.16 16.86 41.44
EAS−G 43.78 16.79 41.07
> 0.2
EAS 46.10 ↓ 18.19 ↓ 43.42 ↓
> 0.1
BART
BASE
50.77 20.23 47.81
EAS−G+RB 44.45 17.55 41.73
EAS+RB 44.48 17.41 41.82
EAS−G 44.67 17.50 42.06
EAS 46.18 18.39 43.51
pre-trained language models due to their environmen-
tal cost and computational requirements. Although
our model only uses BiLSTM and GNN structures,
experimental results show that it still achieves com-
petitive performance. Compared to SOTA generators
like BART, BiLSTM is a lightweight architecture in
terms of inference time and CO2 impact—monitored
with CodeCarbon (Schmidt et al., 2021) (Figure 5).
6.3 Human Evaluation
Table 3 shows the human evaluation results. The aver-
age Kendall’s coefficient among all evaluators’ inter-
EAS
BART
BASE
1
2
sec
Avg. per-summary generation time (sec)
0
2 · 10
−2
4 · 10
−2
6 · 10
−2
8 · 10
−2
0.1
g
Avg. per-document CO2 consumption (g)
Figure 5: Comparison between EAS with BiLSTMs (ours)
and BART
BASE
in terms of inference time and CO2 impact
on the CDSR test set.
rater agreement is 0.61. Kendall’s coefficient ranges
from -1 to 1, indicating low to high association. Con-
sidering the subjectivity of the rating task, this num-
ber indicates an high human agreement. While larger
scale studies are required, this work provides helpful
preliminary evidence. Our model obtains good scores
in fluency and understandability. Deletion and substi-
tution in verbalized facts appear to be the most com-
mon error type, together with repetitiveness. After in-
spection, we find that several utterances with swapped
entities do not belong to event mentions, thus being
not attributable to a non-effectiveness of event injec-
tion. Low hallucinations testify for the benefit de-
riving from leveraging event graph representations.
With a closer look, we observe that human-written
summaries are also discerned to contain a non-trivial
amount of hallucination errors, with humans tending
to include world knowledge not mentioned by the in-
put article. For instance, for a document discussing
about “spironolactone”, the human writer may add
“used since the 1960s” in the summary.
Enhancing Biomedical Scientific Reviews Summarization with Graph-based Factual Evidence Extracted from Papers
175

Table 3: Average human evaluation scores on informative-
ness (Inf.), fluency (Flu.), and understandability (Und.) (1-
to-5), with error percentages for hallucination (Hal.), dele-
tion or substitution (Del./Sub.), and repetitiveness (Rep.).
Inf. Flu. Und. Hal. Del./Sub. Rep.
3.16 3.4 3.44 18% 35% 34%
7 CONCLUSION
We introduced EASUMM, an abstractive lay summa-
rization model with a text-graph tandem architecture
utilizing biomedical event graphs. Our work demon-
strates the importance of event extraction for docu-
ment summarization, allowing a model to better sepa-
rate semantics and lexical surface. By achieving com-
petitive results in terms of ROUGE and readability on
CDSR, we observe a strong link between the sum-
mary quality and (i) a high number of events recog-
nized in the source document, (ii) node features ini-
tialization via domain-specific pre-trained language
models, (iii) the consideration of entity and event
types. Despite being a popular solution character-
ized by reduced inference time, we show that graph-
LSTMs struggle to compete with large pre-trained
language models such as BART, suggesting the need
for architectural improvements in future research.
Future Directions. Based on our findings, we rec-
ognize nine promising future research directions:
1. use of large pre-trained encoder-decoder trans-
formers to replace the most common graph-
LSTMs architectures;
2. increase in the number and size of the events, with
summarization datasets accompanied by event an-
notations (e.g., using metric learning techniques
like in (Moro and Valgimigli, 2021));
3. end-to-end event extraction and document sum-
marization;
4. discovering of new connections between nodes
useful for increasing summarization performance
(i.e., dynamic event graph construction), with
techniques such as random perturbation (Domeni-
coni et al., 2014a) and iterative deep graph learn-
ing (Chen et al., 2020);
5. node relevance scoring supported by term weight-
ing (Domeniconi et al., 2015) and/or perplexity
metrics (Yasunaga et al., 2021);
6. transfer-learning methods (Domeniconi et al.,
2014b; Moro et al., 2018) across multiple biomed-
ical fields;
7. exploitation of continuous edge features within
the graph neural network;
8. additional loss functions based on reinforcement
learning and semantic-driven rewards;
9. interaction and mutual influence between graph
and text encoders.
REFERENCES
Agarwal, O., Ge, H., Shakeri, S., and Al-Rfou, R. (2021).
Knowledge graph based synthetic corpus generation
for knowledge-enhanced language model pre-training.
In NAACL-HLT, pages 3554–3565. Association for
Computational Linguistics.
An, C., Zhong, M., Chen, Y., Wang, D., et al. (2021). En-
hancing scientific papers summarization with citation
graph. In AAAI, pages 12498–12506. AAAI Press.
Angeli, G., Premkumar, M. J. J., and Manning, C. D.
(2015). Leveraging linguistic structure for open do-
main information extraction. In ACL (1), pages 344–
354. The Association for Computer Linguistics.
Arumae, K. and Liu, F. (2019). Guiding extractive sum-
marization with question-answering rewards. arXiv
preprint arXiv:1904.02321.
Bahdanau, D., Cho, K., and Bengio, Y. (2015). Neural ma-
chine translation by jointly learning to align and trans-
late. In Bengio, Y. and LeCun, Y., editors, 3rd Interna-
tional Conference on Learning Representations, ICLR
2015, San Diego, CA, USA, May 7-9, 2015, Confer-
ence Track Proceedings.
Beltagy, I., Lo, K., and Cohan, A. (2019). Scibert: A
pretrained language model for scientific text. arXiv
preprint arXiv:1903.10676.
Bender, E. M., Gebru, T., McMillan-Major, A., and
Shmitchell, S. (2021). On the dangers of stochastic
parrots: Can language models be too big? In Pro-
ceedings of the 2021 ACM Conference on Fairness,
Accountability, and Transparency, pages 610–623.
Brown, T., Mann, B., Ryder, N., Subbiah, M., et al. (2020).
Language models are few-shot learners. In Larochelle,
H., Ranzato, M., Hadsell, R., Balcan, M., et al., edi-
tors, Advances in Neural Information Processing Sys-
tems, volume 33, pages 1877–1901. Curran Asso-
ciates, Inc.
Bui, Quoc-Chinh, Sloot, and M.A., P. (2012). A robust
approach to extract biomedical events from literature.
Bioinformatics, 28(20):2654–2661.
Chen, Y., Wu, L., and Zaki, M. J. (2020). Iterative deep
graph learning for graph neural networks: Better and
robust node embeddings. In NeurIPS.
Coleman, M. and Liau, T. L. (1975). A computer readabil-
ity formula designed for machine scoring. Journal of
Applied Psychology, 60:283–284.
Colon-Hernandez, P., Havasi, C., Alonso, J. B., Hug-
gins, M., et al. (2021). Combining pre-trained lan-
guage models and structured knowledge. CoRR,
abs/2101.12294.
Devlin, J., Chang, M., Lee, K., and Toutanova, K. (2019).
BERT: pre-training of deep bidirectional transform-
ers for language understanding. In NAACL-HLT
DATA 2022 - 11th International Conference on Data Science, Technology and Applications
176

(1), pages 4171–4186. Association for Computational
Linguistics.
Domeniconi, G., Masseroli, M., Moro, G., and Pinoli, P.
(2014a). Discovering new gene functionalities from
random perturbations of known gene ontological an-
notations. pages 107–116. INSTICC Press.
Domeniconi, G., Moro, G., Pagliarani, A., Pasini, K., et al.
(2016a). Job Recommendation from Semantic Sim-
ilarity of LinkedIn Users’ Skills. In ICPRAM 2016,
pages 270–277. SciTePress.
Domeniconi, G., Moro, G., Pasolini, R., and Sartori, C.
(2014b). Iterative Refining of Category Profiles for
Nearest Centroid Cross-Domain Text Classification.
In IC3K 2014, Rome, Italy, October 21-24, 2014,
Revised Selected Papers, volume 553, pages 50–67.
Springer.
Domeniconi, G., Moro, G., Pasolini, R., and Sartori, C.
(2015). A Comparison of Term Weighting Schemes
for Text Classification and Sentiment Analysis with
a Supervised Variant of tf.idf. In DATA (Revised Se-
lected Papers), volume 584, pages 39–58. Springer.
Domeniconi, G., Semertzidis, K., L
´
opez, V., Daly, E. M.,
et al. (2016b). A novel method for unsupervised
and supervised conversational message thread detec-
tion. In DATA 2016 - Proc. 5th Int. Conf. Data Sci-
ence, Technol. and Appl., Lisbon, Portugal, 24-26
July, 2016, pages 43–54. SciTePress.
Dong, L., Yang, N., Wang, W., Wei, F., et al. (2019).
Unified language model pre-training for natural lan-
guage understanding and generation. In Wallach, H.,
Larochelle, H., Beygelzimer, A., d’Alch
´
e-Buc, F.,
et al., editors, Advances in Neural Information Pro-
cessing Systems, volume 32. Curran Associates, Inc.
Fan, A., Gardent, C., Braud, C., and Bordes, A. (2019).
Using local knowledge graph construction to scale
seq2seq models to multi-document inputs. In
EMNLP/IJCNLP (1), pages 4184–4194. Association
for Computational Linguistics.
Fernandes, F. S., da Silva, G. S., Hilel, A. S., Carvalho,
A. C., et al. (2019). Study of the potential adverse ef-
fects caused by the dermal application of dillenia in-
dica l. fruit extract standardized to betulinic acid in
rodents. Plos one, 14(5):e0217718.
Frisoni, G. and Moro, G. (2020). Phenomena Explanation
from Text: Unsupervised Learning of Interpretable
and Statistically Significant Knowledge. In DATA (Re-
vised Selected Papers), volume 1446, pages 293–318.
Springer.
Frisoni, G., Moro, G., and Carbonaro, A. (2020a). Learning
Interpretable and Statistically Significant Knowledge
from Unlabeled Corpora of Social Text Messages: A
Novel Methodology of Descriptive Text Mining. In
DATA 2020 - Proc. 9th Int. Conf. Data Science, Tech-
nol. and Appl., pages 121–134. SciTePress.
Frisoni, G., Moro, G., and Carbonaro, A. (2020b). Towards
Rare Disease Knowledge Graph Learning from So-
cial Posts of Patients. In RiiForum, pages 577–589.
Springer.
Frisoni, G., Moro, G., and Carbonaro, A. (2020c). Unsuper-
vised Descriptive Text Mining for Knowledge Graph
Learning. In IC3K 2020 - Proc. 12th Int. Joint Conf.
Knowl. Discovery, Knowl. Eng. and Knowl. Manage.,
volume 1, pages 316–324. SciTePress.
Frisoni, G., Moro, G., and Carbonaro, A. (2021). A survey
on event extraction for natural language understand-
ing: Riding the biomedical literature wave. IEEE Ac-
cess, 9:160721–160757.
Frisoni, G., Moro, G., Carlassare, G., and Carbonaro, A.
(2022). Unsupervised event graph representation and
similarity learning on biomedical literature. Sensors,
22(1):3.
Gunning, R. e. a. (1952). Technique of clear writing.
Guo, Y., Qiu, W., Wang, Y., and Cohen, T. (2021). Auto-
mated lay language summarization of biomedical sci-
entific reviews. In AAAI, pages 160–168. AAAI Press.
Huang, L., Wu, L., and Wang, L. (2020a). Knowl-
edge graph-augmented abstractive summarization
with semantic-driven cloze reward. In ACL, pages
5094–5107. Association for Computational Linguis-
tics.
Huang, L., Wu, L., and Wang, L. (2020b). Knowl-
edge graph-augmented abstractive summarization
with semantic-driven cloze reward. In ACL, pages
5094–5107. Association for Computational Linguis-
tics.
Ji, X. and Zhao, W. (2021). SKGSUM: abstractive
document summarization with semantic knowledge
graphs. In IJCNN, pages 1–8. IEEE.
Kim, J., Ohta, T., Pyysalo, S., Kano, Y., et al. (2009).
Overview of bionlp’09 shared task on event extrac-
tion. In BioNLP@HLT-NAACL (Shared Task), pages
1–9. Association for Computational Linguistics.
Kim, J., Pyysalo, S., Ohta, T., Bossy, R., Nguyen, N. L. T.,
and Tsujii, J. (2011). Overview of bionlp shared task
2011. In BioNLP@ACL (Shared Task), pages 1–6. As-
sociation for Computational Linguistics.
Kim, J.-D., Wang, Y., and Yasunori, Y. (2013). The
Genia event extraction shared task, 2013 edition -
overview. In Proceedings of the BioNLP Shared Task
2013 Workshop, pages 8–15, Sofia, Bulgaria. Associ-
ation for Computational Linguistics.
Kincaid, J. P., Fishburne, R. P., Rogers, R. L., and Chissom,
B. S. (1975). Derivation of new readability formu-
las (automated readability index, fog count and flesch
reading ease formula) for navy enlisted personnel.
Koncel-Kedziorski, R., Bekal, D., Luan, Y., Lapata, M., and
Hajishirzi, H. (2019). Text generation from knowl-
edge graphs with graph transformers. In NAACL-HLT
(1), pages 2284–2293. Association for Computational
Linguistics.
Landhuis, E. (2016). Scientific literature: Information over-
load. Nature, 535(7612):457–458.
Lewis, M., Liu, Y., Goyal, N., Ghazvininejad, M., Mo-
hamed, A., Levy, O., Stoyanov, V., and Zettlemoyer,
L. (2020). BART: denoising sequence-to-sequence
pre-training for natural language generation, transla-
tion, and comprehension. In ACL, pages 7871–7880.
Association for Computational Linguistics.
Lin, C.-Y. (2004). ROUGE: A package for automatic evalu-
ation of summaries. In Text Summarization Branches
Enhancing Biomedical Scientific Reviews Summarization with Graph-based Factual Evidence Extracted from Papers
177

Out, pages 74–81, Barcelona, Spain. Association for
Computational Linguistics.
Liu, Y. and Lapata, M. (2019). Text summarization with
pretrained encoders. In EMNLP/IJCNLP (1), pages
3728–3738. Association for Computational Linguis-
tics.
Liu, Y., Ott, M., Goyal, N., Du, J., et al. (2019). Roberta:
A robustly optimized BERT pretraining approach.
CoRR, abs/1907.11692.
Manning, C. D., Surdeanu, M., Bauer, J., Finkel, J. R.,
et al. (2014). The stanford corenlp natural language
processing toolkit. In ACL (System Demonstrations),
pages 55–60. The Association for Computer Linguis-
tics.
Maynez, J., Narayan, S., Bohnet, B., and McDonald, R. T.
(2020). On faithfulness and factuality in abstractive
summarization. In ACL, pages 1906–1919. Associa-
tion for Computational Linguistics.
Mihalcea, R. and Tarau, P. (2004). Textrank: Bringing order
into text.
Moradi, M. and Ghadiri, N. (2019). Text summariza-
tion in the biomedical domain. arXiv preprint
arXiv:1908.02285.
Moro, G., Pagliarani, A., Pasolini, R., and Sartori, C.
(2018). Cross-domain & In-domain Sentiment Anal-
ysis with Memory-based Deep Neural Networks. In
IC3K 2018, volume 1, pages 127–138. SciTePress.
Moro, G. and Ragazzi, L. (2022). Semantic Self-
Segmentation for Abstractive Summarization of Long
Legal Documents in Low-Resource Regimes. In
Thirty-Sixth AAAI Conference on Artificial Intelli-
gence, AAAI 2022, Virtual Event, February 22 - March
1, 2022, pages 1–9. AAAI Press.
Moro, G., Ragazzi, L., Valgimigli, L., and Freddi, D.
(2022). Discriminative marginalized probabilistic
neural method for multi-document summarization of
medical literature. In Proceedings of the 60th An-
nual Meeting of the Association for Computational
Linguistics (Volume 1: Long Papers), pages 180–189,
Dublin, Ireland. Association for Computational Lin-
guistics.
Moro, G. and Valgimigli, L. (2021). Efficient self-
supervised metric information retrieval: A bibliogra-
phy based method applied to COVID literature. Sen-
sors, 21(19).
N
´
edellec, C., Bossy, R., Kim, J., Kim, J., Ohta, T., Pyysalo,
S., and Zweigenbaum, P. (2013). Overview of bionlp
shared task 2013. In BioNLP@ACL (Shared Task),
pages 1–7. Association for Computational Linguis-
tics.
Pasunuru, R. and Bansal, M. (2018). Multi-reward rein-
forced summarization with saliency and entailment.
In NAACL-HLT (2), pages 646–653. Association for
Computational Linguistics.
Pyysalo, S., Ohta, T., and Ananiadou, S. (2013). Overview
of the cancer genetics (CG) task of BioNLP shared
task 2013. In Proceedings of the BioNLP Shared Task
2013 Workshop, pages 58–66, Sofia, Bulgaria. Asso-
ciation for Computational Linguistics.
Pyysalo, S., Ohta, T., Miwa, M., Cho, H., et al. (2012).
Event extraction across multiple levels of biological
organization. Bioinform., 28(18):575–581.
Qi, W., Yan, Y., Gong, Y., Liu, D., et al. (2020). Prophetnet:
Predicting future n-gram for sequence-to-sequence
pre-training. In EMNLP (Findings), volume EMNLP
2020 of Findings of ACL, pages 2401–2410. Associa-
tion for Computational Linguistics.
Rothe, S., Narayan, S., and Severyn, A. (2020). Leverag-
ing Pre-trained Checkpoints for Sequence Generation
Tasks. Transactions of the Association for Computa-
tional Linguistics, 8:264–280.
Schmidt, V., Goyal, K., Joshi, A., Feld, B., et al. (2021).
CodeCarbon: Estimate and Track Carbon Emissions
from Machine Learning Computing.
See, A., Liu, P. J., and Manning, C. D. (2017). Get to
the point: Summarization with pointer-generator net-
works. In ACL (1), pages 1073–1083. Association for
Computational Linguistics.
Song, L., Zhang, Y., Wang, Z., and Gildea, D. (2018). A
graph-to-sequence model for amr-to-text generation.
In ACL (1), pages 1616–1626. Association for Com-
putational Linguistics.
Tan, J., Wan, X., and Xiao, J. (2017). Abstractive document
summarization with a graph-based attentional neural
model. In ACL (1), pages 1171–1181. Association for
Computational Linguistics.
Trieu, H., Tran, T. T., Nguyen, A. D., Nguyen, A., et al.
(2020). Deepeventmine: end-to-end neural nested
event extraction from biomedical texts. Bioinform.,
36(19):4910–4917.
Wan, X. (2008). An exploration of document impact on
graph-based multi-document summarization. In Pro-
ceedings of the 2008 conference on empirical methods
in natural language processing, pages 755–762.
Yasunaga, M., Ren, H., Bosselut, A., Liang, P., and
Leskovec, J. (2021). QA-GNN: reasoning with lan-
guage models and knowledge graphs for question an-
swering. CoRR, abs/2104.06378.
Zhang, J., Zhao, Y., Saleh, M., and Liu, P. J. (2019). PE-
GASUS: pre-training with extracted gap-sentences for
abstractive summarization. CoRR, abs/1912.08777.
Zhang, W. E., Sheng, Q. Z., Alhazmi, A., and Li, C.
(2020a). Adversarial attacks on deep-learning mod-
els in natural language processing: A survey. ACM
Trans. Intell. Syst. Technol., 11(3):24:1–24:41.
Zhang, Z., Wu, Y., Zhao, H., Li, Z., et al. (2020b).
Semantics-aware bert for language understanding. In
Proceedings of the AAAI Conference on Artificial In-
telligence, volume 34, pages 9628–9635.
Zhou, C., Neubig, G., Gu, J., Diab, M., Guzm
´
an, F.,
Zettlemoyer, L., and Ghazvininejad, M. (2021). De-
tecting hallucinated content in conditional neural se-
quence generation. In ACL/IJCNLP (Findings), vol-
ume ACL/IJCNLP 2021 of Findings of ACL, pages
1393–1404. Association for Computational Linguis-
tics.
Zhu, C., Hinthorn, W., Xu, R., Zeng, Q., Zeng, M., Huang,
X., and Jiang, M. (2021). Enhancing factual consis-
tency of abstractive summarization. In NAACL-HLT,
DATA 2022 - 11th International Conference on Data Science, Technology and Applications
178
pages 718–733. Association for Computational Lin-
guistics.
A APPENDIX
A.1 Dataset Statistics
We report additional statistics for each source and tar-
get document in CDSR (Table 4). Note: a readability
score is calculated by averaging the results of the met-
rics described in Section 5.4.
Table 4: CDSR average number of words (N. words), sen-
tences (N. sents), and readability.
Document Set N. words N. sents, Readability
Train 644 26 16.43
Val 643 26 16.60Source
Test 653 27 16.45
Target
Train 349 16 15.15
Val 348 16 15.20
Test 353 16 15.22
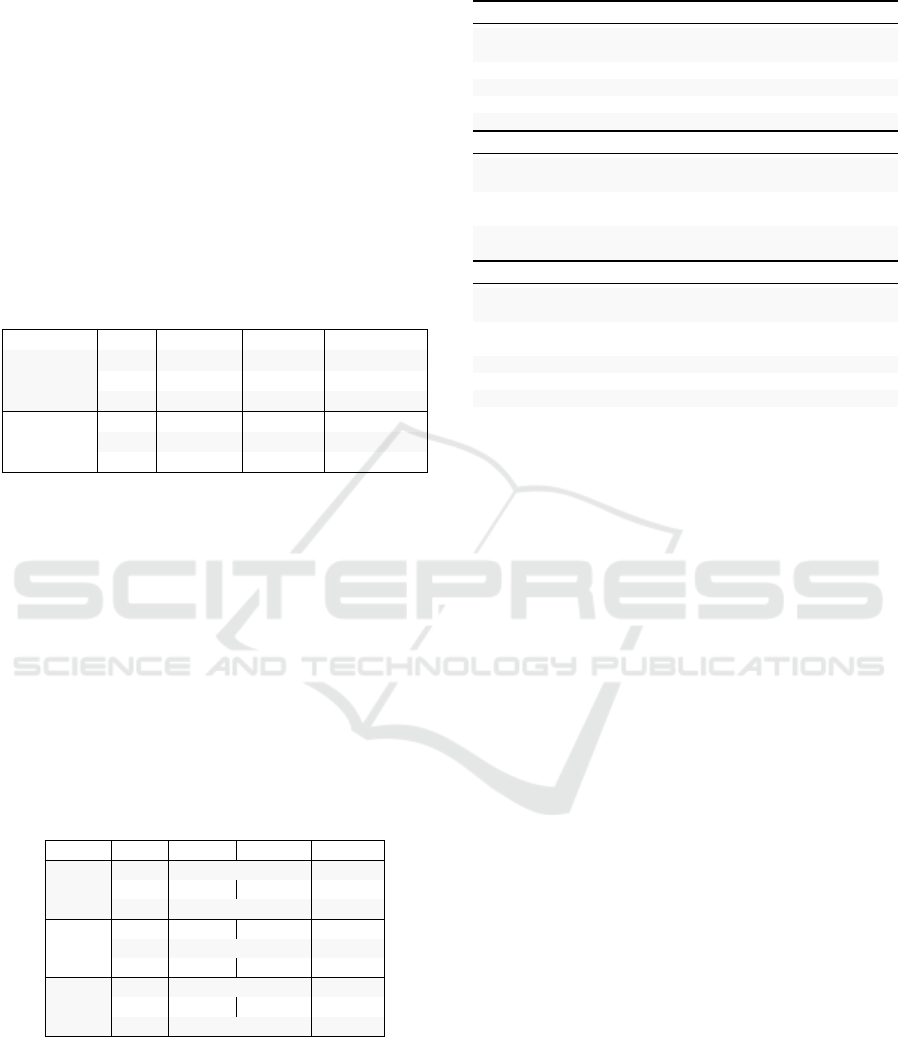
A.2 Event Extraction Dataset Selection
Table 5 provides statistics on the effectiveness of
the three DEM models related to the biomedical EE
dataset with the largest number of annotations and on-
tological targets (Frisoni et al., 2021). MLEE stands
out as the EE task most related to CDSR topics.
Table 5: CDSR event extraction results using different ver-
sions of DeepEventMine pre-trained on MLEE (Pyysalo
et al., 2012), CG13 (Pyysalo et al., 2013) and GE13 (Kim
et al., 2013) tasks. We report the average number of events
(N. evs.), triggers (N.trigs.) and arguments (N. args.) ex-
tracted from training, validation and test samples in each
source document.
Task Set N. evs. N. trigs. N. args.
Train 2.63 2.31 2.78
Val 2.54 2.20 2.73MLEE
Test 2.70 2.42 2.84
CG13
Train 2.13 1.95 2.30
Val 2.02 1.85 2.19
Test 2.12 1.95 2.30
Train 0.05 0.05 0.05
Val 0.06 0.06 0.07GE13
Test 0.07 0.06 0.06
A.3 Human Evaluation Guideline
Table 6 explains each Likert scale score meaning for
the assessed quality criteria. We believe this is impor-
tant to obtain comparable results and work towards an
objective and replicable human evaluation, minimiz-
ing ambiguity and subjectivity.
Table 6: Explanations on human evaluation aspect scales.
Informativeness:
1 Not relevant to the article
2 Partially relevant and misses the main point of the article
3 Relevant, but misses the main point of the article
4 Successfully captures the main point of the article but some
relevant content is missing
5 Successfully captures the main point of the article
Fluency:
1 Summary is full of garbage fragments and is hard to understand
2 Summary contains fragments, missing components but has some
fluent segments
3 Summary contains some grammar errors but is in general fluent
4 Summary has relatively minor grammatical errors
5 Fluent summary
Understandability:
1 Source is easier to understand than the summary
2 Summary is as understandable as the source
3 Summary is easier to understand than the source but it is
partially written in the language of healthcare professionals
4 Summary is easier to understand than the source but contains
some terms from the language of healthcare professionals
5 Summary is easier to understand than the source and is written
in the language of the general public
Enhancing Biomedical Scientific Reviews Summarization with Graph-based Factual Evidence Extracted from Papers
179
