
Metabolism, Metastasis and Drug Resistance in Cancer
Jiawei Liu
a
999 Xuefu Avenue, Honggutan District, Nanchang City, Jiangxi Province, China
Keywords:
Cancer, Metabolism, Metastasis, Drug Resistance.
Abstract:
Cancer is a heterogeneous disease caused by abnormal cell mutation, which has the characteristics of
continuous growth, invasion and metastasis. Despite research advances in cell biology, physiology and
pharmacology over the past decades, the mortality of cancer remains a healthcare issue. Current treatments
are not very effective in treating advanced tumors. Tumor microenvironment (TME) is a complex
environment referring to the surrounding tumor cells, including surrounding blood vessels, immune cells,
fibroblasts, bone marrow-derived inflammatory cells, various signal molecules and extracellular matrix
(ECM). A large number of studies have proved the key role of tumor microenvironment in the development
of cancer. Cancer associated fibroblasts (CAFs) interact with cancer cells to produce growth factors,
inflammatory factors and other factors, inhibit the immune system and promote tumor proliferation and
invasion. Tumor cells provide good conditions for cancer development by remodeling ECM and glycolysis.
This paper has aims and objectives to outline the effects of the interaction between TME and tumor cells on
tumor metabolism, metastasis and drug resistance. The molecular mechanism of TME change promoting
tumor development is discussed and the current therapeutic strategies for targeting tumor drug resistance are
mentioned. Future research with the help of artificial intelligence using large data sets as well as genome
sequencing from cancer patients is required to identify novel targets with fewer side effects in different
individuals for personalized medicine.
1 INTRODUCTION
1
Cancer is the second leading cause of death in the
world. While medical advancements over the past
few decades have increased the survival rate of
cancer, still cancers mortality rate remains. The
World Health Organization International Agency for
research on cancer (IARC) estimated that there were
19.29 million new cancer cases in 2020, including
10.06 million males and 9.23 million females,
causing 9.96 million cancer deaths worldwide,
including 5.53 million males and 4.43 million
females. Although research advances have tackled
some diseases such as infections successfully, cancer
still has a high mortality. While both tumor and
normal tissues are composed of various cell types, the
physiological functions of tumor and normal organs
are different (Egeblad, Nakasone et al. 2010).
One of the most common phenotypes of cancer
cells is uncontrolled cell proliferation. Despite
understanding the mechanism of the cell cycle, many
a
https://orcid.org/0000-0003-3524-3355
treatments are not specific and have severe side
effects with negative consequences on healthy and
rapidly dividing cell. The rapid growth of cancer cells
is mainly due to mutations conferring the ability to
use a wide range of nutrients to adapt to changing
environmental conditions. Current genome
engineering methods such as CRISPR/Cas gene
editing is not specific and validated for use in cancer
(Hanahan, Weinberg 2011). For example, tumor cells
are mainly powered by aerobic glycolysis rather than
glucose oxidative phosphorylation in their
microenvironment, and an increased expression of
fatty acid synthase (FASN) causes elevated fatty acid
synthesis during tumorigenesis of breast and prostate
cancers to support tumor metabolism, maintenance
and growth, or competitively damage anti-tumor
immunity (Lyssiotis, Kimmelman 2017).
While there are plenty of information about
metabolism, metastasis and drug resistance in cancer,
still significant unknown areas are present in our
knowledge. This paper aims to provide a brief
878
Liu, J.
Metabolism, Metastasis and Drug Resistance in Cancer.
DOI: 10.5220/0011311900003443
In Proceedings of the 4th Inter national Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 878-882
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
overview of the existing information, point out to
gaps in our knowledge and propose some ideas for
future research, hoping to provide a summary for
clinicians and researchers working on cancer as well
as opening new avenues for research and discussion.
2 METHODS AND MATERIALS
2.1 Aims and Objectives
This paper aims to investigate existing published and
peer-reviewed literature on metabolism, metastasis
and drug resistance in cancer to identify gaps in our
knowledge about the topic.
2.2 Designed Approach for the
Literature Search
A search strategy was designed and followed to
identify appropriate peer-reviewed articles written in
English from 2005-2020 from the publicly available
database PubMed.
The following terminology was used to identify
papers: cancer AND metastasis AND metabolism
AND drug resistance in the search engine.
2.3 Inclusion and Exclusion Criteria
Papers published in languages other than English and
beyond the date bracket of 2005-2020 were excluded
from the final search result.
3 DISCUSSION
Studying metabolic changes of cancer cells, including
epigenetic processes that may lead to tumorigenesis,
malignancy and cancer stem cell generation can help
us to find more effective treatments. While surgical
resection can treat some tumors during the early
stages of tumorigenesis, metastasis can lead to tumor
recurrence and even death (Steeg 2016). Therefore,
controlling the metastasis of cancer cells is one of the
crucial means to reduce mortality among cancer
patients. The stability of the normal epithelium
structure acts as the internal barrier against the
invasion of cancer cells. Epithelial mesenchymal
transition (EMT) is the key to metastasis and invasion
that is usually defined by the loss of epithelial marker
E-cadherin and the increase of mesenchymal marker
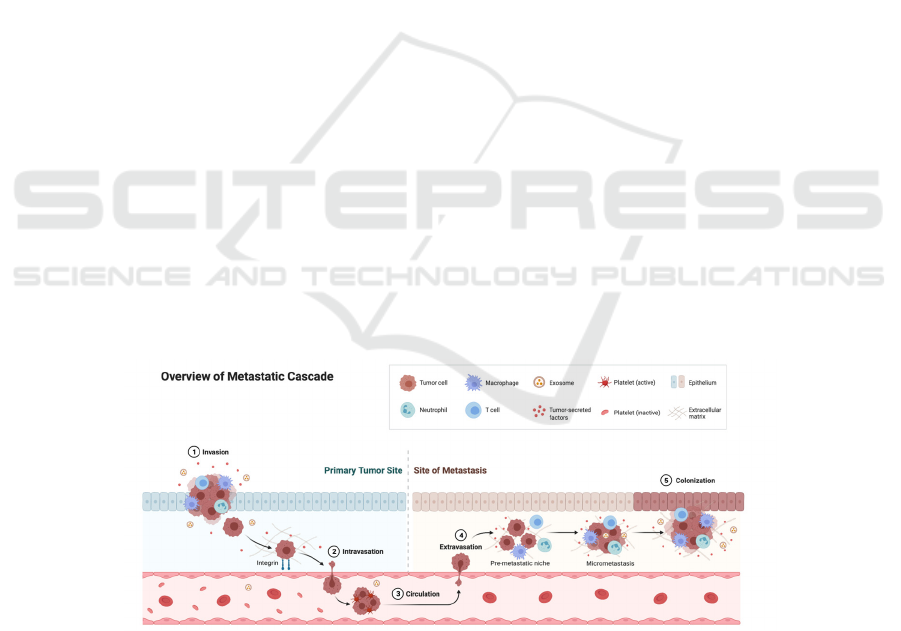
vimentin (Liu, Liu et al. 2018). The initial metastatic
cells usually undergo EMT (Thiery, Acloque et al.
2009), change their shape, transform their
metabolism, enter lymphatic vessels or vascular
lumens , attach to other cells as well as the
extracellular matrix to invade and transfer to other
parts of the body through venous and arterial
circulation (Pantel, Brakenhoff et al. 2008) (Figure
1).
Figure 1:An overview of different stages of cancer metastasis cascade from invasion to colonization. The figure was
generated using Biorender.
Besides surgery, radiotherapy, immunotherapy,
endocrine therapy and gene therapy have been used
in the treatment of various cancers. Chemotherapy is
still a common method in the treatment of cancer
(Figure 2) due to many factors, such as its potential to
destroy cancer cells and ease of administration in the
treatment of inoperable cancer.
Metabolism, Metastasis and Drug Resistance in Cancer
879
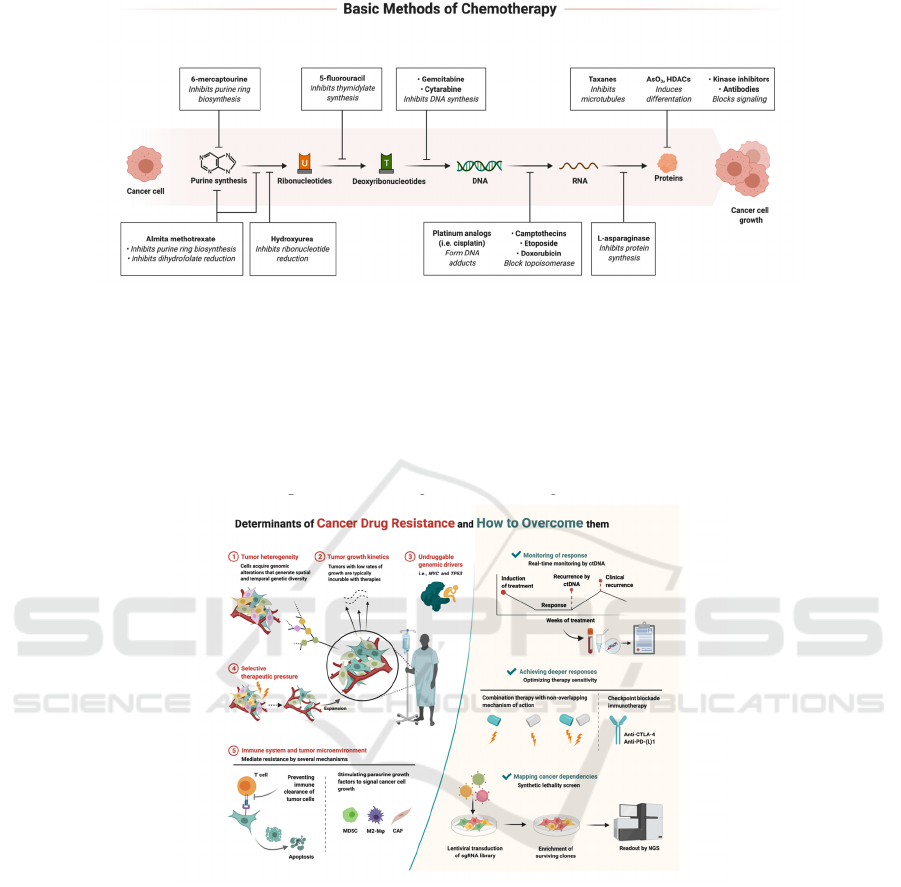
Figure 2: Basic methods of chemotherapy, including different drugs and chemicals used on different components of cancer
cells and their function. The figure was generated using Biorender.
Beyond issues such as side effects to damage
healthy cells, drug resistance can hinder
chemotherapy. Drug resistance can emerge because
of tumor heterogeneity, tumor growth kinetics,
undruggable genomic drivers, selective therapeutic
pressure such as the abnormal expression of drug
transporters as efflux transporters increased and
uptake transporters decreased, immune system and
tumor microenvironment, as well as the presence of
gene mutation (loss of tumor suppressor genes,
abnormal expression of proto-oncogenes) (Luqmani
2005) (Figure 3).
Figure 3. The determinants of cancer drug resistance and several ways to overcome drug resistance. The figure was generated
using Biorender.
3.1 Tumor Microenvironment, Tumor
Growth, and Metastasis
The environment within the tissue strongly affects the
survival and proliferation of tumor cells. Tumor cells
transform their environment to form tumor
microenvironment (TME) to maintain their survival
and proliferation (Reina-Campos, Moscat et al.
2017). Understanding the effect of cellular and non-
cellular components in TME on the metabolism of
cancer cells can provide a new way for the diagnosis
and treatment of cancer. Under normal
circumstances, the main function of activated
fibroblasts is tissue regeneration (Du, Che 2014).
During carcinogenesis, cancer cells produce a loose
microenvironment which is conducive to the further
development and invasion of tumor. The
microenvironment changes its own morphological
characteristics, which not only leads to an increase of
the number of immune and inflammatory cells such
as macrophages, but more importantly, mediates
recruitment of cancer related fibroblasts (CAFs) into
the tumor matrix by the growth factors secreted by
tumor cells. CAF supports the growth, movement and
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
880
invasion of cancer cells, leading to tumor progression,
metastasis and chemoresistance(Du, Che 2014).
CAF acts similar to pro-inflammatory factors in
the early stage of cancer development. Inflammatory
immune cells accumulate in the inflammation sites to
provide soluble growth and survival factors, matrix
remolding enzymes, reactive oxygen species and
other bioactive molecules(Kuzet and Gaggioli 2016).
These components have different effects on the
proliferation, angiogenesis, invasion and metastasis
of cancer cells.
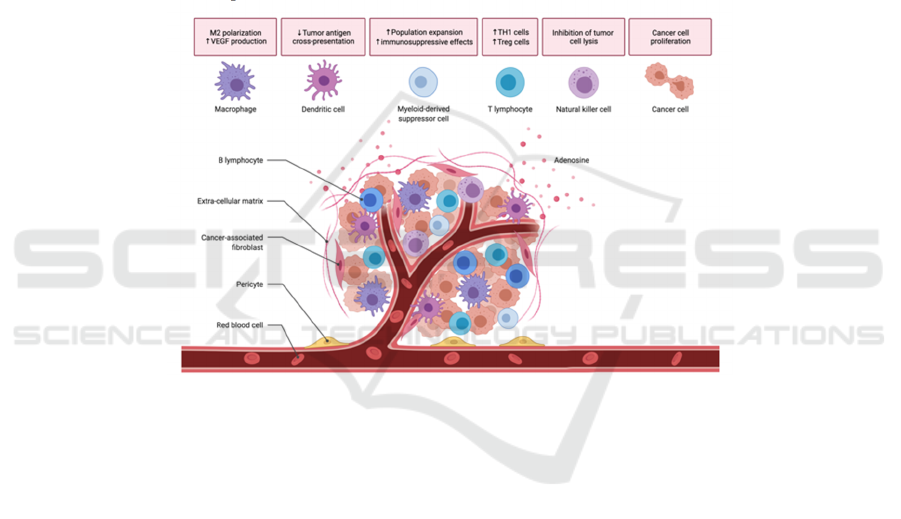
The immune system can prevent the occurrence of
primary tumor (through immune surveillance) and
metastasis by recognizing tumor specific antigen
(Bai, Meng et al. 2019). However, tumor can induce
anti-tumor immune response and immune
suppression mechanism to avoid the attack of
immune system. Macrophages are two phenotypes:
M1 like macrophages and M2 like macrophages. The
development of cancer is closely related to the
transformation of macrophages (Bai, Meng et al.
2019). The differentiation, growth and chemotaxis of
macrophages are regulated by a variety of growth
factors. colony-stimulating factor-1 (CSF-1) induces
macrophages to transform into highly plastic non
polarized (M0) macrophages. NF-κB in TME and p50
form dimer to inhibit NF-κB signal promotes
macrophages to transform from M1 inflammatory
phenotype to M2 trophic phenotype (Bai, Meng et al.
2019). This change will promote the development of
malignant tumors (Figure 4).
Figure 4: The role of different immune cells in the proliferation of tumor cells. The figure was generated using Biorender.
4 CONCLUSION AND OUTLOOK
Complex TME supports the growth, metastasis and
drug resistance of primary tumors. Studying the
mechanism of TME affecting tumor development
may facilitate cancer diagnosis and provides more
effective treatments. Determining the development
stage of tumor by identifying tumor markers can
improve the prognosis of patients. However, the
detection of a single biomarker often cannot
accurately explain the problem. Therefore, the current
research direction is to detect multiple biomarkers to
more accurately judge the development process of
cancer and predict the prognosis of patients. The
detection of some biomarkers for TME mentioned in
this paper provides new approaches for cancer
diagnosis, monitoring and treatment development. At
the same time, it provides guidance for doctors to
formulate appropriate treatment methods. The study
of the mechanism of tumor reprogramming
microenvironment and the development of drugs for
TME has created a new era of cancer medicine. At
present, some targeted therapies have been
developed. Compared with traditional chemotherapy
and radiotherapy, it has better therapeutic effects and
fewer side effects, which brings hope to develop new
treatment methods for cancer that are difficult to treat
by conventional means. Future research with the help
of artificial intelligence utilizing big data sets is
required to establish a robust map of key molecules
in tumor microenvironment for each cancer.
Furthermore, potential molecules identified from
such ‘connectome’ of tumor microenvironment can
be tested for drug responsiveness to design more
effective medication targets with fewer side effects.
In addition, with the availability of genome testing,
Metabolism, Metastasis and Drug Resistance in Cancer
881

tumors genome in different individuals can be
genetically sequenced to identify specific mutations
and provide them with specific treatment for
personalized medicine.
REFERENCES
Bai, Y., L. Meng, L. Han, Y. Jia, Y. Zhao, H. Gao, R. Kang,
X. Wang, D. Tang and E. Dai (2019). "Lipid storage and
lipophagy regulates ferroptosis." Biochem Biophys Res
Commun 508(4): 997-1003.
Du, H. and G. Che (2014). "[Advancement of relationship
between metabolic alteration in cancer-associated
fibroblasts and tumor progression in lung cancer]."
Zhongguo Fei Ai Za Zhi 17(9): 679-684.
Egeblad, M., E. S. Nakasone and Z. Werb (2010). "Tumors
as organs: complex tissues that interface with the entire
organism." Dev Cell 18(6): 884-901.
Hanahan, D. and R. A. Weinberg (2011). "Hallmarks of
cancer: the next generation." Cell 144(5): 646-674.
Kuzet, S. E. and C. Gaggioli (2016). "Fibroblast activation
in cancer: when seed fertilizes soil." Cell Tissue Res
365(3): 607-619.
Liu, Y., B. Liu, G. Q. Zhang, J. F. Zou, M. L. Zou and Z. S.
Cheng (2018). "Calpain inhibition attenuates
bleomycin-induced pulmonary fibrosis via switching
the development of epithelial-mesenchymal transition."
Naunyn Schmiedebergs Arch Pharmacol 391(7): 695-
704.
Luqmani, Y. A. (2005). "Mechanisms of drug resistance in
cancer chemotherapy." Med Princ Pract 14 Suppl 1: 35-
48.
Lyssiotis, C. A. and A. C. Kimmelman (2017). "Metabolic
Interactions in the Tumor Microenvironment." Trends
Cell Biol 27(11): 863-875.
Pantel, K., R. H. Brakenhoff and B. Brandt (2008).
"Detection, clinical relevance and specific biological
properties of disseminating tumour cells." Nat Rev
Cancer 8(5): 329-340.
Reina-Campos, M., J. Moscat and M. Diaz-Meco (2017).
"Metabolism shapes the tumor microenvironment."
Curr Opin Cell Biol 48: 47-53.
Steeg, P. S. (2016). "Targeting metastasis." Nat Rev Cancer
16(4): 201-218.
Thiery, J. P., H. Acloque, R. Y. Huang and M. A. Nieto
(2009). "Epithelial-mesenchymal transitions in
development and disease." Cell 139(5): 871-890.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
882
