
Machine Learning Techniques for Breast Cancer Detection
Karl Hall
1a
, Victor Chang
2b
and Paul Mitchell
1
1
School of Computing, Engineering and Digital Technologies, Teesside University, Middlesbrough, U.K.
2
Department of Operations and Information Management, Aston Business School, Aston University, Birmingham, U.K.
Keywords: Cancer Diagnosis, Machine Learning, Support Vector Machine, Algorithm Tuning.
Abstract: Breast cancer is the second most prevalent type of cancer overall and the most frequently occurring cancer in
women. The most effective way to improve breast cancer survival rates still lies in the early detection of the
disease. An increasingly popular and effective way of doing this is by using machine learning to classify and
analyze patient data to help identify signs of cancer. This paper explores a variety of machine learning
techniques and compares their prediction accuracy and other metrics when using the Breast Cancer Wisconsin
(Original) data set using 10-fold cross-validation methods. Of the algorithms tested in this paper, a support
vector machine model using the radial basis function kernel outperformed all other models we tested and those
previously developed by others, achieving an accuracy of 99%.
1 INTRODUCTION
As the prevalence of big data in the healthcare sector
increases (Ehrenstein et al., 2017), there is an
increasing demand for improving the value and
validity of the methodologies employed to help
clinicians make treatment decisions. Traditional,
manual methods to analyze large databases for
meaningful patterns are becoming more difficult as
the size of the databases grows. Instead, we can
analyze these larger data repositories with technology,
including machine learning techniques and statistical
analysis.
Cancer is a collection of deadly diseases and
involves the uncontrollable growth and reproduction
of cells in a specific organ of the body, developing
into a tumor. Breast cancer makes up roughly 25% of
all cancers in women (Clinton et al., 2020) and is the
most common type of cancer in women altogether. In
2020, there were 2.3 million cases and 685,000 deaths
worldwide (World Health Organization, 2021).
In breast cancer, this mostly occurs in the cells
surrounding the breast ducts. Tumors can take two
forms, either malignant or benign. Malignant tumors
can spread to other parts of the body if left untreated.
If a diagnosis of a malignant tumor is given early, the
a
https://orcid.org/0000-0003-2863-3312
b
https://orcid.org/0000-0002-8012-5852
* Corresponding authors
chance of a full recovery is significantly higher
(Caplan, May, and Richardson, 2000).
Fine needle aspirate (FNA), ultrasound,
mammography, and surgical biopsies (Mu and Nandi,
2007) are some of the popular techniques currently
used to detect breast cancer. The manual detection of
breast cancer is resource-intensive for physicians and
the difficulty of classification can sometimes be
problematic.
Cancer is one of the most feared diseases, with the
mere thought of it often causing stress and anxiety.
Using technological methods, such as machine
learning, allows healthcare professionals to increase
both the speed of diagnosis and the accuracy of
classification. We aim to contribute to breast cancer
research by improving diagnosis times through the
optimization of machine learning algorithms to
improve the success rate of breast cancer treatment.
This was achieved by fine-tuning the parameters of a
support vector machine (SVM) model using the radial
basis function (RBF) kernel, giving better results than
previous work in this domain.
116
Hall, K., Chang, V. and Mitchell, P.
Machine Learning Techniques for Breast Cancer Detection.
DOI: 10.5220/0011123200003197
In Proceedings of the 7th International Conference on Complexity, Future Information Systems and Risk (COMPLEXIS 2022), pages 116-122
ISBN: 978-989-758-565-4; ISSN: 2184-5034
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 RELATED WORKS
Several studies have been carried out to improve
current analysis methods and study breast cancer
survival in general. Many different approaches have
been taken, which have resulted in a high
classification accuracy.
The mathematical method of multi-surface pattern
separation that was applied to the diagnosis of breast
cytology was initially proposed over 30 years ago
(Wolberg and Mangasarian, 1990). They found the
correct separation in 369 out of 370 samples. These
groundbreaking techniques are still used today.
Decision table models have been used to predict
the survival rates of breast cancer (Liu et al., 2009).
They found that the survival rate was 86.5%. They
used C5 node techniques and bagging algorithms to
improve predictive performance.
Logistic regression models, artificial neural
networks, and C5 node decision trees have also been
used (Delen et al., 2005) using 10-fold cross-
validation methods to predict the survival outcome of
over 200,000 breast cancer patients. They found that
their C5 node decision tree gave the highest
predictive accuracy at 93.6% out of the models used.
Investigations of a variety of medical data sets to
determine whether the performance of K-nearest
neighbor (KNN) models is affected by the distance
function have been conducted (Hu et al., 2016). They
looked at different types of data, including mixed,
categorical, and numerical data. They also explored
different types of distance functions, such as
Minkowski, Cosine, Euclidean and Chi-Square. They
concluded that the best type of distance function to
use were Chi-Square functions.
Comparisons of the accuracy of various
supervised learning models, including RBF neural
networks, Decision Trees implementing the Iterative
Dichotomiser 3 (ID3) algorithm, Naïve Bayes and
SVM- RBF kernel, were conducted (Chaurasia and
Pal, 2014a) to determine which models are the most
useful at classifying breast cancer datasets. The most
accurate model they tested was an SVM model using
a radial basis function kernel (SVM-RBF) which
achieved a score of 96.8%.
In another study, they also explored the use of data
mining algorithms and their effectiveness at
diagnosing heart disease (Chaurasia and Pal, 2014b).
They concluded that the Classification and Regression
Trees (CART) algorithm gave the highest accuracy.
The effectiveness of SVM, KNN and probabilistic
neural networks at detecting breast cancer was
explored (Osareh and Shadgar, 2010). They combined
this with rankings of signal-to-noise ratio features and
other techniques. The highest accuracy they achieved
was 98.80% by using an SVM-RBF classifier.
A study to compare the effectiveness of Bayesian
models, KNN, SVM, Multilayer Perceptron (MP),
Random Forest (RF) and Logistical Regression (LR)
was conducted using the same Breast Cancer
Wisconsin (Original) data used in this paper (Erkal
and Ayyildiz, 2021). They found their Bayesian
Network performed the best, returning an accuracy of
97.1%.
3 DATASET
The machine learning algorithms demonstrated in this
paper were trained and tested using the Breast Cancer
Wisconsin (Original) data set. The information was
obtained by digitizing images of the FNA of a breast
mass. This data set can be used to predict whether the
cancer cells among numerous patients are benign or
malignant. The data set is made up of nine attributes
of the cell nucleus, along with a benign or malignant
cancer type classification:
•
Clump thickness 1-10
•
Uniformity of cell size: 1-10
•
Uniformity of cell shape: 1-10
•
Marginal adhesion: 1-10
•
Single epithelial cell size: 1-10
•
Bare nuclei: 1-10
•
Bland chromatin: 1-10
•
Normal nucleoli: 1-10
•
Mitoses: 1-10
•
Predicted class: 2 for benign, 4 for malignant
In the digitized images of the breast tissue,
malignant tumor images show the cell nuclei to be
inconsistently sized and asymmetrical. Conversely,
benign tumor cells are usually uniform in their shape
and size. This can be seen in Figure 1, with benign
cells on the left next to the malignant cells on the
right.
Figure 1: Medical image of benign and malignant cancer
cells. (Sizilio et al., 2012).
Machine Learning Techniques for Breast Cancer Detection
117
4 METHODOLOGY
This paper uses three types of supervised learning
classifiers: various kernels of Support Vector Machine,
K-nearest Neighbor and an Artificial Neural
Network. The SVM and KNN models were implemented
using the “Sklearn” library in Python, while the ANN
model was implemented using the “neuralnet” package in
R. Supervised learning models were chosen because
the data set contains information about the cancer
cells, allowing this information to be used as inputs
and corresponding outputs. Supervised models
perform particularly well when they are given
classification tasks with labeled data.
Data cleaning was undertaken before the
implementation of any algorithms. Incomplete or
duplicate entries were removed from the data set and
other checks to ensure the data was consistent. For
example, users should ensure that all data for each
attribute fell within the valid ranges (1-10).
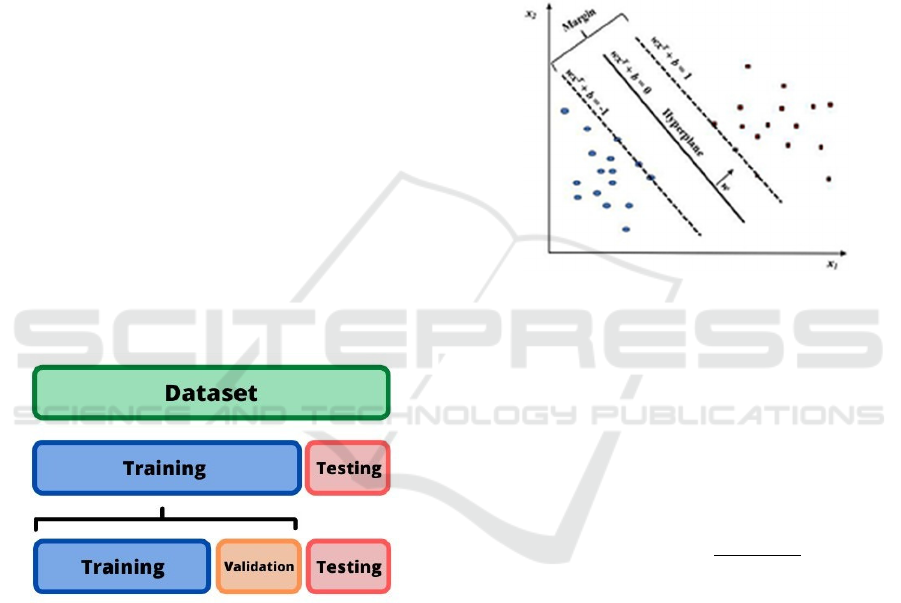
The data was split into segments for training,
testing and validation. Analytics for each model were
obtained, including accuracy, precision, and recall
scores to compare the effectiveness of the models.
The ratio of training data to testing data used was
80:20. A visualization of this process can be seen in
Figure 2.
Figure 2: Visualization of data partition segments.
4.1 Support Vector Machine
SVMs possess the capability of processing multiple
and continuous categorical variables. The SVM
constructs a hyperplane that corresponds to the
training data. The testing data is then classified
alongside the training data on this hyperplane. The
maximum marginal hyperplane is then calculated.
The margin of an SVM is defined as the distance
between the two nearest support vectors relating to the
different classes. In this case, the class refers to benign
and malignant cancer classifications. The larger the
margin, the stronger the classification. A visualization
of this process can be seen in Figure 3. The SVM
algorithm employs cross-fold validation techniques.
The optimal value for this was determined to be 10,
which results in estimates with moderate variance and
low bias (Chaves et al., 2009). In summary, this
process was repeated ten times to improve
consistency. There are different kernels that can be
used in SVM classifiers, such as RBF and polynomial,
with linear being the default kernel. In this case,
kernels define the functions that define the decision
boundaries between classes. These kernels were
compared to optimize the SVM model further.
Figure 3: SVM hyperplane and margins (Huang et al.,
2018).
4.1.1 Radial Basis Function Kernel
In contrast to the linear kernel, the RBF kernel is a
non-linear kernel often used when class boundaries
are hypothesized to be curve-shaped. By given two
samples x and x’, as feature vectors, the RBF kernel
is defined as:
𝐾
(
𝑥,𝑥
)
𝑒𝑥𝑝
|
|
𝑥𝑥
|
|
2𝜎
(1)
where |
|
𝑥𝑥
|
|
is defined as the squared Euclidian
distance between the feature vectors (Vert, Tsuda and
Schölkopf, 2004). The performance of the RBF
kernel is primarily affected by the adjustment of the
parameter 𝜎 and the cost value.
4.1.2 Polynomial Kernel
The polynomial kernel, more commonly known as
SVM-Poly, is another non-linear kernel representing
the similarity of vectors in the feature space over
polynomials of the original variables. For
polynomials of degree d, the kernel can be defined as:
𝐾
(
𝑥,𝑦
)
(𝑥𝑇𝑦+𝑐)
(2)
COMPLEXIS 2022 - 7th International Conference on Complexity, Future Information Systems and Risk
118
where x and y are vectors of features from testing and
training samples and c is the cost value.
4.2 K-nearest Neighbor
Instead of using training data like most supervised
algorithms, the lazy learning of KNN updates in real-
time as new data points are added. When this occurs,
the classification of k pre-existing data points is
considered to determine the new data classification
depending on proximity. The k-value is the number of
nearby data points considered for this classification.
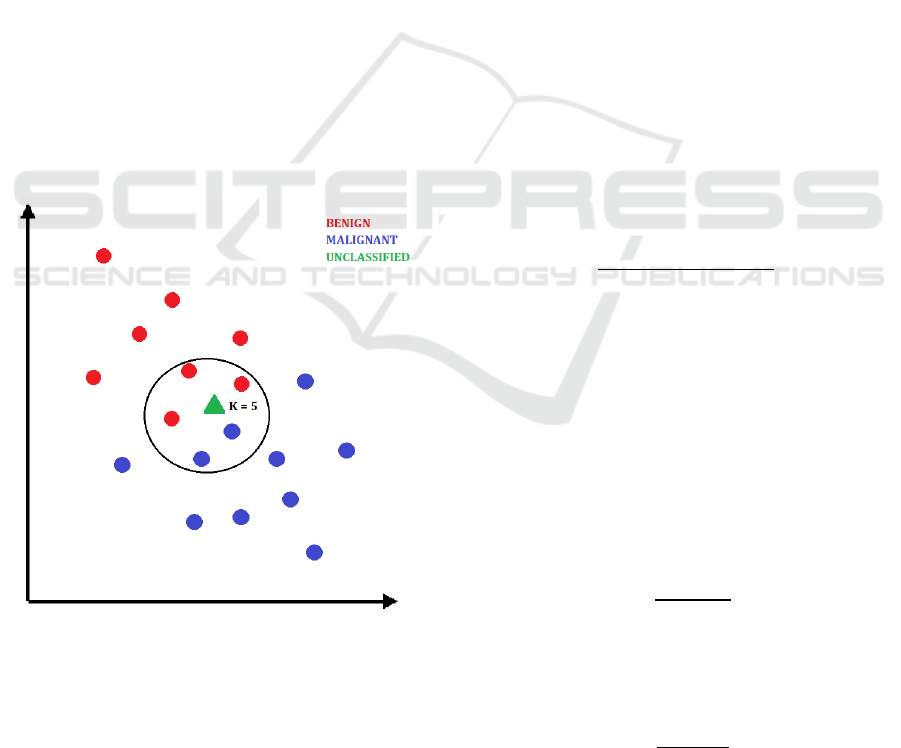
This process is visualized in Figure 4. In this case, the
new data point visualized as a green triangle would be
classified as benign.
The square root of the number of observations was
calculated to determine the optimal k-value for our
data set. This gives a value of 21.8. Therefore,
initially, two KNN models with k-values of 21 and 22
were running.
Aiming to ensure that 21 and 22 were the best k-
values, the accuracy percentage was calculated for
both, and confusion matrices were produced. A loop
was then created to run this process for all values
between 1 and 28. Finally, a graph was plotted to find
any correlation between the k-values and the accuracy
percentages.
Figure 4: Classification of a new data point with KNN.
4.3 Artificial Neural Network
The ANN uses the seed setting function to produce a
sample for the training segments. The independent
variable x was used as the starting point for the input
layer vector.
Two hidden layers were used for the network, with
each consisting of six neurons. The output layer
consists of the dependent variable y, corresponding to
the cancer class. The output layer outputs either
malignant or benign as response variables. Each value
of x was used to calculate the most likely value of y.
This was calculated by using regression algorithms
that output the corresponding values of y based on a
finite number of noisy x measurements
(Gholamrezaei and Ghorbanian, 2007).
The measurements for the nine independent
variables are obtained by the ANN as weights as they
pass through the ANN from the input layer towards
the output layer. A bias, b, is added to the neurons and
the weight values are modified as the data travels
through the hidden layers. This bias forms the net
input, n, by summing with the weighted inputs using
an activation function (Demuth and Beale, 2000).
This sum is defined as the argument of the transfer
function f (Landis and Koch, 1977).
5 RESULTS AND DISCUSSION
For each of the models, three main metrics were
obtained to assess their performance. The accuracy
score quantifies the percentage of predictions the
classifier got right and is defined in (3).
𝑇𝑃+𝑇𝑁
𝑇𝑃+𝑇𝑁+𝐹𝑃+𝐹𝑁
(3)
where TP = true positive, TN = true negative, FP =
false positive and FN = false negative. A true positive
refers to when the model correctly classifies a positive
result, a true negative is when the model correctly
classifies a negative result, a false positive is when a
positive result is incorrectly predicted, and a false
negative is when a negative result is incorrectly
predicted,
Recall, or sensitivity measures the number of
correct positive predictions made from all possible
positive predictions and is defined in (4).
𝑇𝑃
𝑇𝑃+𝐹𝑁
(4)
The F1 score combines precision (P) and recall
(R), considering both false positives and negatives and
is defined in (5).
2(𝑃∗𝑅)
𝑃+𝑅
(5)
Machine Learning Techniques for Breast Cancer Detection
119
5.1 Support Vector Machine
The c-value, or cost value to be used, was determined
using linear grids. The SVM was trialed using
different c-values ranging from 0.01 to 2.5. The best
c-value was shown to be 0.05 (Table 1). By using this
c-value, linear, RBF and polynomial kernels were
tested to compare their accuracy, recall and F1 scores.
A comparison of results is shown in Table 2. From
these results, it can be concluded that SVM-RBF
performs the best compared to SVM-Linear and
SVM-Poly.
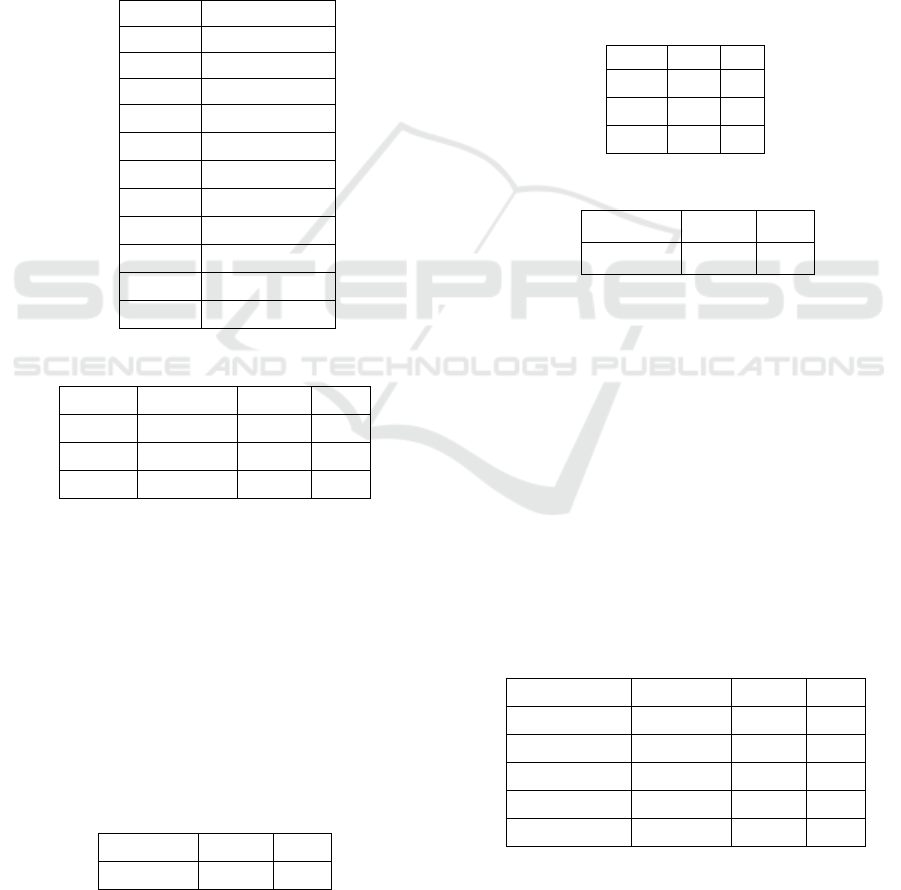
Table 1: Comparison of SVM c-values.
c-value
Accuracy (%)
0.01
96.67
0.05
97.22
0.10
96.94
0.25 97.01
0.50 97.01
0.75 97.01
1.00 96.94
1.25 96.94
1.50 96.87
1.75 96.87
2.00 96.94
Table 2: Comparison of SVM kernels.
Kernel Accuracy Recall F1
Linear 97.3 97.8 97.5
RBF 99.0 97.8 98.3
Poly 95.7 94.8 95.2
5.2 K-nearest Neighbor
When the KNN model was tested with the previously
determined optimal k-values of 21 and 22, both
returned an accuracy percentage of 93.55%.
Simulations were run for other k-values to confirm
that 21 and 22 were the optimal k-values. These were
not the optimal values, however. K-values of 3, 6 and
8 all returned higher accuracy percentages of over
94%. Using a k-value of 3 produced the results shown
in Table 3.
Table 3: KNN classifier results.
Accuracy Recall F1
94.5 92.9 93.6
5.3 Artificial Neural Network
The ANN confusion matrix (Table 4) shows the
resulting frequency of the individual data points. The
numbers 2 and 4 correspond to the malignant and
benign classifications, respectively. Of the 123
malignant data points, the network successfully
identified all but one, resulting in 122 true positives
and 1 false negative. Of the 69 benign data points, 66
were true negatives and 3 were false positives.
Overall, the neural network successfully classified
188 out of 192 cancer diagnoses, resulting in an
accuracy score of 97.92%. Table 5 shows the accuracy
along with the other metrics.
Table 4: Neural network confusion matrix.
Ref
Pre
d
2
4
2
122
1
4
3
66
Table 5: Neural network classifier results.
Accuracy Recall F1
98.0 96.2 96.7
5.4 Model Comparisons
When looking at all the models in this paper (Table 6),
it can be concluded that the SVM using the RBF
classifier outperformed all the other models across all
metrics used, except for SVM-Linear matching its
recall ability. Furthermore, our SVM- RBF model
outperformed all the models implemented by
previous research, as discussed in Section 2.
Comparisons between our results and those conducted
by other research outlined in Section 2 are shown in
Table 7. The models developed in this paper are
denoted with an asterisk (*) and are shown alongside
models implemented by others.
Table 6: Comparison of all models.
Models
Accuracy Recall
F1
SVM-RBF
99.0 97.8 98.3
ANN
98.0 96.2 96.7
SVM-Linea
r
97.3 97.8
97.5
SVM-Pol
y
95.7 94.8 95.2
KNN
94.5 92.9 93.6
COMPLEXIS 2022 - 7th International Conference on Complexity, Future Information Systems and Risk
120

Table 7: Comparison of our models to past research.
Models
Accuracy Recall
F1
SVM-RBF*
99.0 97.8 98.3
SVM-RBF
(Osareh and Shadgar,
2010)
98.8 N/A N/A
ANN*
98.0 96.2 96.7
SVM-Linear*
97.3 97.8
97.5
SVM-RBF (Chaurasia
and Pal, 2014)
96.8 N/A N/A
SVM-Pol
y
*
95.7 94.8
95.2
KNN*
94.5 92.9 93.6
DT-C5
(Delen, Walker and
Kadam, 2005)
93.6 N/A N/A
6 ETHICS
We have demonstrated a high level of ethics as
follows. First, all the data has been kept anonymous.
We do not reveal any patients’ identities. Second, the
work we do is fully GDPR compliant. We are only
allowed to analyze data that we have the permission
to use, which does not enclose any sensitive data at
all. Third, our results and analyses do not reveal any
patient identities or any sensitive information. Fourth,
we follow strict privacy regulations and data
governance to ensure the integrity and ethics of our
work. We retain a high level of professionalism and
responsibility towards the ethical use of the data and
the ethical requirements in performing data
processing, analysis and visualization.
Our research follows an ethical framework for
breast cancer detection. In other words, we design,
deploy and validate our algorithms that follow ethical
requirements. Our analyses do not reveal or leak any
sensitive information. Any scientific work, including
machine learning algorithm and development, are
only used on top of following ethical requirements
and compliance.
7 CONCLUSION
With the increasing popularity of big data in the
healthcare sector, larger data sets are becoming
available. One concern throughout this project was
the size of the data set used. The Breast Cancer
Wisconsin (Original) data set consists of less than 700
entries. Different machine learning algorithms
perform better or worse depending on the size of the
data set and the number of data features that can
impact the outcome. SVM algorithms traditionally
perform very well at binary classification problems
with pre-labeled data, which can explain why SVM-
based models outperform other models when using this
data set, in both this paper and work conducted by
others.
Other machine learning techniques for
classification problems could prove even more
accurate than those explored in this paper when
implemented with a more modern approach. Neural
networks and other deep learning algorithms tend to
perform well on very large data sets. When given a
larger and more complex data set, it can be
hypothesized that neural networks would see an
increase in performance compared to other models.
Therefore, one suggestion for future direction in this
area is to explore how different sizes of data sets impact
on the performance of machine learning algorithms.
Random Forest, XGBoost, LightGMB and
CatBoost are examples of increasingly popular
algorithms that can be utilized for handling
classification problems as part of future research to
aid early disease diagnosis. These algorithms fall
under the category of ensemble learning algorithms,
whereby multiple models are integrated
simultaneously and often achieve better performance
than singular models. Additionally, our work is fully
ethical and GDPR compliant and follows strict
privacy and data protection.
It is also possible that a similar approach of
adapting and improving machine learning models for
uses on binary-class data sets can be utilized to
improve medical outcomes for other diseases in the
future, such as COVID-19 and diabetes.
ACKNOWLEDGEMENTS
The Breast Cancer Wisconsin (Original) data set used
in this paper was obtained from the University of
Wisconsin Hospitals, Madison, from Dr. William H.
Wolberg. This work is partly supported by VC
Research (VCR 0000174) for Prof. Chang.
REFERENCES
Caplan, L. S., May, D. S., & Richardson, L. C. (2000). Time
to diagnosis and treatment of breast cancer: results from
the National Breast and Cervical Cancer Early
Detection Program, 1991-1995. American journal of
public health, 90(1), 130.
Machine Learning Techniques for Breast Cancer Detection
121

Chaurasia, V., & Pal, S. (2014a). Data mining techniques:
to predict and resolve breast cancer survivability.
International Journal of Computer Science and Mobile
Computing IJCSMC, 3(1), 10-22.
Chaurasia, V. and Pal, S. (2014b). Performance analysis of
data mining algorithms for diagnosis and prediction of
heart and breast cancer disease. Review of research,
3(8).
Chaves,
R.,
Ram´ırez,
J.,
Go´rriz,
J.,
Lo´pez,
M.,
Salas-Gonzalez, D., Alvarez, I., and Segovia, F. (2009).
Svm-based computer-aided diagnosis of the
Alzheimer’s disease using t-test nmse feature selection
with feature correlation weighting. Neuroscience
let- ters, 461(3):293–297.
Clinton, S. K., Giovannucci, E. L., and Hursting, S. D.
(2020). The world cancer research fund/american
institute for cancer research third expert report on diet,
nutrition, physical activity, and cancer: impact and
future directions. The Journal of nutrition, 150(4):663–
671.
Delen, D., Walker, G., and Kadam, A. (2005). Predicting
breast cancer survivability: a comparison of three data
mining methods. Artificial intelligence in medicine,
34(2):113–127.
Demuth, H. and Beale, M. (2000). Neural network toolbox
user’s guide.
Ehrenstein, V., Nielsen, H., Pedersen, A. B., Johnsen, S. P.,
and Pedersen, L. (2017). Clinical epidemiology in the
era of big data: new opportunities, familiar challenges.
Clinical epidemiology, 9:245.
Erkal, B., & Ayyıldız, T. E. (2021, November). Using
Machine Learning Methods in Early Diagnosis of
Breast Cancer. In 2021 Medical Technologies Congress
(TIPTEKNO) (pp. 1-3). IEEE.
Gholamrezaei, M. and Ghorbanian, K. (2007). Rotated
general regression neural network. In 2007
International Joint Conference on Neural Networks,
pages 1959– 1964. IEEE.
Hu, L.-Y., Huang, M.-W., Ke, S.-W., and Tsai, C.F. (2016).
The distance function effect on k-nearest neighbor
classification for medical datasets. Springer Plus,
5(1):1–9.
Huang, S., Cai, N., Pacheco, P. P., Narrandes, S., Wang, Y.,
and Xu, W. (2018). Applications of support vector
machine (svm) learning in cancer genomics. Cancer
genomics & proteomics, 15(1):41–51.
Landis, J. R. and Koch, G. G. (1977). The measurement of
observer agreement for categorical data. Biometrics,
pages 159–174.
Liu, Y.-Q., Wang, C., and Zhang, L. (2009). Decision tree
based predictive models for breast cancer survivability
on imbalanced data. In 2009 3rd international
conference on bioinformatics and biomedical
engineering, pages 1–4. IEEE.
Mu, T. and Nandi, A. K. (2007). Breast cancer detection
from fna using svm with different parameter tuning
systems and som–rbf classifier. Journal of the Franklin
Institute, 344(3-4):285–311.
Osareh, A. and Shadgar, B. (2010). Machine learning
techniques to diagnose breast cancer. In 2010 5th
international symposium on health informatics and
bioinformatics, pages 114–120. IEEE.
Sizilio, G. R., Leite, C. R., Guerreiro, A. M., and Neto, A.
D. D. (2012). Fuzzy method for pre-diagnosis of breast
cancer from the fine needle aspirate analysis.
Biomedical engineering online, 11(1):1–21.
Vert, J. P., Tsuda, K., & Schölkopf, B. (2004). A primer on
kernel methods. Kernel methods in computational
biology, 47, 35-70.
WHO (2021, Mar. 26). Breast cancer [Online]. Available:
https://www.who.int/news-room/fact-
sheets/detail/breast-cancer
Wolberg, W. H. and Mangasarian, O. L. (1990). Multi-
surface method of pattern separation for medical
diagnosis applied to breast cytology. Proceedings of the
national academy of sciences, 87(23):9193–9196.
COMPLEXIS 2022 - 7th International Conference on Complexity, Future Information Systems and Risk
122
