
A Platform to Generate FAIR Data for COVID-19 Clinical Research
in Brazil
Vânia Borges
a
, Natalia Queiroz de Oliveira
b
, Henrique F. Rodrigues
c
,
Maria Luiza Machado Campos
d
and Giseli Rabello Lopes
e
Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
Keywords: VODAN Brazil, FAIRification, Platform, ETL4FAIR, COVID-19 Clinical Research.
Abstract: The COVID-19 pandemic and the global actions to address it have highlighted the importance of clinical care
data for more detailed studies of the virus and its effects. Extracting and processing such data, in terms of
confidentiality issues, is a challenge. In addition, the mechanisms necessary for their publication are aimed at
reuse in research to better understand the effects of this pandemic or other viral outbreaks. This paper
describes a modular, scalable, distributed, and flexible platform, based on a generic architecture, to promote
the publication of FAIR clinical research data. This platform collects heterogeneous data from Electronic
Health Records, transforms these data into interconnected and interoperable (meta)data that are processable
by software agents, and publishes them through technological solutions such as repositories and FAIR Data
Point.
1 INTRODUCTION
Due to the COVID-19 pandemic, global actions have
been conceived to produce and deploy information to
support decision making on combating the virus and
its effects. Although the amount of information
available on the Web concerning COVID-19 has
grown far beyond expectation, most of them are
aggregated data, i.e., totals of people infected,
hospitalized, recovered, and deaths. Data regarding
clinical conditions, patients treatment, and their
outcome are, though, complementary to aggregate
data and essential for more detailed studies in clinical
research. However, access to these data outside the
Hospital Unit (HU) is often limited (Hallock et al.,
2021).
In order to accelerate and promote cooperation
among different initiatives referring to COVID-19
research results, the Virus Outbreak Data Network
(VODAN) (GO FAIR, 2020a) was created. VODAN
is an Implementation Network (IN) carried out by
CODATA (Committee on Data International Science
a
https://orcid.org/0000-0002-6717-1168
b
https://orcid.org/0000-0001-8371-142X
c
https://orcid.org/0000-0002-6777-3935
d
https://orcid.org/0000-0002-7930-612X
e
https://orcid.org/0000-0003-2482-1826
Council), RDA (Research Data Alliance), WDS
(World Data System), and the GO FAIR Initiative.
This IN was created under the goal to establish a
federated data infrastructure to support the capture
and use of data, following the FAIR data principles,
not only during this pandemic but also on future
disease outbreaks (Mons, 2020). This federated data
infrastructure is targeted for both human and machine
exploration, fostering reuse and reproducibility of
scientific resources (GO FAIR, 2020b).
The VODAN Brazil (VODAN BR) project is
responsible for the implementation of a pilot of this
federated data infrastructure in Brazil. It is
coordinated by the Oswaldo Cruz Foundation
(FIOCRUZ) in partnership with the Federal
University of Rio de Janeiro (UFRJ), the Federal
University of the State of Rio de Janeiro (UNIRIO),
and the University of Twente (FIOCRUZ, 2021). The
first phase of the implementation plan aims at
COVID-19 clinical cases, with data being collected
through a set of Brazilian hospitals, such as the Albert
Einstein hospital, the Gaffrée Guinle University
218
Borges, V., Queiroz de Oliveira, N., Rodrigues, H., Campos, M. and Lopes, G.
A Platform to Generate FAIR Data for COVID-19 Clinical Research in Brazil.
DOI: 10.5220/0011066800003179
In Proceedings of the 24th International Conference on Enterprise Information Systems (ICEIS 2022) - Volume 1, pages 218-225
ISBN: 978-989-758-569-2; ISSN: 2184-4992
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Hospital, and the São José State Hospital of Duque de
Caxias. These data include clinical features about
anonymized patients related to COVID-19 cases,
following the World Health Organization (WHO)
Rapid Core Case Report Form (CRF).
VODAN BR is establishing an IT platform to
manage these data, addressing practical challenges
from the hospital partners, such as collecting data
from different Electronic Health Records (EHR) and
integrating these data with semantic-oriented CRF,
which is based on the WHO semantic model
developed by the VODAN IN. This platform
addresses requirements for an analytical environment
that is able to cope with distinct data sources formats
and semantics, e.g., CSV format and triplestore
Application Programming Interface (API). It also
includes a defined licensing authorization schema and
metadata management supported by FAIR Data
Points (FAIR DPs).
This position paper aims to present an overview
of a scalable, distributed, flexible and modular
platform, based on a generic architecture, with its
processes and computational assets developed to
support the VODAN BR project. This generic
architecture will attend an intensive data collection
process with high heterogeneity, turning the FAIR
data available on repositories. In these repositories,
interoperable data and metadata could be processed
by software agents, supporting the discovery of other
resources that can be linked with them. As a result,
the VODAN BR platform promotes greater agility in
discovery and knowledge generation through the
efficient reuse of research results.
This paper is organized as follows: Section 2
presents some background; Section 3 describes the
proposed platform for the VODAN BR; Section 4
presents a discussion about the challenges identified
in this work; and Section 5 concludes with final
comments and future works.
2 BACKGROUND
2.1 Semantic Web and FAIR Principles
The Semantic Web proposes that data on the Web can
be defined and connected in a way that allows
interpretation by both humans and machines,
stimulating sharing and reuse by applications,
companies, and communities. To achieve this goal, a
set of standards and best practices for publishing and
linking data on the Web has been defined (W3C,
2017). These best practices are based on annotating
data with controlled vocabularies and ontologies,
facilitating the identification of new connections
among items from different data sources, thus
forming a global data space, the so-called Web of
Data (Heath and Bizer, 2011).
The FAIR principles aim to make data (and, more
recently, digital objects in general) Findable,
Accessible, Interoperable, and Reusable (Wilkinson
et al., 2016). In essence, the principles emphasize the
importance of using metadata to facilitate data
discovery and understanding, especially by machines
(software agents), to the standards established by the
World Wide Web Consortium (W3C). It should be
noted that the FAIR principles neither establish
standards nor supporting technologies, but, rather,
guide the creation of FAIR data and metadata. Recent
initiatives consider it vital to ensure that data and
other associated resources are FAIR, in the original
sense of the acronym, and also in the sense of
“Federated and Artificial Intelligence (AI)-Ready
data”, therefore readable and actionable by machines
(GO FAIR, 2020b).
Metadata standards and content annotations are
established to promote common understanding about
the meaning of the data, ensuring correct
interpretation and proper usage. Metadata need to be
findable and structured to be interpreted by machines,
i.e., machine-actionable. These machine-actionable
metadata, essential to the FAIR principles, have
fostered discussion of Metadata for Machine (M4M),
stimulating the creation and reuse of metadata
components and metadata templates for machine
processing. In the VODAN implementation, M4M
has been active in standardizing metadata regarding
catalogs and datasets that will be made available via
FAIR DPs, as well as the services associated with
them (GO FAIR, 2021).
The formalism and flexibility required for
creating and making data and metadata available are
provided by the Resource Description Framework
(RDF) model, including in this context RDF Schema
(RDFS). The Web Ontology Language (OWL),
developed to create robust ontologies, also meets
these criteria. The set of statements represented by
RDF triples constitutes an RDF Knowledge Graph.
2.2 FAIRification Process
The process of making data FAIR is called
FAIRification. It is in fact a complex process,
requiring several areas of expertise and data
stewardship knowledge. In order to facilitate this
process, Jacobsen et al. (2020) proposed a generic
workflow comprising three defined phases: pre-
FAIRification, FAIRification, and post-
A Platform to Generate FAIR Data for COVID-19 Clinical Research in Brazil
219

FAIRification. The phases are further divided into
seven steps: 1) identify the FAIRification objective;
2) analyze data; 3) analyze metadata; 4) define
semantic model for data (4a) and metadata (4b); 5)
make data (5a) and metadata (5b) linkable; 6) host
FAIR data; and 7) assess FAIR data. Each step
describes how data and metadata can be processed,
which knowledge is required, and which procedures
and tools can be used to obtain FAIR (meta)data. This
FAIRification workflow is applicable to any kind of
data and metadata.
2.3 FAIR Data Point
One of the main components of a FAIR Ecosystem is
the FAIR DP. FAIR DP is a software that works as an
infrastructure for (meta)data storage and accessibility
with the goals of: (i) allowing data owners to expose
datasets in a FAIR manner; (ii) facilitating the
information discovery about FAIR DP by data users,
in a network of FAIR DPs; (iii) establishing
mechanisms that handle consumer access, according
to the licenses and restrictions imposed on the data by
their managers; (iv) providing access indicators on
the (meta)data made available for data owners; and
(v) providing data access for humans, through a
Graphical User Interface (GUI), and for software
agents, using API (Santos et al., 2016).
3 VODAN BRAZIL PLATFORM
In VODAN BR, the starting point is the COVID-19
patients’ clinical data, which are processed and
transformed into linked data according to the Rapid
Core Case Report Form - named in this paper by
“WHO-CRF” (WHO, 2021). This CRF was
developed by the WHO to standardize data collection
of clinical features of COVID-19 among hospitalized
patients. The generic FAIRification workflow was
extended to guide the processes on this platform,
ensuring FAIR data and metadata (Oliveira et al.,
2021). In addition, these processes obey the Semantic
Web standards and comply with established licensing
and anonymization criteria.
3.1 Desiderata and Requirements
Desiderata were established to guide the development
of the platform, with emphasis on data management
and FAIR (meta)data. They are intended to provide a
readily adjustable structure, i.e., one that significantly
reduces the impact of changes to each evolution and
versioning of CRFs or the semantic artifacts
(vocabularies, thesaurus, ontologies etc). Some of
these desiderata are highlighted below:
creating an infrastructure capable of
implementing and making available a digital
CRF (application) for health care professionals,
meeting the epidemic episodes of COVID-19
pandemic or other viral outbreaks;
storing the information established in the
CRFs, in an anonymized way, considering that
their versioning can include, alter or exclude
elements;
allowing the creation of national CRFs or the
inclusion of specific additional questions. This
requirement emerged evaluating the different
CRF types used in Brazil, which, besides the
elements established by the WHO-CRF,
consider additional information for research,
such as participation in vaccination campaigns
and date of the last dose;
developing a conceptual model that aligns CRF
elements with ontologies (semantic models),
contributing to the data FAIRification process;
providing a flexible, modular, scalable, and
agile infrastructure to support software and
database adaptations;
transforming the collected data, i.e., "non-
FAIR data" into linked data by mapping them
to machine-readable formats using RDF,
making them available in datasets, and
publishing the associated metadata also in
RDF, in a FAIR DP;
providing a public FAIR DP configured to meet
the privacy requirements agreed by
participants, allowing access to data through
controlled queries rather than traditional
downloads
.
The main requirements of the platform
infrastructure are to be modular, scalable,
distributed, and flexible:
Modular because the planned activities are
organized in the form of modules that interact
in a sequential manner, with the result of a
module being the input for the subsequent
module;
Scalable and distributed because the idea is that
a staging database will be made available in
each HU, as well as repositories and/or
triplestores. These components will host the
structured data according to the WHO-CRF in
their different distributions or formats. This
means that as more hospitals join the project,
more IT infrastructure will be integrated,
leading to a natural horizontal scaling up;
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
220
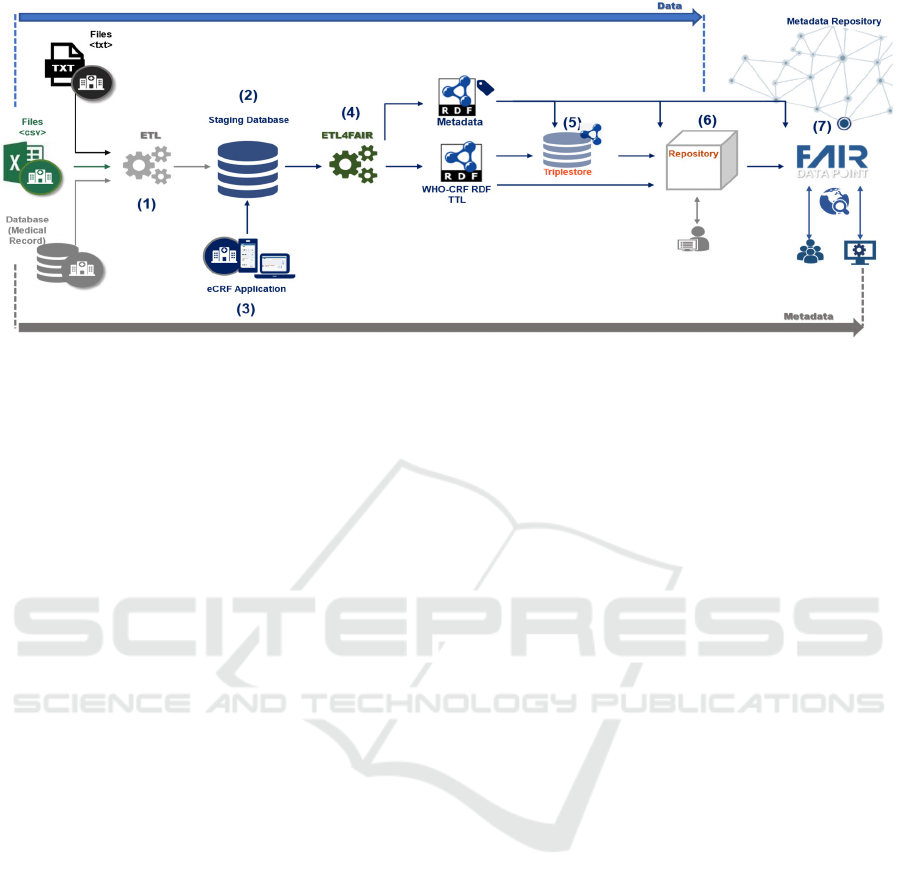
Figure 1: Schematic view of the VODAN BR architecture.
Flexible platform because the heterogeneous
data produced by the HUs EHR are treated and
transformed into an RDF graph representation,
which is one of the formats that facilitate data
linkage (interconnection).
3.2 Generic Architecture and the
VODAN BR Platform
Figure 1 depicts the architecture designed for the
VODAN BR Project, considering the requirements
and the desiderata. It covers the process from
collection of clinical data provided by HUs to
metadata publishing in the FAIR DP.
The computational assets considered for this
architecture are: a staging database (2) to store data
processed and transformed using a CRF format;
automated solutions to handle ETL processes (1) and
(4); a triplestore (5) for publishing and querying RDF
triples; a repository (6) for datasets and their
associated publications; and a FAIR DP (7), as a
public metadata repository, for visibility and access
for the queries. To serve hospitals without access to
their EHR data, it makes available an eCRF
application (3). The(meta)data flow is from left to
right as described in the Figure 1.
Initially, the architecture captures data that can be
in several formats, such as text, CSV, or in the format
used in each HU. An Extract-Transform-Load (ETL)
(1) process is employed to clean and transform the
data, storing them in a staging database (2). The data
in the staging database are then transformed into
linked data (4) and annotated in vocabularies and
ontologies to satisfy the interoperability principle.
They are loaded into a triplestore (5) and/or made
available for download in a repository, as an RDF
dataset (6). The associated metadata is also treated (4)
and then loaded and published in a FAIR DP (7).
The VODAN BR platform is based on the
proposed architecture with data and services
treatment guided by an adapted FAIRification
workflow (Oliveira et al., 2021). The actions outlined
in this workflow allowed the analysis of solutions to
automate process steps. The actual version of this
platform adopts these components: (i) a MySQL
relational database as staging database; (ii) Pentaho
Data Integration (PDI) solution for HUs data ETL
process; (iii) the ETL4FAIR approach for generating
and publishing FAIR data into triplestores and
repositories; (iv) GraphDB as triplestore; (v)
Dataverse as data repository; and (vi) FAIR DP as
metadata repository.
Some mechanisms must be developed for the
platform to play its role. Among them, we shall
highlight, due to their relevance in the project and the
challenges in (meta)data management: (i)
mechanisms to capture and process data, that
contemplate different requirements and EHR of the
HUs; (ii) the approach for processing, transformation,
and annotation of linked (meta)data, i.e.,
FAIRification of data; and (iii) the alternatives for
publishing the (meta)data. The main tasks associated
with these mechanisms are described in the following
subsections.
3.2.1 Extracting and Processing Data
As presented before, the project foresees different
forms of collecting clinical research data on COVID-
19: an application (eCRF) developed to capture
information according to the WHO-CRF; an ETL
solution to load and process anonymized data from
files in text or CSV formats made available by the
A Platform to Generate FAIR Data for COVID-19 Clinical Research in Brazil
221

UHs; and an ETL solution that connects both
databases, staging and from HUs, collecting and
treating data from the EHR in the format established
by the WHO-CRF.
Extracting data from an existing EHR poses an
additional challenge, despite the use of the staging
database as a transition database to the WHO-CRF
format. Existing EHRs frequently allow some aspects
of treatment to be recorded in textual fields. Due to
the lack of standardization in these entries and the
significant amount of unstructured data, the collection
and transformation processes become difficult and
complex. In these cases, interpretation and coding
support by healthcare professionals is essential. This
problem is not new and has been a constant in health
care data interoperability studies (Santos, 2020).
3.2.2 FAIRification
Based on the recommendations of the original
FAIRification workflow, VODAN BR has been using
an adapted and extended version proposed by
(Oliveira et al., 2021). The adaptation follows the
phases and steps of the generic FAIRification
workflow. Although steps 6 and 7 have been renamed
to 6) host FAIR data and metadata and 7) assess FAIR
data and metadata, emphasizing the importance of
storing, publishing, and evaluating both FAIR data
and metadata. The adaptation followed an approach
of associated actions for the FAIRification process in
a delimited and specific way, justifying
implementation choices to support the transformation
and publishing of FAIR (meta)data.
The FAIRification phase occurs after processing
the raw data, generating the FAIR (meta)data
associated with the semantic models in RDF. The
semantic data model COVIDCRFRAPID (BioPortal,
2020) was adopted for the data representation
according to the WHO-CRF. This model associates
the questionnaire questions with a set of entities from
the health domain which refers to other existing and
well-documented ontologies, providing quality and
additional information for data reuse.
The reference metadata follow the specifications
established for the FAIR DP metadata schemas.
These schemas define a set of standardized metadata
that describe information such as licenses, access
conditions, context, and provenance (da Silva Santos,
2020).
The ETL4LOD+ tool (GRUPO-GRECO, 2019),
adopted in this phase, consists of a set of plugins
developed in JAVA that extend the functionalities of
PDI, which is a widely used ETL solution. In this
architecture, ETL4LOD+ provides the transformation
of data from different sources and formats into linked
(meta)data and their publication in Semantic Web
technologies such as triplestores and FAIR DP.
Potential solutions have been tested in the
VODAN BR project to support the FAIRification
phase (Oliveira et al., 2021). They contribute to
automating some of the established actions integrated
with ETL4LOD+. Some of them are presented below.
The ETL4LinkedProv approach was tested to
collect provenance metadata associated with an ETL
workflow. The approach uses ETL workflows and
employs the Provenance Collector Agent (PCA)
component to capture prospective and retrospective
provenance metadata at different granularity levels.
The approach also supports the assessment of the
quality and reliability of FAIR provenance metadata
(Mendonça et al., 2016). It is currently being
reengineered to be aligned to the FAIRification
process.
The CEDAR Workbench was analyzed with
respect to metadata schemas established for the FAIR
DP. Through CEDAR, it is possible to create
metadata schemas as templates (Gonçalves et al.,
2017). These templates must be instantiated with the
metadata for the dataset and distribution to be
generated.
3.2.3 Publishing FAIR Data
According to the established desiderata, research
(meta)data should be made available in linked data
format, using the RDF standard. Following trends in
research data management, these data can be
published in institutional or thematic repositories,
which become responsible for storing the RDF
dataset and its metadata.
In addition to RDF datasets, triplestores and FAIR
DP are used to improve data reuse. Both provide
graph stores for querying the data. The triplestores
can be accessed from the FAIR DP or the repository,
via API, by algorithms that collect the data of interest.
The FAIR DP facilitates transparent and
controlled access to metadata. This access is made
through four different hierarchical layers: beginning
with the metadata of the FAIR DP itself, followed by
the metadata of the catalog, datasets, and
distributions. Publishing metadata about COVID-19
data in the VODAN BR FAIR DP allows access to
these data by software agents and humans (Santos et
al., 2016) and their integration into the VODAN
federation.
It is important to note that, after the FAIRification
process, a set of data and metadata is obtained,
compliant with the FAIR principles. These well-
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
222
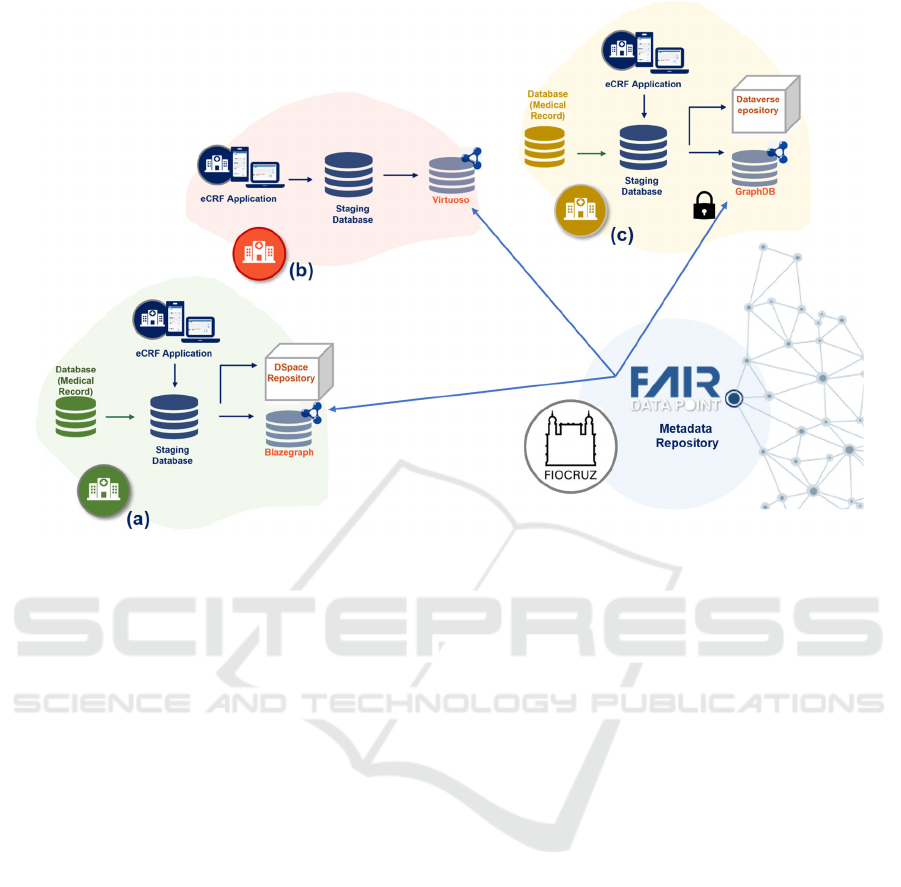
Figure 2: Overview of the scalable and distributed platform for VODAN BR.
structured (meta)data can be exploited through
machine learning techniques, and other artificial
intelligence (AI) approaches. These techniques can
contribute to the discovery of significant patterns in
epidemic outbreaks, supporting decisions and actions
to address them.
As stated by the VODAN network, the datasets
must be "visited" by algorithms, respecting the access
established by the HUs. Therefore, the associated
metadata, such as information about the origin of the
existing data, distribution types, and access policy,
will be available and accessible in the FAIR DP.
4 DISCUSSION AND
CHALLENGES
The development of a platform to disseminate viral
outbreaks research data during a pandemic is a
challenge per se. The VODAN BR team is working
on implementing such a platform to ensure
appropriate FAIR data management. This
management includes data from the moment of their
capture to their publication in the FAIR technological
supporting solutions (triplestores, repositories, and
FAIR DP).
The proposed platform aims at a federated
infrastructure. Therefore, it respects the
independence of HUs in managing their data and IT
resources. However, it demands HUs compliance
with rules for the metadata publication/dissemination
established by FIOCRUZ. Figure 2 shows an example
of the implementation of this platform contemplating
three HU. In the figure, hospitals (a) and (c) process
clinical data from the EHR database and publish the
results in triplestore and data repository. The former
uses Blazegraph and DSpace, and the latter GraphDB
and Dataverse. Hospital (b) uses the eCRF
application to register the survey and publishes it in
the Virtuoso triplestore. The metadata are published
in the VODAN BR FAIR DP hosted by FIOCRUZ.
Data access is performed through this FAIR DP,
respecting the authorization rules established by each
hospital. In the depicted example, hospital (c)
requires authorization for access.
The main challenges encountered during the
development and deployment stages of this flexible
platform are related to (Campos et al., 2021): (i) an
extraction and collection strategy for the
heterogeneous EHR available at the HU; (ii) the
creation and maintenance of the staging relational
database adherent to the WHO-CRF, enriched with a
reference ontology and other associated standard
vocabularies; (iii) the development of an ETL4FAIR
A Platform to Generate FAIR Data for COVID-19 Clinical Research in Brazil
223

approach, supporting a process-oriented
FAIRification, presenting automated steps in order to
reduce human intervention and mitigating possible
errors caused by it; (iv) the technical
qualification/competence and the computing
infrastructure required to support local triplestores at
each participating HU; and (v) expertise to install,
configure and employ a FAIR DP.
During the implementation of the architecture, it
was observed the lack of solutions to support the
entire FAIRification process. FAIRification steps can
be automated, improving the process and making the
resulting platform more stable to meet new
challenges. The collection of provenance metadata
according to the granularity established by the
process manager is an example.
Other initiatives to improve FAIR data and
metadata management are being developed, usually
for specific domains, such as the Collaborative Open
Omics (COPO) platform (Shaw et al. 2020). It was
developed for researchers to publish their assets,
providing metadata annotation and mediation for data
submission to appropriate repositories. VODAN
AFRICA was the first VODAN IN. It is funded by the
Philips Foundation and aims to promote distributed
access to CRF data from African countries, serving
African universities, hospitals, and research
institutions. VODAN AFRICA proposes an
integrated architecture with clinical and research data
(Van Reisen et al., 2021). In this architecture, data are
made available in closed dashboards. Two levels of
dashboards are available: the first with data from each
clinic and the second with aggregated data from the
VODAN community. In contrast to the initiatives
presented, the VODAN BR project proposes a
generic architecture, which allows the establishment
of scalable, distributed, and flexible domain-oriented
platforms for the generation and publication of FAIR
(meta)data processable by software agents.
5 CONCLUSIONS AND FUTURE
WORKS
Data for more detailed clinical research studies are
highly valuable to the scientific community, but are
not always available (Hallock et al., 2021). Among
the main problems in extracting and collecting this
type of data are: (i) privacy protection issues
concerning personal data in the EHR, aligned to the
Brazilian Protection Law for Personal Data; (ii)
complexity in processing free text fields from EHRs,
hampering data extraction; (iii) the challenges of
publishing FAIR health data, with respect to
developing and deploying a federated infrastructure
to support this process; and (iv) the difficulty in
providing linked (meta)data with different semantic
artifacts to facilitate reuse by researchers.
The experience in the project reinforces the
importance of the FAIR DP as an essential
component for federated access points, as well as for
research and (re)use mechanisms for FAIR
(meta)data. In addition, it also supports sensitive
research data that require some level of privacy, such
as patient data. According to its specification, the
FAIR DP provides an appropriate authentication and
authorization infrastructure in distributed scenarios.
Hence, sensitive data are only accessible when
authorized, allowing for "data to be as open as
possible and as closed as necessary" (Mons, 2020).
This project is carried out with the support of
undergraduate, master, and doctoral students from the
universities involved. Currently, there are ongoing
studies to provide: a high availability FAIR DP; the
development of new clinical trials using eCRF
application; and automatic capture of provenance
metadata throughout the FAIRification process, with
the granularity established by the data stewards. Also,
the ETL4FAIR approach itself has been tested on the
VODAN BR project with an emphasis on the
FAIRification phase. It contributes to FAIRification
steps automatization, collaborating on the integration
of different solutions, and improving the federated
FAIR data ecosystem.
Once the platform defined by the project is
completely implemented, it will enable the country
participation in the federated data network of
epidemiological FAIR DPs. Once accessible to
researchers, it will contribute to future research
related to the COVID-19 pandemic or other potential
future outbreaks.
ACKNOWLEDGEMENTS
This work has been partially supported with students
grants from CAPES (Process numbers
223038.014313/2020-19, and 88887.613048/2021-
00), CNPq (Process number 158474/2020-1) and
UNIRIO university funding.
REFERENCES
BioPortal. (2020). WHO COVID-19 Rapid Version CRF
semantic data model. https://bioportal.bioontology.org/
ontologies/COVIDCRFRAPID.
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
224

Campos, M.L.M., Borges, V.; Lopes, G. R.; Cavalcanti, M.
C.; Moreira J.; Cruz, S. M. S. (2021). VODAN BR – uma
plataforma de apoio para dados COVID-19 seguindo
os princípios FAIR. In: SALES, Luana Farias; VEIGA,
Viviane dos Santos; HENNING, Patrícia; SAYÃO,
Luís Fernando (org.). Princípios FAIR aplicados à
gestão de dados de pesquisa. Rio de Janeiro: Ibict, 2021.
p. 253 - 270. DOI: 10.22477/9786589167242.cap18.
FIOCRUZ. (2021). VODAN Brazil. https://portal.fio
cruz.br/en/vodan-brazil.
GO FAIR. (2020a, March). Data Together COVID-19
Appeal and Actions. https://go-fair.org/wp-
content/uploads/2020/03/Data-Together-COVID-19-
Statement-FINAL.pdf .
GO FAIR. (2020b, March). VODAN IN Manifesto.
https://www.go-fair.org/wp-
content/uploads/2020/03/VODAN-IN-Manifesto.pdf.
GO FAIR. (2021). Metadata for Machines. https://www.
go-fair.org/how-to-go-fair/metadata-for-machines/.
Gonçalves, R. S., O’Connor, M. J., Martínez-Romero, M.,
Egyedi, A. L., Willrett, D., Graybeal, J., & Musen, M.
A. (2017, October). The CEDAR workbench: an
ontology-assisted environment for authoring metadata
that describe scientific experiments. In International
Semantic Web Conference (pp. 103-110). Springer,
Cham.
GRUPO-GRECO. (2019). ETL4LOD+. https://github.com/
Grupo-GRECO/ETL4LODPlus.
Hallock, H., Marshall, S., t Hoen, P. A., Nygård, J. F.,
Hoorne, B., Fox, C., & Alagaratnam, S. (2021).
Federated networks for distributed analysis of health
data. Frontiers in Public Health, 1316.
Heath, T., Bizer, C. (2011). Linked data: Evolving the web
into a global data space. Synthesis lectures on the
semantic web: theory and technology, 1(1), 1-136.
Jacobsen, A., Kaliyaperumal, R., Santos, L. O. B. S., Mons,
B., Schultes, E., Roos, M., & Thompson, M. (2020). A
generic workflow for the data FAIRification process.
Data Intelligence, 2(1-2), 56-65.
Mendonça, R. R., Cruz, S. M. S., & Campos, M. L. M.
(2016). Etl4linkedprov: Managing multigranular
linked data provenance. Journal of Information and
Data Management, 7(2), 70-85.
Mons, B. (2020). The VODAN IN: support of a FAIR-based
infrastructure for COVID-19. European Journal of
Human Genetics; 28. 1-4.
Oliveira, N. Q., Borges V., Rodrigues, H. F., Campos, M.
L. M., Lopes, G. R. (2021, December). A Practical
Approach of Actions for FAIRification Workflows.
Metadata and Semantic Research Conference.
Santos, L. B. S., Wilkinson, M. D., Kuzniar, A.,
Kaliyaperumal, R., Thompson, M., Dumontier, M., &
Burger, K. (2016). FAIR data points supporting big
data interoperability. Enterprise Interoperability in the
Digitized and Networked Factory of the Future. ISTE,
London, 270-279.
Santos, L. B. S. (2020). FAIR Data Point Metadata
Specification. https://github.com/FAIRDataTeam/
FAIRDataPoint-Spec/blob/master/spec.md.
Shaw, F., Etuk, A., Minotto, A., Gonzalez-Beltran, A.,
Johnson, D., Rocca-Serra, P., ... & Davey, R. P. (2020).
COPO: a metadata platform for brokering FAIR data
in the life sciences. F1000Research, 9(495), 495.
van Reisen, M., Oladipo, F., Stokmans, M., Mpezamihgo,
M., Folorunso, S., Schultes, E., ... & Musen, M. A.
(2021). Design of a FAIR digital data health
infrastructure in Africa for COVID‐19 reporting and
research. Advanced Genetics, e10050.
W3C. (2017). Data on the Web Best Practices.
https://www.w3.org/TR/dwbp/.
WHO. (2021). Global COVID-19 Clinical Platform: Rapid
core case report form (CRF). https://www.who.int/
publications/i/item/WHO-2019-nCoV-Clinical_CRF-
2020.4.
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J.,
Appleton, G., Axton, M., Baak, A., ... & Mons, B.
(2016). The FAIR Guiding Principles for scientific data
management and stewardship. Scientific data, 3(1), 1-
9.
A Platform to Generate FAIR Data for COVID-19 Clinical Research in Brazil
225
