
Privacy-preservation and the Use of Data for Research: A COVID-19 Use
Case in Randomly Generated Healthcare Records
Madalena Lopes E. Silva
1 a
, Maria Claudia Cavalcanti
1 b
and Maria Luiza M. Campos
2 c
1
Instituto Militar de Engenharia, Prac¸a General Tib
´
urcio 80, CEP 22290-270,
Rio de Janeiro, RJ, Brazil
2
Universidade Federal do Rio de Janeiro, Av. Athos da Silveira Ramos, 274 - CCMN - Ilha do Fund
˜
ao, CEP 21941-90,
Keywords:
Web of Data, Research Data Management, Law, Legal Compliance, COVID-19 Pandemic, Anonymous Data.
Abstract:
The provision of clinical data for research purposes has become central to monitoring and understanding the
COVID-19 outbreak. In such a pandemic scenario, obtaining new research results is an imperative and urgent
requirement. However, nowadays, personal data are protected by different legal regulations, to which all these
data must comply, especially those related to the health of individuals. Then, a tough challenge arises in the
academic sphere: how to provide a large amount of detailed clinical data for research and, simultaneously,
guarantee the privacy of the individuals involved? Thus, this article discusses how the biomedical community
may face this challenge and it presents the main ongoing initiatives and available emergent technologies that
are useful to meet such urgent demand. Moreover, it also shows, through a use case, how it is possible to deal
with this challenge, presenting the applicability of privacy-preserving techniques over a randomly generated
typical dataset of COVID-19 health records.
1 INTRODUCTION
There are several articles (Hutchings et al., 2020) that
discuss some reasons why researchers do not share
data in the health domain, such as the concern that
other researchers take their results, loss of opportu-
nities or funding, and having data misinterpreted or
misused. However, the health crisis caused by the
COVID-19 pandemic highlighted the urgent need to
share clinical data for research purposes and to sup-
port decision-making in the definition of public poli-
cies.
On the other hand, clinical data sharing makes it
possible: (i) to achieve greater transparency and con-
fidence in research conducted in this sector; (ii) to
more quickly identify the behaviour of diseases aim-
ing at controlling actions (Smaradottir, 2018); (iii) to
positively impact decisions on treatments to which
patients will be submitted (Smaradottir, 2018); and
(iv) to support health research and the definition of
public policies.
In line with this new scenario of high demand for
a
https://orcid.org/0000-0001-7024-667X
b
https://orcid.org/0000-0003-4965-9941
c
https://orcid.org/0000-0002-7930-612X
research data, many initiatives emerged to monitor
COVID-19 outbreak evolution and general profile of
patients, such as analytical panels
1
. More specifically,
clinical data about COVID-19 patients have also been
shared, such as the FAPESP COVID-19 Data Shar-
ing/BR repository
2
, in Brazil.
Complementary to other descriptive metadata that
support data reuse, provenance metadata about the
used privacy-preserving techniques is important for
researchers when it is necessary to track back ag-
gregated and/or anonymized data to reach an origi-
nal clinical data. Provenance metadata is information
about entities, activities, and people involved in pro-
ducing a piece of data or thing, which can be used to
form assessments about its quality, reliability, or trust-
worthiness and can be applied in several domains.
Although there are works that discuss the legis-
lation around the issue of privacy of health data (Car-
valho et al., 2020; Ferreira, 2020; Bondel et al., 2020),
they did not take into account scientific research de-
mands for accessing original data. Therefore, the
main contributions of this article are: (i) to discuss
the issue of privacy guarantees, addressed by the lat-
1
https://covid19.who.int/
2
https://repositorio.uspdigital.usp.br/handle/item/243
Silva, M., Cavalcanti, M. and Campos, M.
Privacy-preservation and the Use of Data for Research: A COVID-19 Use Case in Randomly Generated Healthcare Records.
DOI: 10.5220/0011057900003179
In Proceedings of the 24th International Conference on Enterprise Information Systems (ICEIS 2022) - Volume 2, pages 317-324
ISBN: 978-989-758-569-2; ISSN: 2184-4992
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
317

est regulations and protection codes, in contrast to the
need to make data available for scientific research in
critical moments such as the pandemic of COVID-19;
and (ii) to demonstrate, through a use case, that it is
possible to anonymize health records for research and
still track back an individual identification if neces-
sary, with the help of provenance metadata.
The article is organized as follows: In Section 2,
concepts and facts related to the preservation of pri-
vacy are presented to contextualize the reader; in Sec-
tion 3 the main policies and initiatives for research
data are explored; in Section 4, health systems in
Brazil are addressed as well as examples of actions
aimed at making clinical data available for research;
in Section 5, a use case of randomly generated health
records is discussed; and, finally, the last section con-
cludes the paper.
2 BACKGROUND
Advances in automation and digitization of health
systems have brought agility in service, and facili-
tated the retrieval of information and exams, but, on
the other hand, they have also made patient data more
exposed to privacy violations and security breaches.
It is necessary to point out that data security and pri-
vacy are two related concepts, but they should not be
confused. Security refers to aspects of protecting a
system from unauthorized use, including user authen-
tication, information encryption, access control, fire-
wall policies, and intrusion detection. Privacy refers
to ensuring those entitled to control over the avail-
ability and use of their data, through data governance
mechanisms.
To understand policies and initiatives to provide
data security and preserve privacy, it is also neces-
sary to know the already consolidated legislation. The
Health Insurance Portability and Accountability Act
(HIPAA
3
) in USA, the General Data Protection Regu-
lation (GDPR
4
) in Europe, and the General Data Pro-
tection Law (Lei Geral de Protec¸
˜
ao de Dados LGPD
5
,
in portuguese) in Brazil define many requirements
and qualify possible penalties for cases of their viola-
tion. Among these requirements there are guarantees
such as the right to be forgotten (data exclusion) and
the need to collect user consent for the use of their
data (Ferreira, 2020).
3
https://www.hhs.gov/hipaa/for-professionals/index.ht
ml
4
https://gdprinfo.eu/
5
http://www.planalto.gov.br/ccivil\ 03/\ ato2015-201
8/2018/lei/l13709.htm
Pioneeringly, the HIPAA regulation has defined,
in a comprehensive manner, which protective mea-
sures should be used to treat data related to indi-
vidual clinical data. The aforementioned regulation
also defines a group of 18 sensitive attributes con-
sidered identifiers, which may uniquely identify an
individual. These attributes are known as Protected
Health Information (PHI), such as name, date, regis-
tration numbers, IP addresses, photos, and biometric
data, among other demographic information. More
recently, GDPR and LGPD defined the set of personal
data that can lead to the identification of a particular
individual, directly or indirectly. There are also at-
tributes that are classified as semi-identifiers (Brito
and Machado, 2017), such as race, age, schooling,
among others, which may indirectly identify an indi-
vidual when combined with external information. Al-
though there are already many anonymization tools
available to cope with these law requirements and
minimize the risks of identification, these tools still
need to be improved (Carvalho et al., 2020).
On the other hand, according to GDPR Art. 9,
items (h) and (i), and LGPD Art. 7, item (IV ), and
Art. 13, data used for health research and academic
activities are exempt from consent collection. Ad-
ditionally, as a result of the waiving of consent for
research purpose, projects are also exempt from pro-
viding guarantees of data exclusion, since, in prin-
ciple, the data used in research may remain per-
petually available for reuse. Therefore, while pre-
processing personal data using anonymization tech-
niques is strongly recommended, an important re-
quirement for the pre-processing tools is to apply
anonymization in such a way that it should be possi-
ble to have access to the original data when requested
by a restricted group of researchers.
Nowadays, there are different types of anonymiza-
tion techniques, which are usually applied to identi-
fier attributes. It is worth mentioning the technique
known as pseudoanonymization. It consists of any
process of transformation of personal data, carried
out in such a way that these data cannot be associ-
ated with the individual without the use of additional
data, which must be kept separately. It is a process
of desidentification that removes or replaces identi-
fying attributes such as names and identification keys
(IDs) of a given dataset but keeping in a separate place
the data that can directly identify the individual. In
pseudoanonymization, different ids must be used for
each existing domain, such as research, administra-
tive or medical. In this way, the possibility of re-
identification of a given patient is guaranteed when
necessary and by duly authorized persons.
On the other hand, for semi-identifier attributes,
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
318

the simplest anonymization strategy is data suppres-
sion, a mechanism in which defined attributes are re-
moved from the dataset. The suppression can be ap-
plied as a filter over a certain value, to remove just a
cell of data or an entire record.
Other anonymization techniques are also usually
applied: generalization, masking and disturbance.
Generalization consists in replacing semi-identifier
attribute values by more generic ones, increasing the
uncertainty regarding that data. Masking is widely
used in the generation of datasets for testing or train-
ing. An example of data disturbance is the addition
of noise, usually applied to numerical attributes that
receive their original disturbed values by adding or
multiplying by a value. In general, it preserves the
statistical properties of the data although it can gener-
ate meaningless values (Brito and Machado, 2017).
The ideal point to be reached is where there is
a sufficient degree of anonymity in the dataset, with
a calculated and acceptable risk of re-identification,
but which does not compromise the usefulness of
the data. The appropriate strategy chosen should
combine pseudonymization for identifying attributes
and anonymization techniques for semi-identifying
attributes (Sauermann et al., 2020).
3 INITIATIVES AND POLICIES
FOR RESEARCH DATA
Within the new scenario of open science, specifically
in the health domain, the reuse of data collected at the
patient care level can enable transparency, credibil-
ity and reproducibility of research. Thus, initiatives
have emerged to facilitate data publication and shar-
ing, standardization of processes, establishing sets of
good practices. The Resource Data Alliance (RDA
6
)
and GO-FAIR
7
are important examples of these ini-
tiatives that are detailed in the next subsections.
3.1 Resource Data Alliance (RDA)
RDA is a global initiative that brings together re-
searchers and those interested in discussing and seek-
ing solutions on sharing and reuse of open research
data. It is structured through many subject-oriented
interest groups (IG) and working groups (WG), which
discuss and deliberate on good practices and recom-
mendations in this scope.
One of those groups that focused on discussions
about COVID-19 is the RDA COVID-19 Working
6
https://www.rd-alliance.org/about-rda
7
https://www.go-fair.org/
Group which, after meetings held in 2020, released
recommendations related to the sharing of COVID-
19 data and preserving the privacy of individuals in-
volved (COVID-19-Workgroup, 2020).
In the aforementioned paper, the authors classify
attributes in two main groups: (i) direct identifiers
and (ii) indirect identifiers. Direct identifiers should
be treated with ”hashing with key”, i.e., using a hash
function on the target value with the addition of a con-
stant (salt). For example, one may apply the SHA-3
function on the name of an individual, using a key
concatenated with a constant string ”215” (salt). For
the same identifier, several different pseudonyms can
be produced, according to the choice of the salt. In
this approach, there must be a person who is the secret
key owner (usually the original data administrator).
Besides the key, he also must keep track of the hash
function and the ”salt” used for a given anonymized
dataset. This way, he will be able to identify the
pseudonyms through a simple decryption process. In-
direct identifiers such as gender, age, schooling, loca-
tion, race/ethnicity should be treated with encrypting
using a key that provides a cipher-text. The same se-
cret key is needed for the decryption.
3.2 GO-FAIR Initiative
Proposed in (Wilkinson et al., 2016), the FAIR prin-
ciples aim to guarantee some data characteristics that
are often not found and that make it difficult to dis-
cover, obtain, and reuse data for research purposes.
FAIR is an acronym for Findable, Accessible, Inter-
operable, and Reusable; each letter represents a group
of characteristics. Data sharing based on these princi-
ples means that the data is standardized and described
in a way that it can be easily reused by humans and
machines
8
.
GO FAIR is an international initiative that has
as its objective to disseminate and implement FAIR
principles and aims to implement the vision of the
European Open Science Cloud. This initiative pro-
poses implementation network structures, which aim
to establish a community to exchange information on
these principles and the implementation of a FAIR
infrastructure on the Internet, through the GO FAIR
Implementation Networks (IN) - community-led and
self-governed working groups across disciplines and
countries.
The Implementation Networks involve people, in-
stitutions, and organizations. The implementation
of an IN also provides gains in the preservation of
privacy as it keeps metadata in public repositories,
8
https://researchdata.springernature.com/posts/51916-
fair-data-7-initiatives-you-should-know-about
Privacy-preservation and the Use of Data for Research: A COVID-19 Use Case in Randomly Generated Healthcare Records
319

separately from data, that can remain in institutional
repositories, and the attribution of Creative Commons
licensing, which restricts access to the data. The
GO BUILD strand mediates communication between
researchers and the institution that wishes to have
greater control over their data.
Although anonymization techniques reduce but do
not eliminate the risk of re-identification, how much
should the guarantee of the privacy of individuals
weigh against research results for the development of
vaccines and medicines? Is it possible to find a bal-
ance spot where both goals are achieved? The GO-
FAIR initiative attempts to address these issues by
expanding alternatives to ensure privacy preservation
through the use of Creative Commons licensing for
published datasets for reuse in research. This type
of licensing offers flexibility, allowing the institution
that captures or generates the data to define in advance
which data will be published open, which will have
restricted access, and which researchers will have ac-
cess to those data.
4 BRAZILIAN HEALTH SYSTEM
AND RESEARCH DATA
In response to the pandemic, the WHO created
and released the COVID-19 Clinical Platform, an
anonymized data platform that enables member states
of International Health Regulations (IHR
9
) to share
clinical data of SARS-COV-2 infection suspected or
confirmed patients. To this end, the WHO provided
a standard Clinical Report Form (CRF
10
), defined to
collect data from exams, consultations, and follow-up
of these patients.
Hospital and administrative systems in primary,
secondary, and tertiary areas capture about 80Mb of
clinical data per patient per day (Huesch and Mosher,
2017). Thus, to reuse as much of these data as possi-
ble, several hospital units have been making efforts
to adapt data collection to the CRF format. How-
ever, this change was not possible in a short period
of time, as the pandemic scenario makes it difficult
for the medical staff to adapt to the evolution of the
system.
The next subsections will briefly address the ex-
isting health systems in Brazil and two initiatives for
the publication and sharing of clinical data collected
by hospital systems that are also used for research.
9
https://bityli.com/OqwIB
10
https://bityli.com/fbPFR
4.1 Unified Health System
Universal health coverage, the goal of the member
states of WHO, has been achieved in Brazil with the
implementation of the Unified Health System (Sis-
tema
´
Unico de Sa
´
ude (SUS) in Portuguese). It was
created in 1991 by the Health Ministry (Minist
´
erio da
Sa
´
ude (MS) in portuguese) to coordinate the devel-
opment of Health Information Systems (SIS in Por-
tuguese) meeting the needs in this area, according to
constitutional guidelines (Cunha and Vargens, 2017).
The MS is also responsible for consolidating and
making data supplied by municipal health secretari-
ats available on the website of Datasus
11
. e-SUS epi-
demiological surveillance system (e-SUS-VE)
12
re-
ceives notification of suspicious, probable, and con-
firmed cases of COVID-19, starting on March 2020,
which was previously managed by the Redcap plat-
form.
4.2 FAPESP COVID-19 Data
Sharing/BR Repository
An important initiative was also undertaken by
FAPESP (S
˜
ao Paulo research funding agency), re-
sponsible for the COVID-19 DataSharing/BR Repos-
itory, in which data were made available from patients
who underwent any diagnostic exam (related or not to
COVID-19), as of November 1st, 2019, even for those
who did not obtain a positive result on the exam. The
participating institutions that established a coopera-
tion agreement are the Clinic Hospital of Medicine
University of S
˜
ao Paulo, the Syrian-Lebanese Hospi-
tal, the Israeli Hospital Albert Einstein, the Fleury In-
stitute, and Portuguese Beneficence of S
˜
ao Paulo, up
to now, being the captors and publishers of datasets in
this repository.
The guarantee of privacy preservation was defined
as the responsibility of each institution and is part
of the agreement with FAPESP. Anonymization al-
gorithms have been developed that comply with the
requirements demanded by the legislation that de-
fines HIPAA. Identifying attributes such as name,
Cadastro de Pessoa F
´
ısica (CPF), date of birth have
been deleted from the datasets. As a treatment to be
able to disclose the residence area code - C
´
odigo de
Enderec¸amento Postal (CEP), the granularity of only
the first five digits was adopted.
The data made available by FAPESP include a set
of attributes not identical to those contained in the
CRF of the WHO and a reduced quantity, due to the
11
http://plataforma.saude.gov.br/coronavirus/covid-19/
12
https://covid.saude.gov.br
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
320

anonymization techniques employed. Although with
losses, the published dataset is significant and still
proves useful for a considerable number of data an-
alyzes.
4.3 A Virus Outbreak Data Network:
VODAN
One of the implementation networks currently active
within the GO FAIR initiative
13
is the Virus Outbreak
Data Network
14
(VODAN), jointly with the Commit-
tee on Data (CODATA), RDA, and World Data Sys-
tems (WDS). Dedicated to the publication and sharing
of data on epidemics and pandemics, the IN initially
aims at capturing data about the SARS-COV-2 virus,
materialized in VODAN networks
15
. A second objec-
tive is to provide metadata associated with these data
in FAIR Data Points (FDP
16
), an architecture com-
posed of repositories and services necessary for shar-
ing, publishing, and reusing those data.
To add a higher level of preservation of privacy
and security, the VODAN network proposes that the
federation of FDP nodes publish and allow access to
metadata, maintaining the data to be shared in their
local infrastructure. To make it easier and faster to im-
plement FDP, the VODAN initiative created and made
available ”VODAN in a box” (VIAB
17
).
In the Brazilian scenario, VODAN BR is con-
ducting efforts to perform de-identification experi-
ments. Also, hospitals are already applying specific
de-identification techniques according to each insti-
tution implementation decision. Section 5 presents an
use case that illustrates a typical de-identification pro-
cess that may be used in those hospitals. Furthermore,
it also illustrates that re-identification of patients may
eventually be necessary, and that with the help of re-
served provenance data, this becomes possible.
5 USE CASE IN EHR
To highlight how anonymization techniques may be
applied in Electronic Health Record (EHR) and make
it possible to reidentify individuals, we discuss a use
case based on randomly generated EHR. For the pur-
pose of making this use case, we used a sample of
13
https://www.go-fair.org/wp\-content/uploads/2020/0
3/data\-together\ march\-2020.pdf
14
https://www.go-fair.org/implementation-networks/ov
erview/vodan/
15
https://www.vodan-totafrica.info/about-vodan-africa/
about-vodan-africa-asia
16
https://www.fairdatapoint.org/
17
https://docs.vodan.fairdatapoint.org/en/latest/
randomly generated data (Table 1), extracted from the
dataset named COVID-DS1, since this avoids expos-
ing real data. This dataset has as identifier attributes
person id and name, and as semi-identifiers gender,
birth date, address and phone. In this use case there
are also sensitive personal data (that are attributes that
have meaning for researchers) named temperature,
apdiastolic (diastolic arterial pressure) and apsystolic
(systolic arterial pressure). It is worth to mention that
from a variety of available anonymization techniques
(Fung et al., 2011), some of them have been arbitrar-
ily chosen for this use case, since it is not in the scope
of this paper to compare them.
For the pseudoanonymization process, as was
published by RDA (Sauermann et al., 2020), it is
necessary to apply different privacy-preserving tech-
niques according to the privacy category of the at-
tribute. As one can see in the anonymized dataset in
Table 2, firstly it was applied a ”SHA-3 hash 256” al-
gorithm over person id with different keys for each
domain to access the data, such as Health, Research
and Admin. On the name attribute a suppression tech-
nique was applied, where all names were removed.
The gender attribute was sustained for analytical pur-
poses. Birth date was transformed in age range, con-
taining only a reference to the age range, featuring a
generalization. In the case of the address attribute,
part of it was deleted, remaining only the ZIP Code.
The phone attribute was also removed. The other sen-
sitive attributes, temperature, apdiastolic and apsys-
tolic were maintained to be further used also for ana-
lytical purposes.
As highlighted before, provenance metadata plays
an important role on these processes, specially when
it is necessary to reidentify individuals for further in-
vestigation. To illustrate such a situation, consider
the pandemic scenario. Analytical panels of the pan-
demic usually show aggregated numbers, i.e., they do
not show individual data. This is due to the anonymity
requirement for sensitive data, but also because pan-
els are used to provide a broad view of the pandemic
behavior. However, while analyzing such behavior, in
order to understand the reasons why a certain region
has more cases than others, or why it has a worse
death rate than others, it is necessary to look more
closely, and reach patient detailed clinical data. In or-
der to obtain such data it is necessary to trace back the
transformations applied so far, one by one. The meta-
data that supports patient reidentification is reserved
and available only for data curators. It is necessary
to request special permission to obtain them.
Therefore, to make reidentification possible,
provenance metadata should be captured, up to the
attribute level of the original dataset. A metadata
Privacy-preservation and the Use of Data for Research: A COVID-19 Use Case in Randomly Generated Healthcare Records
321

structure, inspired on workflow ontologies (Rauten-
berg et al., 2015; de Mendonc¸a, 2016), was used in
this use case. This metadata structure is presented in
Table 3, where: (i) attribute id represents the unique
identification keys of attributes, (ii) dataset id rep-
resents the unique identification keys of datasets in
which attributes are linked, (iii) attribute cat id rep-
resents the suggested attribute privacy category keys
of each one of the attributes (e.g. 1-Identifier, 2-
Quasi-identifier, 3-Sensible, etc.) (iv) step id repre-
sents the keys of the executed steps in the workflow
(e.g. 3-Anonymization), (v) execution seq represents
the unique keys of the steps execution sequence, (vi)
pptechnique id represents the unique keys of the pri-
vacy preservation technique used in step execution
(e.g. 1-Hash Function, 2-Hash Function + Salt, etc.),
(vii) domain id represents the unique keys of domains
involved in anonymization process (e.g. 1-Health, 2-
Research, 3-Admin, etc.), (viii) hash key and salt rep-
resent the hash and salt used in the anonymization
process for that specific attribute, (ix) label shows an
attribute processed name in the workflow execution,
and (x) type represents the type of involved attributes
(e.g. string, date, number, etc.). More details can be
found in the GitHub
18
repository.
Analyzing the first row of Table 3, for exam-
ple, the captured values mean that this record refers
to the anonymization process applied to the per-
son id attribute (attribute id = 000001) of COVID-
DS1 dataset (dataset id = 000001). This at-
tribute was categorized as an Identifier Attribute
(attribute cat id = 0001). Metadata about the work-
flow execution was captured. First, it informs the
workflow step category, i.e., the generic action that
was applied on the mentioned attribute, which was
an Anonymization action (step id = 003). Then, it
also indicates the step execution sequence number
(execution seq = 003). Then, it informs the spe-
cific anonymization process used, and in this case
it was applied a specific Pseudonymization Tech-
nique (pptechnique id = 0002), using as parame-
ters a hash key value (hash key = jqD9d fV) and
a salt value (salt = 500) for the Health domain
(domain id = 0001). Thus, it is possible to apply
the decryption function over the pseudonymized iden-
tifier (person id = d1b2d2347eb4364463e8... in Ta-
ble 2) with the parameterized hash and salt values
(available in Table 3), in order to re-identify the in-
dividual represented by person id value (person id =
ea5d0c10 − d816... in Table 1). Once the patient is
identified, it becomes possible to request additional
clinical data to deepen the analysis on specific cases.
18
https://github.com/madalenals/privacy-preserving-c
ovid-19
6 FINAL CONSIDERATIONS
Although there are many initiatives to provide data
for reuse and research, there is still a lack of use of
anonymity techniques incorporated into the publish-
ing processes of these data. In the literature as well,
there are few papers that are concerned with the bal-
ance between the preservation of privacy and the de-
mand for rich clinical data for analysis. The COVID-
19 scenario is a typical case where this kind of bal-
ance is required for a detailed understanding of the
pandemic. Also, the GO FAIR initiative has brought
an important contribution through the use of license
attribution, establishing access control levels and al-
lowed data usage.
This work sought to present the initiatives and
technologies available to meet this challenge. Ini-
tially, it presented an overview of related initiatives
in the current international and domestic scenarios, in
the direction of anonymized clinical data sharing for
research purposes. It also presented concepts about
privacy, sensitive data, and data protection, as well as
a summary of recent and relevant legislation. Besides,
this work presented a use case that illustrates a typi-
cal scenario, evidencing the use of privacy preserving
techniques while making possible individual reidenti-
fication, through provenance metadata capture.
As future work, we are already performing new
experiments on real data, applying different tech-
niques, specially on scenarios where reidentification
is needed. The idea is to compare such techniques
and verify their effectiveness on anonymization and
deanonymization tasks. Moreover, it is also inter-
esting to explore the combined use of license and
anonymization, similarly to what is proposed by the
Data Use Ontology (DUO) (Delgado and Llorente,
2020) and by an extension of the UsablePrivacy
Project (Pandit et al., 2018). Finally, another ongo-
ing study is on the combination of existing ontologies
to enrich provenance metadata for the reidentification
of individuals.
ACKNOWLEDGEMENTS
Special thanks to Programa INOVA UNIRIO
PROPGPI/DIT 2020 who partially supported the
VODAN BR project.
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
322

REFERENCES
Bondel, G., Garrido, G., Baumer, K., and Matthes, F.
(2020). The use of de-identification methods for se-
cure and privacy-enhancing big data analytics in cloud
environments. In Proc. 22nd Int. Conf. on Enterprise
Inf. Syst.
Brito, F. and Machado, J. (2017). Preservac¸
˜
ao de privaci-
dade de dados: Fundamentos, t
´
ecnicas e aplicac¸
˜
oes. In
Jorn. de Atual. em Inform
´
atica, pages 91–130. SBC.
Carvalho, A., Canedo, E., Carvalho, F., and Carvalho, P.
(2020). Anonymisation and compliance to protection
data: Impacts and challenges into big data. In Proc.
22nd Int. Conf. on Enterprise Inf. Syst.
COVID-19-Workgroup (2020). Recommendations and
Guidelines on Data Sharing, final release 30. Re-
search Data Alliance (RDA).
Cunha, E. and Vargens, J. (2017). Sist. de informac¸
˜
ao do
sistema
´
Unico de sa
´
ude. In Gondim, G., Christ
´
ofaro,
M., and Miyashiro, G., editors, T
´
ecnico de vigil
ˆ
ancia
em sa
´
ude: fundamentos, volume 2. EPSJV.
de Mendonc¸a, R. R. (2016). Etl4linkedprov: Managing
multigranular linked data provenance. JOURNAL
OF INFORMATION AND DATA MANAGEMENT -
JIDM, 7(2):16.
Delgado, J. and Llorente, S. (2020). Security and privacy
when applying fair principles to genomic information.
Studies in Health Techn. and Inform., 275:37–41.
Ferreira, A. (2020). Gdpr: What’s in a year (and a half)? In
Proc. 22nd Int. Conf. on Enterprise Inf. Syst.
Fung, B. C. M., Wang, K., Fu, A. W.-C., and Yu, P. S.
(2011). Introduction to privacy-preserving data pub-
lishing: concepts and techniques. Chapman and
Hall/CRC data mining and knowledge discovery se-
ries. Chapman and Hall/CRC.
Huesch, M. and Mosher, T. (2017). Using it or losing it?
the case for data scientists inside health care. Nejm
Catalyst 3.3.
Hutchings, E., Loomes, M., Butow, P., and Boyle, F. (2020).
A systematic literature review of researchers’ and
healthcare professionals’ attitudes towards the sec-
ondary use and sharing of health administrative and
clinical trial data. Syst Rev., 9(240):1–27.
Pandit, H. J., O’Sullivan, D., and Lewis, D. (2018). Ex-
tracting provenance metadata from privacy policies.
In Belhajjame, K., Gehani, A., and Alper, P., edi-
tors, Provenance and Annotation of Data and Pro-
cesses, pages 262–265, Cham. Springer International
Publishing.
Rautenberg, S., Ermilov, I., Marx, E., Auer, S., and Ngomo,
A.-C. N. (2015). Lodflow: a workflow management
system for linked data processing. In Proc. 11th Int.
Conf. on Semantic Syst., pages 137—-144, Vienna,
Austria. ACM.
Sauermann, S., Kanjala, C., Templ, M., Austin, C., and
RDA-COVID19-WG (2020). Preservation of in-
dividuals’ privacy in shared covid-19 related data.
In COVID-19 Data sharing in epidemiology, ver-
sion 0.054. Research Data Alliance RDA-COVID19-
Epidemiology WG.
Smaradottir, B. (2018). Security management in electronic
health records: Attitudes and experiences among
health care professionals. In Int. Conf. on Comp. Sci-
ence and Comp. Intell. (CSCI), pages 715–719. IEEE.
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J. J., and
et al. (2016). The fair guiding principles for scientific
data management and stewardship. Sci Data, 3.
Privacy-preservation and the Use of Data for Research: A COVID-19 Use Case in Randomly Generated Healthcare Records
323
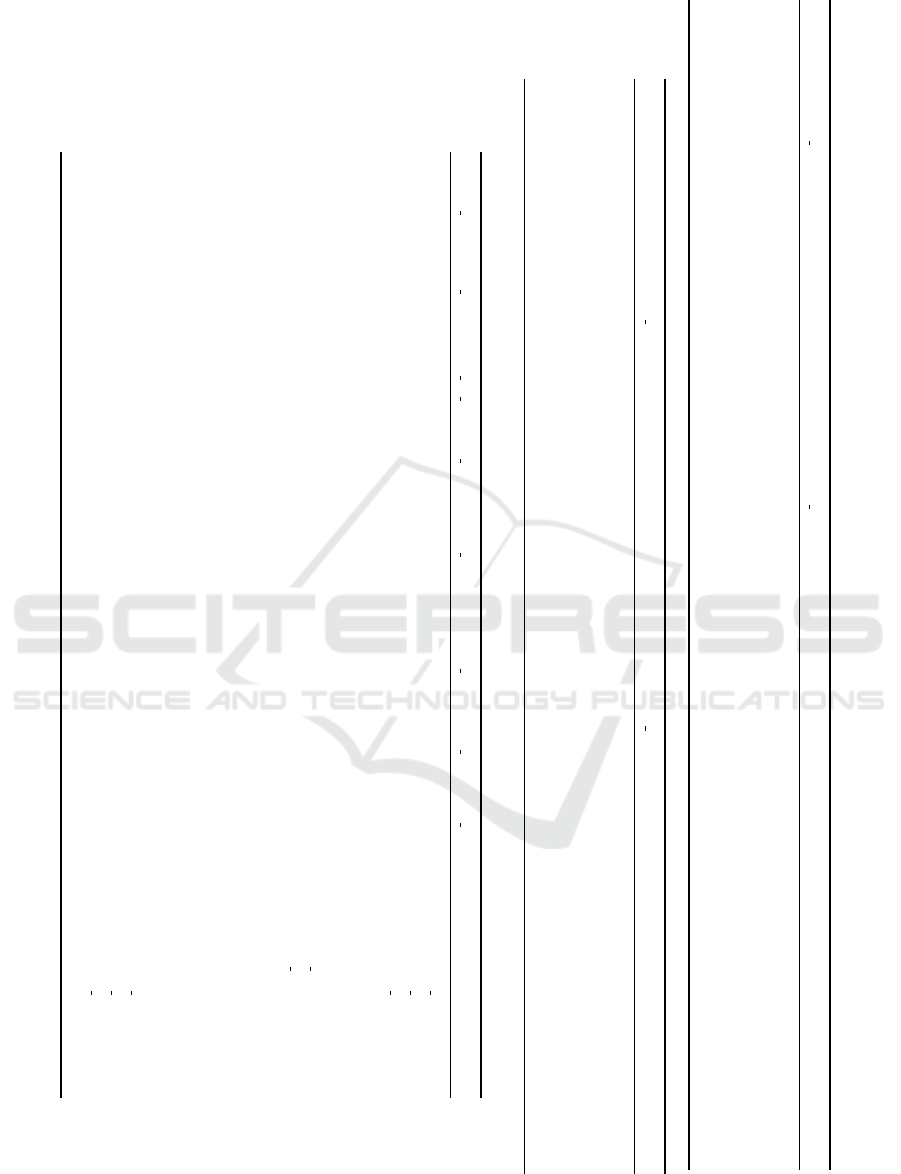
Table 1: Original Data (Dataset COVID-DS1).
person id name gender birth date address phone temperat. apdiast. apsyst.
ea5d0c10-d816-4981-ab34-92d61ea5ac1a Ellynn Philcock Male 15/08/1925 7386 Mayfield Junction ZCode 2287945 +86 (685) 614-1409 35 10,2 19,7
00fc771f-29aa-45d0-8942-1375c1d0f5cb Teodoro Filipchikov Female 06/08/2016 493 Norway Maple Crossing ZCode 2287312 +66 (837) 896-5022 35,6 11,8 19,7
e2c5f2dd-69b9-4deb-bc16-31bdad1aa73b Yolane Bursnoll Male 30/06/2007 25 Crest Line Plaza ZCode 5198625 +34 (881) 908-5963 39,1 9,6 20,8
326f7609-60a0-4d32-80a4-df20459f7822 Molly Lownes Male 02/12/1982 28115 Bartelt Drive ZCode 5198568 +62 (916) 666-7495 36,9 8,7 17,5
d38f0849-3077-4907-af22-68ae4f874c25 Gard Selway Male 29/09/1990 1 North Drive ZCode 4823533 +48 (487) 736-4522 40,4 10,3 22,6
Table 2: Anonymized Data (Dataset COVID-DS1).
person id name gender age range address phone temper. apdiast. apsyst.
d1b2d2347eb4364463e8f0c961c36bb0268696b516cc6ca5d99f3a68147a7433 REMOVED Male 80 - ZCode 2287945 REMOVED 35 10,2 19,7
c9ee2d5bcf7b5564d3a586d7a571a7a222a894c38c67990f17571e9bf1fc05c8 REMOVED Female 0 - 20 ZCode 2287312 REMOVED 35,6 11,8 19,7
7a16ca0622309d8c45b8b3e3f57eeb484d4a11436135ea2f46e680d563a8dd9c REMOVED Male 0 - 20 ZCode 5198625 REMOVED 39,1 9,6 20,8
826f3866ae955f801ddcfddfce2d10433160b9dff1baf2cba1ce5a005d8fdfd4 REMOVED Male 20 - 40 ZCode 5198568 REMOVED 36,9 8,7 17,5
746efa474010d80c4c11f341a02a226dff56ceb79d9d02f6f2eddba78c2a2f8f REMOVED Male 20 - 40 ZCode 4823533 REMOVED 40,4 10,3 22,6
Table 3: Attribute Metadata.
attribute id dataset id attribute cat id step id execution seq pptechnique id domain id hash key salt label type
000001 000001 0001 0003 0003 0002 0001 jqD9dfV 500 person id uid
000001 000001 0001 0003 0003 0002 0002 jqD9dfV 750 person id uid
000001 000001 0001 0003 0003 0002 0003 jqD9dfV 235 person id uid
000002 000001 0001 0003 0003 0004 0002 NULL NULL name string
000002 000001 0001 0003 0003 0004 0003 NULL NULL name string
000003 000001 0002 0003 0003 NULL NULL NULL NULL gender string
000004 000001 0002 0003 0003 0003 0002 NULL NULL birth datetime date
000004 000001 0002 0003 0003 0003 0003 NULL NULL birth datetime date
000005 000001 0002 0003 0003 0004 0002 NULL NULL address string
000005 000001 0002 0003 0003 0004 0003 NULL NULL address string
000006 000001 0002 0003 0003 0004 0002 NULL NULL phone string
000006 000001 0002 0003 0003 0004 0003 NULL NULL phone string
000007 000001 0003 0003 0003 NULL NULL NULL NULL temperature number
000008 000001 0003 0003 0003 NULL NULL NULL NULL apdiastolic number
000009 000001 0003 0003 0003 NULL NULL NULL NULL apsystolic number
000001 000001 0001 0003 0007 0001 0001 ObpTD3iM NULL person id uid
000001 000001 0001 0003 0007 0001 0002 ObpTD3iM NULL person id uid
000001 000001 0001 0003 0007 0001 0003 ObpTD3iM NULL person id uid
.... .... .... .... .... .... .... .... .... .... ....
ICEIS 2022 - 24th International Conference on Enterprise Information Systems
324
