
Learning Embeddings from Free-text Triage Notes using Pretrained
Transformer Models
Émilien Arnaud
1
, Mahmoud Elbattah
2,3
,
Maxime Gignon
1
and Gilles Dequen
2
1
Emergency Department, Amiens-Picardy University, Amiens, France
2
Laboratoire MIS, Université de Picardie Jules Verne, Amiens, France
3
Faculty of Environment and Technology, University of the West of England, Bristol, U.K.
Keywords: Natural Language Processing, BERT, Transformers, Clustering, Healthcare Analytics.
Abstract: The advent of transformer models has allowed for tremendous progress in the Natural Language Processing
(NLP) domain. Pretrained transformers could successfully deliver the state-of-the-art performance in a myriad
of NLP tasks. This study presents an application of transformers to learn contextual embeddings from free-
text triage notes, widely recorded at the emergency department. A large-scale retrospective cohort of triage
notes of more than 260K records was provided by the University Hospital of Amiens-Picardy in France. We
utilize a set of Bidirectional Encoder Representations from Transformers (BERT) for the French language.
The quality of embeddings is empirically examined based on a set of clustering models. In this regard, we
provide a comparative analysis of popular models including CamemBERT, FlauBERT, and mBART. The study
could be generally regarded as an addition to the ongoing contributions of applying the BERT approach in
the healthcare context.
1 INTRODUCTION
Artificial Intelligence (AI) is being intensively used
for a multitude of tasks in the healthcare arena.
Healthcare is typically delivered in data-rich settings
where abundant amounts of data are generated
continuously. In this respect, Machine Learning (ML)
solutions could provide high benefits to help develop
strategies for improving the quality of services or
curbing costs, for example.
Natural Language Processing (NLP) has received
particular attention since clinical data are largely
accumulated into unstructured notes made by
physicians or nurses along the patient journey. Novel
approaches of NLP have made a translational impact
for a variety of healthcare applications (e.g. Ambrosy
et al. 2021; Viani et al. 2021; Arnaud et al. 2021).
Several studies discussed the role of NLP in this
regard (e.g., Elbattah et al. 2021; Hao et al 2021).
With advances in Deep Learning, large-scale
language models have been developed to achieve the
state-of-the-art performance. Deep architectures of
Convolutional Neural Networks (CNNs) have
allowed for learning feature representations from raw
data automatically (LeCun et al. 1989; LeCun,
Bottou, Bengio, and Haffner, 1998).
Moreover, Transfer Learning (TL) is being
increasingly adopted for a variety of healthcare and
medical applications. The TL concept has deemed as
an attractive path in situations where data paucity and
imbalance inherently exist. Pretrained models allow
for transferring knowledge from one or more source
tasks towards the application to another target task
(Pan, and Yang, 2009). In this regard, recent studies
utilized the Bidirectional Encoder Representations
from Transformers (BERT), a state-of-the-art NLP
model (Devlin et al. 2019). The BERT approach
brings the advantage of allowing pre-trained models
to tackle a broad set of NLP tasks.
The present study seeks to employ transformer
models to extract contextual embeddings from free-
text triage notes. The study is based on a large-scale
dataset collected by the Amiens-Picardy University
Hospital in France. The dataset contained about 260K
ED records collected over more than four years.
The study aims to contribute towards providing a
comparative analysis of popular BERT models as a
mechanism for learning embeddings from clinical
notes. We present an exemplary case of triage notes
in the French language, whereas the literature still
generally lacks similar studies using languages other
than English.
Arnaud, É., Elbattah, M., Gignon, M. and Dequen, G.
Learning Embeddings from Free-text Triage Notes using Pretrained Transformer Models.
DOI: 10.5220/0011012800003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 835-841
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All r ights reserved
835

2 RELATED WORK
A growing body of studies seeks to implement
transformer models for a variety of healthcare-related
NLP tasks. This section aims to explore the recent
developments in this regard. In general, the review
sheds insights into the potential applications of
transformers in the healthcare domain. Thus, the
review is largely selective rather than exhaustive.
One study developed a BERT-based framework for
transforming free-text descriptions into a standardized
form based on the Health Level 7 (HL7) standards
(Peterson, Jiang, and Liu, 2020). They utilized a
combination of domain-specific knowledgebases
along with BERT models. It was demonstrated that the
BERT-based representation of language contributed
significantly to improving the model performance.
(Rasmy et al., 2021) introduced the Med-BERT
model, which adapted the BERT approach to the EHR
data. Med-BERT provided contextualized embeddings
pretrained on an EHR dataset including more than 28M
patients. Their experiments demonstrated that fine-
tuning Med-BERT could improve the prediction
accuracy in two disease prediction tasks from two
clinical databases. They reported improvement in the
area under the receiver operating characteristics (ROC)
curve by 1.21–6.14%. As well, (Cai et al., 2021)
proposed another BERT-based model, named as
EMBERT, for text mining of Chinese medical data.
Other studies implemented transformer-based
approaches to perform the tasks of entity recognition
and relation extraction from medical text. For
instance, (Xue et al., 2019) integrated the BERT
language model to extract relations from medical text
in Chinese. They used a dataset related to coronary
angiography collected from the Shuguang Hospital in
Shanghai. Their results were claimed to outperform
the state-of-the-art methods for entity recognition and
relation classification by 1.65% and 1.22%,
respectively. Likewise, (Kim and Lee, 2020) utilized
a BERT model to extract clinical entities from an QA
dataset for medical diagnosis in the Korean language.
In addition, BERT models were used to extract
embeddings from clinical documents for developing
predictive models. For example, (Tahayori, Chini-
Foroush, and Akhlaghi, 2021) used a retrospective
cohort of ED triage notes from St Vincent's Hospital
in Melbourne. The BERT embeddings were utilized
to develop a Deep Learning model to predict the
disposition of patients. By the same token, (Chang,
Hong, and Taylor, 2020) used a BERT model to
extract embeddings from ED notes.
Furthermore, part of the recent contributions has
been positioned within the COVID-19 context. For
example, (Wang et al., 2020) utilized a BERT model
for sentiment analysis of posts on Sina Weibo, a
popular social media platform in China. About 1M
COVID-related posts were analyzed. The literature
includes many studies that used BERT models for
different NLP tasks such as (Chintalapudi, Battineni,
and Amenta, 2021), and (Liu et al. 2021).
The present work attempts to add to the ongoing
contributions towards making use of BERT models in
the healthcare domain. We focus on analyzing the
performance of popular pretrained transformers to
extract embeddings from triage notes in the French
language. To the best of our knowledge, the literature
still lacks the application of BERT to triage notes,
especially for languages rather than English.
3 DATA DESCRIPTION
The dataset under consideration was provided by the
Amiens-Picardy University Hospital in France. The
authors had obtained the necessary permissions for
accessing the dataset and conducting research. The
study has been specifically approved by the Hors Loi
Jarde Committee (PI2019_843_0066 reference).
Moreover, all personal information was fully
anonymized to ensure the privacy of patients.
The dataset contained more than 260K ED records
spanning more than four years from January 2015 to
June 2019. Each record was associated with a binary
label representing the outcome at triage (i.e.
hospitalization or discharge). The average length of
stay (LOS) in ED was 4h23m. While the average LOS
of discharged patients was around 4h04m.
The dataset included a variety of numeric and
categorical variables, and textual notes. Numeric
variables mainly described vital signs such as
temperature, heart rate, etc. While the categorical
variables described general information about
patients such as gender, origin, family status, etc. In
addition, a set of four textual fields provided the
triage observations, surgical history, psychiatric
history, and medical history of patients. The free-text
notes were input in the French language by nurses or
physicians at the ED stage. Our focus was exclusively
on the textual fields as to be elaborated later.
As well, the data records indicated the specialty
assigned to hospitalized patients. The majority of
specialties can be grouped into three broad categories
including short-term hospitalization unit, surgery, or
medical specialties (e.g., cardiology, neurology).
Table 1 provides statistics of specialties among
hospitalized patients in the dataset.
Scale-IT-up 2022 - Workshop on Scaling-Up Health-IT
836
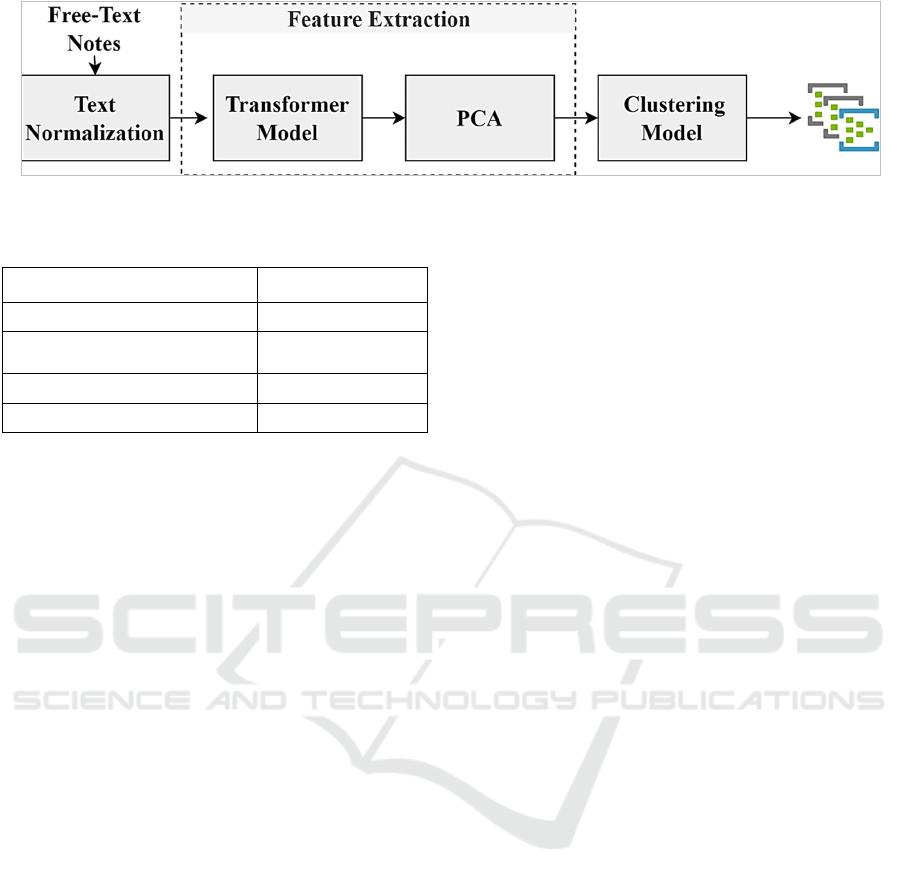
Figure 1: The text preprocessing pipeline.
Table 1: Summary of medical specialties.
Specialty / Label Hospitalization %
Surgery / CHIR
19.7%
Short-Term Hospitalization
Unit / UHCD
42.4%
Medical Specialty / MED
33%
Other
4.9%
4 FEATURE EXTRACTION
4.1 Experimental Set-up
The BERT experiments were conducted using a
Nvidia GPU, which was essential for implementing
Deep Learning in a timely manner. While the
clustering experiments were implemented using a
standard Xeon processor of 2.00GHz.
4.2 Text Normalization
The initial step included the pre-processing and
anonymization of patient records. The process of text
normalization was conducted over a set of stepwise
procedures as follows. Initially, we had to exclude the
dataset records that contained blank triage notes.
More specifically, about 70K records did not include
any textual information at all.
Secondly, the four columns of triage notes were
merged into a single text field. The merged text can
be regarded as a unique document including
observations, surgical history, psychiatric history,
and medical history for each patient.
Eventually, typical procedures of text
normalization were applied to clean and standardize
the textual notes. The procedures included case
conversion, removing stop words, and spell checking.
The normalization process was implemented using
standard Python libraries including NLTK (Bird,
Klein, and Loper, 2009). The spell checking was
facilitated thanks to the pyspellchecker library
(Barrus, 2021), which supports the French language.
4.3 Transformer Models
A set of pretrained transformer models were
experimented for the feature extraction process. The
models were originally pretrained using large-scale
text datasets in French. Specifically, our experiments
included the following models:
• CamemBERT (Martin et al. 2019): One of
the state-of-the-art language models for
French. It is based on the RoBERTa
architecture (Liu et al. 2019). It was pretrained
on the French corpus, which is part of OSCAR
dataset, a huge multilingual corpus data
(Suárez, Sagot, and Romary, 2019). The
CamemBERT currently includes six variants
with varying number of parameters, amount of
pretraining data, and pretraining source
domains. In our case, we used the camembert-
base model.
• FlauBERT (Le et al. 2019): a BERT-based
model pretrained on a very large and
heterogeneous French corpus. The FlauBERT
includes different four versions, which were
trained using the Jean Zay supercomputer in
France. We specifically used the flaubert-base-
uncased version.
• mBART (Liu et al. 2020): mBART is based
on the Bidirectional and Auto-Regressive
Transformer approach (Lewis et al. 2019). It is
a multilingual encoder-decoder sequence-to-
sequence model, primarily intended for
translation tasks. The mBART includes 12
layers of encoder and decoder trained on
monolingual corpuses of 25 languages.
All models were accessed through the
HuggingFace repository (Wolf et al. 2019), which is
widely used for the distribution of pretrained
transformer models. The SentenceTransformers
library was utilized as well (Reimers, and Gurevych,
2019), which facilitated the usage of models.
We adopted a Transfer-Learning approach for the
feature extraction process. As such, the default
parameters were fully transferred into the target task.
Learning Embeddings from Free-text Triage Notes using Pretrained Transformer Models
837

The mBART model included the largest number of
parameters and embedding dimension, and
accordingly the longest runtime. Table 2 describes the
transformer models used in our experiments.
Table 2: Summary of experimental models.
Model Params
Embedding
Dimension
Runtime
CamemBERT 110M 768 31 min
FlauBERT 137 M 768 32 min
MBART 610M 1024 64 min
Eventually, Principal Component Analysis (PCA)
was applied to the feature maps extracted by each
model. The PCA provided a compact transformation
of features, which was more amenable for developing
clustering models as discussed in the next section.
The Scikit-Learn library (Pedregosa et al. 2011) was
used to perform the PCA transformation. Figure 1
summarizes the pipeline of feature extraction.
5 CLUSTERING EXPERIMENTS
5.1 K-Means Clustering
The goal of this empirical part was to cluster patient
records exclusively based on the embeddings
extracted from the triage notes. In this regard, the
feature sets learned by transformers were utilized to
develop standard K-Means clustering models.
The dataset included the whole dataset excluding
the records of blank notes. Roughly, about 194K
samples were used for clustering. The dataset was
relatively imbalanced, about 63% of hospitalization.
The clustering models were experimented with
different values of K ranging from 2 to 10. The
models were implemented using the Scikit-Learn
library (Pedregosa et al. 2011). Table 3 gives the
parameters used in the experiments.
Table 3: K-Means parameters.
Parameter Value
Number of Clusters (K) 2–10
Centroid Initialisation k-means++
Similarity Metric Euclidian Distance
Number of Iterations 200
5.2 Evaluation of Clusters
The quality of clusters was examined based on two
metrics including the Silhouette score (Rousseeuw,
1987), and the Fowlkes-Mallows (FM) score
(Fowlkes, and Mallows, 1983).
The Silhouette score has been widely used in
clustering-related studies as an objective means to
measure the robustness of cluster membership. It is
represented as a continuous range of [-1, 1], where
scores near +1 positively indicate that a point is far
away from neighboring clusters. In contrast, values
closer to 0 can be interpreted as being on or very close
to the decision boundary between neighboring
clusters, while negative scores would strongly
suggest that the point may have been assigned to the
wrong cluster. The score can be calculated for each
point as follows:
𝑺
(
𝒊
)
=
(
𝒃(𝒊) − 𝒂(𝒊)
)
𝐦𝐚𝐱 (𝒂(𝒊), 𝒃(𝒊))
Where a(i) is the average distance of point i to all
other points in containing cluster, and b(i) is the
smallest average distance of point i to points in
another cluster.
While the FM score can be used when ground-
truth labels are known, which applied to our case. It
is defined as the geometric mean of the pairwise
precision and recall as below:
FM SCORE =
TP
(TP + FP)(TP + FN)
Where TP is the number of True Positives, FP is
the number of False Positives, and FN is the number
of False Negatives.
On the one hand, Figure 2 compares the Silhouette
scores achieved by the clustering models for K=2:10.
As it appears, the highest score could be achieved
when K=2 in all cases. However, the quality of
clusters declined while applying further partitioning
of clusters (i.e. K=3, 4). This goes well with the fact
that we had a binary grouping of patients (i.e.
hospitalized or discharged). In addition, it should be
noted that the mBART-based features could provide
the highest coherence of clusters while K=2 and 3.
The FlauBERT performed generally better for
K>3.While the quality of CamemBERT-based
clusters was inferior in general.
On the other hand, Figure 3 analyzes the FM
scores. The mBART-based clusters notably achieved
a high score, around 0.62 for K=2. As well, the
FlauBERT and CamemBERT provided a comparable
performance of about 0.56 and 0.57, respectively.
This generally translates that the clusters were largely
coherent compared to the original binary grouping of
patients. However, the quality of clusters declined
significantly by increasing K towards 10.
Scale-IT-up 2022 - Workshop on Scaling-Up Health-IT
838
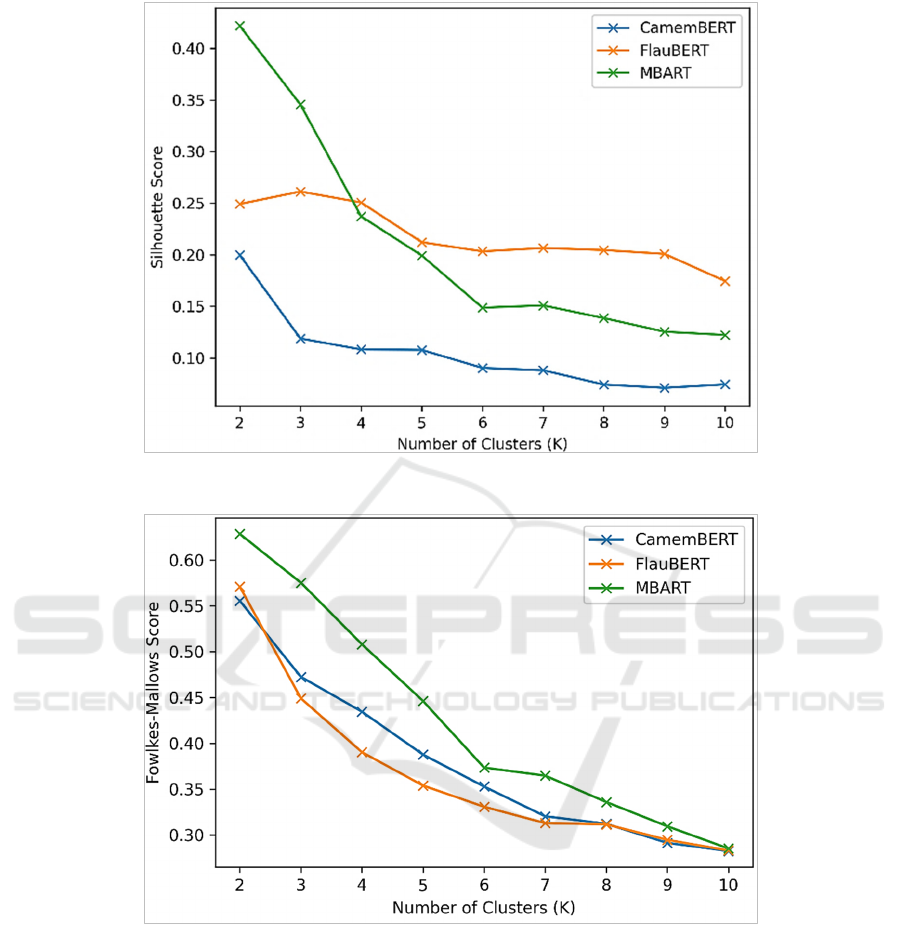
Figure 2: Silhouette score of clusters.
Figure 3: Fowlkes-Mallows score of clusters.
6 CONCLUSIONS
This study experimented a set of pretrained BERT
models to learn contextual embeddings from free-text
triage notes. The embeddings were utilized to develop
multiple K-Means clustering models. The BERT-
based embeddings could be employed for developing
clusters of good coherence in general.
Accordingly, the empirical experiments could
largely validate the suitability of Transfer Leaning in
this context. Pretrained transformers can serve as an
effective mechanism for learning contextual
embeddings from a variety of free-text notes, which
exist ubiquitously in the healthcare environment. The
present work could open several avenues for further
investigation as well. For example, the BERT-based
embeddings can also be used to develop predictive
models, such as predicting patient hospitalization or
medical specialties at the triage stage.
Learning Embeddings from Free-text Triage Notes using Pretrained Transformer Models
839

REFERENCES
Ambrosy, A. P., Parikh, R. V., Sung, S. H., Narayanan, A.,
Masson, R., Lam, P. Q., ... & Go, A. S. (2021). A
Natural Language Processing–Based Approach for
Identifying Hospitalizations for Worsening Heart
Failure Within an Integrated Health Care Delivery
System. JAMA Network Open, 4(11), e2135152-
e2135152.
Arnaud, É., Elbattah, M., Gignon, M., & Dequen, G. (2021).
NLP-Based Prediction of Medical Specialties at
Hospital Admission Using Triage Notes. In
Proceedings of the 9th International Conference on
Healthcare Informatics (ICHI) (pp. 548-553). IEEE.
Barrus, T. (2021). GitHub Repo:
https://github.com/barrust/pyspellchecker
Bird, S., Klein, E., & Loper, E. (2009). Natural language
processing with Python: analyzing text with the natural
language toolkit. O'Reilly Media, Inc.
Cai, Z., Zhang, T., Wang, C., & He, X. (2021). EMBERT:
A Pre-trained Language Model for Chinese Medical
Text Mining. In Proceedings of Joint International
Conference on Web and Big Data (pp. 242-257).
Springer, Cham.
Chang, D., Hong, W. S., & Taylor, R. A. (2020). Generating
contextual embeddings for emergency department chief
complaints. JAMIA Open, 3(2), 160-166.
Chintalapudi, N., Battineni, G., & Amenta, F. (2021).
Sentimental Analysis of COVID-19 Tweets Using
Deep Learning Models. Infectious Disease Reports,
13(2), 329-339.
Devlin, J., Chang, M., Lee, K., & Toutanova, K. (2019).
BERT: Pre-training of Deep Bidirectional
Transformers for Language Understanding. In
Proceedings of the Annual Conference of the North
American Chapter of the Association for
Computational Linguistics (NAACL-HLT).
Elbattah, M., Arnaud, E., Gignon, M., & Dequen, G.
(2021). The role of text analytics in healthcare: A
review of recent developments and applications. In
Proceedings of the 14th International Joint Conf. on
Biomedical Engineering Systems and Technologies
(BIOSTEC).
Fowlkes, E. B., & Mallows, C. L. (1983). A method for
comparing two hierarchical clusterings. Journal of the
American Statistical Association, 78(383), 553-569.
Hao, T., Huang, Z., Liang, L., Weng, H., & Tang, B. (2021).
Health Natural Language Processing: Methodology
Development and Applications. JMIR Medical
Informatics, 9(10), e23898.
Kim, Y. M., & Lee, T. H. (2020). Korean clinical entity
recognition from diagnosis text using BERT. BMC
Medical Informatics and Decision Making, 20(7), 1-9.
Le, H., Vial, L., Frej, J., Segonne, V., Coavoux, M.,
Lecouteux, B., ... & Schwab, D. (2019). Flaubert:
Unsupervised language model pre-training for French.
arXiv preprint arXiv:1912.05372.
LeCun, Y., Boser, B. E., Denker, J. S., Henderson, D.,
Howard, R. E., Hubbard, W. E., and Jackel, L. D.
(1989). Handwritten digit recognition with a back-
propagation network. In Proceedings of Advances in
Neural Information Processing Systems (NIPS) (pp.
396-404).
LeCun, Y., Bottou, L., Bengio, Y., & Haffner, P. (1998).
Gradient-based learning applied to document
recognition.
In Proceedings of the IEEE, 86(11), 2278-
2324.
Lewis, M., Liu, Y., Goyal, N., Ghazvininejad, M.,
Mohamed, A., Levy, O., ... & Zettlemoyer, L. (2019).
Bart: Denoising sequence-to-sequence pre-training for
natural language generation, translation, and
comprehension. arXiv preprint arXiv:1910.13461.
Liu, Y., Ott, M., Goyal, N., Du, J., Joshi, M., Chen, D., ...
& Stoyanov, V. (2019). Roberta: A robustly optimized
bert pretraining approach. arXiv preprint
arXiv:1907.11692.
Liu, Y., Gu, J., Goyal, N., Li, X., Edunov, S., Ghazvininejad,
M., ... & Zettlemoyer, L. (2020). Multilingual denoising
pre-training for neural machine translation.
Transactions of the Association for Computational
Linguistics, 8, 726-742.
Liu, G., Liao, Y., Wang, F., Zhang, B., Zhang, L., Liang,
X., ... & Cui, S. (2021). Medical-vlbert: Medical visual
language BERT for COVID-19 CT report generation
with alternate learning. IEEE Transactions on Neural
Networks and Learning Systems, 32(9), 3786-3797.
Martin, L., Muller, B., Suárez, P. J. O., Dupont, Y., Romary,
L., de La Clergerie, É. V., ... & Sagot, B. (2019).
Camembert: a tasty French language model. arXiv
preprint arXiv:1911.03894.
Pan, S. J., & Yang, Q. (2009). A survey on transfer learning.
IEEE Transactions on Knowledge and Data
Engineering, 22(10), 1345-1359.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., ... & Duchesnay, E. (2011).
Scikit-learn: Machine learning in Python. Journal of
Machine Learning Research, 12, 2825-2830.
Peterson, K. J., Jiang, G., & Liu, H. (2020). A corpus-driven
standardization framework for encoding clinical
problems with HL7 FHIR. Journal of Biomedical
Informatics, 110, 103541.
Rasmy, L., Xiang, Y., Xie, Z., Tao, C., & Zhi, D. (2021).
Med-BERT: pretrained contextualized embeddings on
large-scale structured electronic health records for
disease prediction. NPJ Digital Medicine, 4(1), 1-13.
Reimers, N., & Gurevych, I. (2019). Sentence-BERT:
Sentence embeddings using siamese bert-networks.
arXiv preprint arXiv:1908.10084.
Rousseeuw, P. J. (1987). Silhouettes: a graphical aid to the
interpretation and validation of cluster analysis. Journal
of Computational and Applied Mathematics, 20, 53-65.
Suárez, P. J. O., Sagot, B., & Romary, L. (2019).
Asynchronous pipeline for processing huge corpora on
medium to low resource infrastructures. In Proceedings
of the 7th Workshop on the Challenges in the
Management of Large Corpora (CMLC-7).
Tahayori, B., Chini-Foroush, N., & Akhlaghi, H. (2021).
Advanced natural language processing technique to
predict patient disposition based on emergency triage
Scale-IT-up 2022 - Workshop on Scaling-Up Health-IT
840

notes. Emergency Medicine Australasia, 33(3), 480-
484.
Viani, N., Botelle, R., Kerwin, J., Yin, L., Patel, R., Stewart,
R., & Velupillai, S. (2021). A natural language
processing approach for identifying temporal disease
onset information from mental healthcare text.
Scientific Reports, 11(1), 1-12.
Wang, T., Lu, K., Chow, K. P., & Zhu, Q. (2020). COVID-
19 sensing: Negative sentiment analysis on social
media in China via BERT model. IEEE Access, 8,
138162-138169.
Wolf, T., Debut, L., Sanh, V., Chaumond, J., Delangue, C.,
Moi, A., ... & Rush, A. M. (2019). Huggingface's
transformers: State-of-the-art natural language
processing. arXiv preprint arXiv:1910.03771.
Xue, K., Zhou, Y., Ma, Z., Ruan, T., Zhang, H., & He, P.
(2019). Fine-tuning BERT for joint entity and relation
extraction in Chinese medical text. In Proceedings of
the IEEE International Conference on Bioinformatics
and Biomedicine (BIBM) (pp. 892-897). IEEE.
Learning Embeddings from Free-text Triage Notes using Pretrained Transformer Models
841
