
A Comprehensive and Scientifically Accurate Pharmaceutical
Knowledge Ontology based on Multi-source Data
Pengfei Wang
#a
, Yiqing Mao
#
, Wei Song
b
, Wenting Jiang
c
, Yang Liu
d
, Liumeng Zheng
e
,
Bin Ma
f
, Qingqing Sun
g
and Sheng Liu
*h
Beijing MedPeer Information Technology Co., Ltd., Beijing, China
Keywords: Pharmaceutical Knowledge, Ontology, Drug, Knowledge Graph.
Abstract: Recently, knowledge graphs have been applied by large pharmaceutical companies to improve the efficiency
of drug discovery. Specifically, knowledge graphs based on drug ontology have been used for many purposes.
Current drug ontologies have different scopes, but mainly focus on the description of basic drug information.
Here, we describe a comprehensive pharmaceutical knowledge ontology, including information of active
ingredients, indications, inactive ingredients, drugs, clinical trials, organs and tissues, literature, patents,
targets, therapeutics, and biomolecules. Using multiple data sources, we apply a seven-step method for
ontology modelling using Protégé software. A comprehensive pharmaceutical knowledge ontology model is
established to complete the knowledge representation of drug information. By means of ontology theory, the
pharmaceutical knowledge is modelled, standardized and networked, so as to clarify the knowledge structure
and quickly acquire related knowledge and logical relationships. In the future, knowledge graphs based on
this ontology could be helpful to deal with the dispersion, heterogeneity, redundancy and fragmentation of
medical big data, to share and integrate pharmaceutical data, and to provide a set of solutions for the
networked development of pharmaceutical knowledge.
1 INTRODUCTION
Drug discovery is a complex process with a
development cycle of 10-15 years and an average
research and development cost of $2.6 billion
(Wouters et al., 2020). In recent years, knowledge
graphs have been applied by large pharmaceutical
companies to improve the efficiency of drug
discovery; for example, AstraZeneca is applying
BenevolentAI for drug development for chronic
kidney disease (CKD) and idiopathic pulmonary
fibrosis (IPF), showing the application and
development prospect of knowledge graph
#
Contributed equally to this work.
a
https://orcid.org/0000-0003-0956-6556
b
https://orcid.org/0000-0002-4596-5303
c
https://orcid.org/0000-0002-0900-7220
d
https://orcid.org/0000-0003-0679-2275
e
https://orcid.org/0000-0003-3280-8445
f
https://orcid.org/0000-0003-0235-3419
g
https://orcid.org/0000-0002-2442-4735
h
https://orcid.org/0000-0002-1054-6440
*
Address correspondence to: Sheng Liu
technology in such tasks. Knowledge graphs based on
drug ontology have been used for many purposes,
such as comparative effectiveness research (Hanna et
al., 2013), adverse drug reactions (Cai et al., 2015;
Hur et al., 2018), and clinical data warehousing
(Podchiyska et al., 2010). RxNorm was created to
address a lack of standardization of drug names and
to make them interoperable by integrating drug terms
into a reference system (Nelson et al., 2020). RxNorm
currently integrates terminology information from
most drug knowledge base vendors (e.g., First
DataBank, Multum, Micromedex, Gold Standard), as
well as drug ingredients from standard terminology
168
Wang, P., Mao, Y., Song, W., Jiang, W., Liu, Y., Zheng, L., Ma, B., Sun, Q. and Liu, S.
A Comprehensive and Scientifically Accurate Pharmaceutical Knowledge Ontology based on Multi-source Data.
DOI: 10.5220/0011012100003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 3: BIOINFORMATICS, pages 168-175
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

(e.g., SNOMED CT, MeSH). RxNorm focuses on
drug names and codes; however, clinical information
and administrative information are out of scope
(Bodenreider et al., 2018). Based on the RxNorm
drug terminology and the Chemical Entities of
Biological Interest ontology (ChEBI), Hanna et al.
(2013) built Drug Ontology (DrOn), a modular,
extensible body of drugs, ingredients, and biological
activities, originally created to enable comparative
effectiveness. OCRx is a Canadian drug ontology
system built to provide a normalized and standardized
description of drugs authorized to be marketed in
Canada. OCRx is focused on clinical drug description
(i.e., substance, strength, route of administration,
pharmaceutical form) (Nikiema et al., 2021). Sharp
(2017) created a drug-indication database (DID), a
database of structured drug-indication relations
intended to facilitate building practical,
comprehensive, and integrated drug ontologies.
Current drug ontologies have different scopes, for
example, RxNorm and OCRx describe drugs
available in the U.S. and Canada, respectively, and
mainly focusing on the description of basic drug
information. However, the current lack of
comprehensive pharmaceutical knowledge ontology
covering drug targets, adverse drug reactions, clinical
trials, patents, literature and other classes limits the
application of drug ontology.
In this paper, we describe a comprehensive drug
knowledge ontology, including active ingredients,
indications, inactive ingredients, drugs, clinical trials,
organ and tissue targets, literature, patents,
therapeutics, and biomolecules. We use ontology
theory to model, standardize and network
pharmaceutical knowledge, which is convenient for
clarifying the knowledge structure, quickly obtaining
relevant knowledge and logical relationships, and
helping to deal with the dispersion, heterogeneity,
redundancy and fragmentation of medical big data.
The purpose of this ontology is to aid the sharing and
interaction of pharmacy data and provides a set of
solutions for the development of pharmacy
knowledge network.
2 METHODS
Ontology theory has been widely used for knowledge
presentation; common building methods are seven-
step method, Skeletal Methodology, IDEF5, and
METHONTOLOGY. Based on a comprehensive
analysis of the existing approaches to the construction
of drug ontology, we applied a seven-step method
developed by Stanford University School of
Medicine to construct a pharmaceutical knowledge
ontology using Protégé software (Musen et al., 2015).
The construction process of pharmaceutical
knowledge ontology is shown in Figure 1. Ontology
modelling is based on hierarchical and structured
biomedical thesauri such as MeSH and ICD,
combined with authoritative data sources such as
Drugbank and PubChem, to develop a standardized
glossary and glossary of pharmaceutical ontologies to
unify and integrate multi-source data and facilitate
inter-term relationships. The next step is to refer to
the results of the analysis of the web site data, record
the conceptual level, format, and data type of the
entity in the data source for the pharmacy
terms/entities identified in the data. For relevant text
of an entity that cannot be directly identified, entity
name recognition is carried out through the biological
named entity recognition tool and the standardized
terminology. After the entity is identified, the
relationship between entities is recognized by the
machine learning method, summarizing the
properties and inter-entity relations of the new
entities, and classifying the entities to determine the
hierarchical structure of the data. Finally, experts
examine and verify the entities, properties and
relationship information, and then incorporate newly
identified entities and relationships into existing
models. The final realization uses the standard
terminology of ontology, annotates the entity, and
completes the knowledge representation.
2.1 Data Sources
Data sources are the basis of pharmaceutical
knowledge ontology modelling. Data acquired from
multiple databases and platforms is listed in Table 1.
MeSHDrugbank ICD PubChem
ATC FDA
ClinicalTrials
Registry
ę
Data Source
ObjectpropertiesClasses Dataproperties
Knowledge
extraction
DrugOntology
Knowledge
Presentation
Figure 1: Construction process of pharmaceutical
knowledge ontology.
A Comprehensive and Scientifically Accurate Pharmaceutical Knowledge Ontology based on Multi-source Data
169
The following section provides an overview of the
multiple data sources used here.
Medical Subject Headings (MeSH) is an
authoritative thesaurus compiled by The United
States National Library of Medicine (NLM). MeSH
is a standardized and expandable dynamic thesaurus
of medical concepts and is used for indexing,
cataloging, and searching of biomedical and health-
related information. It includes the subject headings
appearing in MEDLINE/PubMed, the NLM Catalog,
and other NLM databases.
The International Classification of Diseases and
Related Health Problems (ICD) is a tool for
recording, reporting, and grouping conditions and
factors that influence health. It contains categories for
diseases, health-related conditions, and external
causes of illness or death. The ICD is used to translate
disease diagnoses into an alphanumeric code, which
allows data storage, retrieval, and analysis.
DrugBank is a comprehensive, free-to-access
online database containing information on drugs and
drug targets (Wishart et al., 2006). The latest release
(version 5.1.8, released 2021-01-03) contains 14,589
entries. 5,263 non-redundant protein (i.e., enzyme,
transporter, carrier) sequences are linked to these
entries. Each entry contains more than 200 data fields
with half of the information devoted to drug/chemical
data and the other half devoted to drug target or
protein data (Wishart et al., 2018).
PubChem is an open chemistry database by the
National Institutes of Health (NIH). It contains both
small and larger molecules such as nucleotides,
carbohydrates, lipids, peptides, and chemically
modified macromolecules. PubChem collects
information on chemical structures, identifiers,
chemical and physical properties, biological
activities, patents, health, safety, and toxicity (Kim et
al., 2019).
The Anatomical Therapeutic Chemical (ATC)
system is the official drug classification of the World
Health Organization (WHO). In the ATC system,
active substances are classified in a five-level
hierarchy.
China Medical Information Platform is an expert-
certified information platform for all types of medical
information, including disease and symptom queries,
drug introduction, drug instructions, hospital queries,
and expert queries.
Clinical Pathways were released by the National
Health Commission of the People's Republic of China
and contains clinical pathways for 224 disease species
in 19 disciplines.
U.S. Clinical Trials is a web-based resource
maintained by the NLM and NIH. Each record
presents summary information about a study protocol
and includes information such as disease or condition,
intervention, title, description, and study design. The
European Union Clinical Trials Register, Chinese
Clinical Trial Registry, China Drug Trials, and WHO
International Clinical Trials Registry Platform are
similar platforms for the registration of clinical trials.
Table 1: Data sources for ontology modelling.
No. Data Source URL of Data Source
1 MeSH
https://www.nlm.nih.gov
/databases/download/me
sh.html
2 ICD-10
https://icd.who.int/brows
e10/2019/en
3 ICD-11
https://icd.who.int/brows
e11/l-m/en
4 DrugBank
https://go.drugbank.com/
releases/latest
5 PubChem
https://ftp.ncbi.nlm.nih.g
ov/
p
ubchem
/
6 ATC
https://www.whocc.no/at
c_ddd_index
/
7
China Medical
Information Platform
https://www.dayi.org.cn/
8 Clinical Pathways
http://www.nhc.gov.cn/y
zygj
/
9 US Clinical Trials
https://www.clinicaltrial
s.gov/ct2/resources/dow
nloa
d
10
European Union
Clinical Trials Register
https://www.clinicaltrial
sregister.eu/ctr-search/
search
11
Chinese Clinical Trial
Registr
y
http://www.chictr.org.cn
/searchproj.aspx
12
China Drug Trials
http://www.chinadrugtri
als.or
g
.cn
/
13
International Clinical
Trials Re
g
istr
y
Platfor
m
https://trialsearch.who.in
t/
14 FDA Orange Book
https://www.accessdata.f
da.gov/scripts/cder/ob/in
dex.cf
m
15 Japanese Orange Book
http://www.jp-orange
book.gr.jp/cgi-bin/
search_h/search_e.cgi
16
List of Reference
Preparations for Generic
Dru
g
s
(
China
)
https://www.nmpa.gov.c
n/xxgk/ggtg/qtggtg/inde
x.html
17
FDA Inactive
Ingredients Database
https://www.fda.gov/dru
gs/drug-approvals-and-
databases/inactive-
ingredients-database-
downloa
d
The publication Approved Drug Products with
Therapeutic Equivalence Evaluations (commonly
known as the Orange Book) identifies drug products
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
170
approved on the basis of safety and effectiveness by
the Food and Drug Administration (FDA) under the
Federal Food, Drug, and Cosmetic Act and related
patent and exclusivity information. It contains
information regarding active ingredient, proprietary
name, applicant, application number, dosage form,
route of administration or patent number of approved
drug products. The Japanese Orange Book is a similar
guide published in Japan.
The List of Reference Preparations for Generic
Drugs published by the National Medical Products
Administration of China is the basis for registration
application and consistency evaluation of generic
drugs in China. Inquiry contents include generic
name, English name or trade name, license holder,
specifications, dosage form, remarks, sources and
other information.
The Inactive Ingredients Database provides
information on inactive ingredients present for FDA-
approved drug products. It contains information on
route of administration, dosage and dosage form,
CAS Number, UNII, potency amount and potency
unit on inactive ingredients.
2.2 Modelling of Classes
Based on hierarchical and structured biomedical
lexicon such as MeSH and ICD, combined with
relevant terms from authoritative data sources such as
Drugbank and PubChem, we developed a
standardized glossary and term annotations using a
pharmaceutical knowledge ontology.
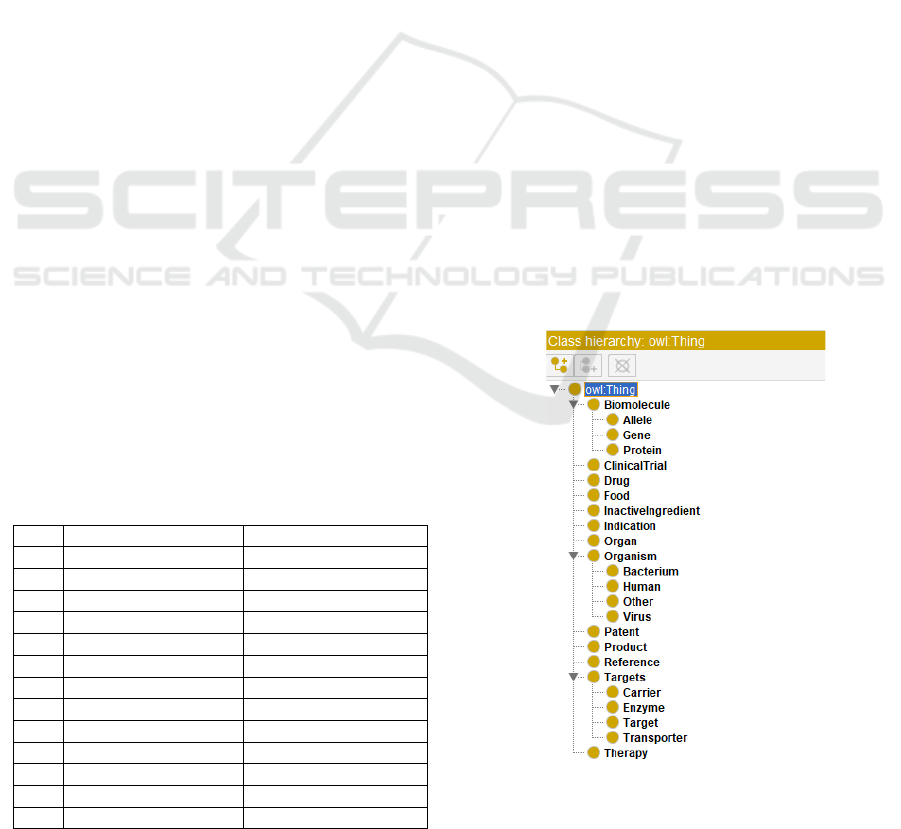
13 concepts were extracted as first-level semantic
types (classes), named Drug, Indication, Therapy,
ClinicalTrial, Product, Food, InactiveIngredient,
Patent, Reference, Organ, Targets, Biomolecule and
Organism. Each class was extracted from one or more
of the corresponding data sources, shown in Table 2
(data source numbers are from Table 1).
Table 2: Extraction of the classes from data sources.
No. Class Data Source No.
1 Drug 1, 4, 5, 6
2 Indication 1, 2, 3, 4, 7
3 Thera
py
1, 8
4 ClinicalTrial 9, 10, 11, 12, 13
5 Product 4, 14, 15, 16
6 InactiveIngredient 17
7 Foo
d
4
8 Patent 4
9 Reference 4
10 Or
g
an 7
11 Targets 4
12 Biomolecule 4
13 Organism 1, 4
2.3 Modelling of Object Properties
and Data Properties
Sentence and word segmentation were executed on
data collected from multi-source heterogeneous
sources to generate pre-processed data. Name
information of a plurality of entities defined in the
concept layer was extracted from the pre-processed
data by the named entity recognition, and the name
information was contained in the entity statement of
the data. The relationship extraction method based on
the depth neural network identifies the relationships
between different entities defined in the concept layer
from the entity statements.
Specifically, with words as the basic units, the
entity statements were characterized by word and
position vectors, and the vector splicing results
corresponding to the entity statements are obtained.
These results were input into a deep neural network,
and the relationship classification were distinguished,
which comprises an attention model, a fully
connected neural network layer and a convolutional
neural network model. Focused on the deep neural
network, the entity sentences with relationships are
identified. Syntax was analyzed, and related words
were extracted according to the dependency relation
among entities, thus obtaining the cause-effect tuples,
which include a relationship between different
entities.
The property information of each entity was
modelled as data properties, and the relationship
between the entities is modelled as object properties.
Figure 2: Classes of pharmaceutical knowledge ontology.
A Comprehensive and Scientifically Accurate Pharmaceutical Knowledge Ontology based on Multi-source Data
171
3 RESULTS
3.1 Classes
After data cleansing and parsing, with the help of
experts, we extracted 13 classes from multiple data
source species. These classes are listed in Table 2.
With further analysis of the data sources, a total of 11
second-level semantic types (i.e., subclasses), were
identified in three classes: Targets, Biomolecules and
Organism. Finally, the classes and subclasses were
built in Protégé, shown as Figure 2.
3.2 Object Properties and Data
Properties
Using analysis of data sources, and verification by
experts, 11 object properties and 5 subproperties, as
well as 39 data properties and 43 subproperties were
determined. The detailed object properties and data
properties are listed in Tables 3 and 4, respectively.
Object properties are used to describe the
relationships between pharmaceutical knowledge
entities, each of which defines the subject and scope
of application, as shown in Table 3. Drugs, for
example, includes the following relationships: drug-
literature-citation, drug-clinical trial-effective effect,
drug-indication-active effect, drug-drug-interaction,
drug-drug-active ingredient, drug-target-effective
effect, drug-biomolecular-benign/adverse effect. In
addition, each entity includes the same basic
properties: hasName, hasDescription, hasEntityClass,
hasSynonyms, hasSource, and hasID.
3.3 Model of the Pharmaceutical
Knowledge Ontology
As with the classes, object properties and data
properties were determined. The data level, scope,
type and definition of each type of entity were
formulated to achieve a structured, standardized and
normalized description of pharmaceutical entities.
The ontology model of pharmaceutical knowledge
based on Protégé is shown in Figure 3. Relationships
between classes are represented by lines with arrows.
At this point, the basic architecture of the
ontology is complete, and entities can be added into
the ontology as individuals in Protégé manually, or by
using data association between multi-source data and
ontology model by means of technology.
Table 3: Object properties and subproperties defining the relationships between pharmaceutical entities.
No. Object property Subproperty Domains Ranges
1 isAIRelationOn
2 isCitationOf
Patent
Reference
Biomolecule
ClinicalTrial
Drug
InactiveIngredient
Indication
Product
Targets
3 isClinicalTrialOn
ClinicalTrial
Drug
Indication
Product
Therap
y
4 isDrugActionOn
Drug
Product
Indication
Thera
py
5 isIndicationLocationIn
Indication Organ
6 isInteractionWith
Drug
Biomolecule
Drug
7 isProductIngredientOf
isProductActiveIngredientOf Drug Product
isProductInactiveIngredientOf Inactive Ingredient Product
8
isProductAdverseReaction
On
Product
ClinicalTrial
Indication
Therap
y
9 isProductReferenceOf
Product Product
10 isTherapyOn
Therapy Indication
11 isTargetActionsOn
isTargetActionOn Drug Targets
isTargetOrganismOf Drug Organ
isTargetPharmacologicalActionOn Drug Targets
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
172
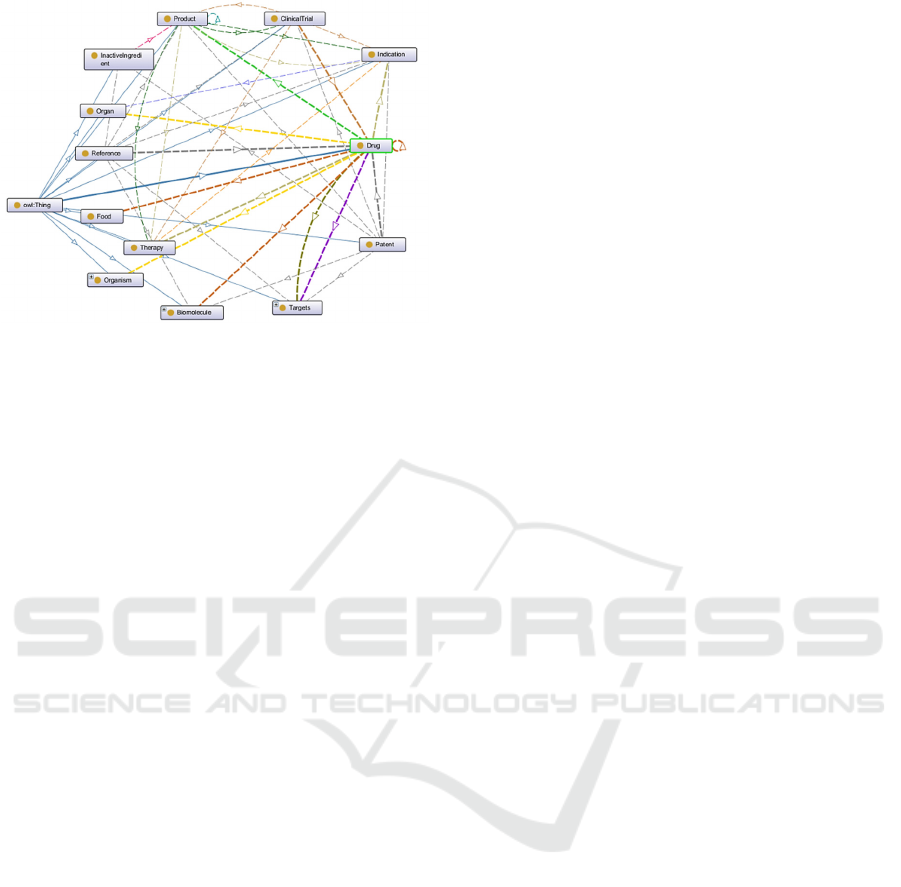
Figure 3: Model of the pharmaceutical knowledge ontology.
4 DISCUSSION
Pharmaceutical knowledge ontology helps describe
and organize pharmaceutical information, and forms
a network of pharmaceutical knowledge about
pharmaceutical terms and the relationships between
them. Combined with computer technology,
pharmaceutical-related data can be shared and
exchanged in the network. Through the
standardization of terms in pharmaceutical
knowledge ontology, metadata from different data
sets can be unified to eliminate heterogeneity and
realize the integration of pharmaceutical data. At the
same time, through the relationship between the
standardized terms in the ontology, metadata in the
data set can also construct the semantic association
and realize the index of the metadata content, in order
to achieve deeper level conformity, annotation,
analysis and mining of the original data.
Knowledge base is a structured, operable and
organized cluster in knowledge engineering. With the
common demand of solving problems in a particular
domain, it is a set of interrelated knowledge slices that
are stored, organized, managed, and used in computer
memory in a certain knowledge representation
manner.
Ontology provides a basic architecture for the
establishment of knowledge base. It describes the
domain with a set of concepts and terms, and obtains
the essential conceptual structure of the domain.
Ontology constitutes the core of the domain
knowledge representation system. The knowledge
base uses these terms to represent information.
Ontology-based knowledge base can help users to
acquire knowledge most suitable to their needs
through these relationships and properties, thus
avoiding irrelevant information during knowledge
acquisition. Pharmaceutical knowledge ontology can
help realize the standardized description and
structured organization of pharmaceutical knowledge
and information, promote efficient use of
pharmaceutical data, and lay a foundation for
knowledge graph.
In building this ontology model, we have reached
a milestone. However, there are still much more to do
in the future. To reach the goal of knowledge graph,
further than the ontology model, we need to optimize
the data model and realize the data mapping from
ontology to relational data and graph database.
5 CONCLUSIONS
Based on the basic concepts and knowledge system
of pharmacy, using the idea of ontology modelling,
and referring to existing pharmaceutical, biological
and medical knowledge ontology models and
resource databases, we sort out the concept, scope,
classification, hierarchy and structure of
pharmaceutical knowledge ontology. We have built a
basic ontology model, which can be gradually
integrated and updated by analysing and sorting the
data content of authoritative global data sources.
Finally, a relatively complete pharmaceutical
knowledge ontology model was established to
complete the knowledge representation of drug
information. With this tool, we can not only show
pharmaceutical entities and their relationships, and
systematically describe pharmaceutical knowledge,
but also can integrate and supplement existing
biomedical ontologies, in order to further realize the
standardization, normalization and structurization of
pharmaceutical data in the form of knowledge graph.
ACKNOWLEDGEMENTS
This work was conducted using Protégé, which is
supported by grant GM10331601 from the National
Institute of General Medical Sciences of the United
States National Institutes of Health.
A Comprehensive and Scientifically Accurate Pharmaceutical Knowledge Ontology based on Multi-source Data
173

Table 4: Data properties and subproperties defining the basic attributes of pharmaceutical entities.
No. Data property Subproperty
1 hasApproved
2 hasCategory
3 hasClinicalTrial
hasClinicalTrialDrug
hasClinicalTrialIndication
hasClinicalTrialStatus
hasClinicalTrialThera
py
4 hasCountryName
5 hasDates
6 hasDeleted
7 hasDescription
8 hasDescription
hasDetailDescription
hasSummar
y
9 hasDrugChemicalIdentifier
hasDrugCas
hasDrugInChI
hasDrugInChIKey
hasDrugIupacName
hasDru
g
Unii
10 hasDrugPharmacology
11 hasDrugDissolution
12 hasDrugProperty
13 hasDrugSpectra
14 hasEntityClass
15 hasFunctions
16 hasGene
17 hasNames
hasGenericName
hasName
hasNonProprietaryName
hasPreferredName
hasS
y
non
y
ms
18 hasID
19 hasIndicationDiagnosis
hasIndicationCause
hasIndicationCheck
hasIndicationChiefComplaint
hasIndicationDiagnosis
hasIndicationDiagnosisBasis
hasIndicationS
y
m
p
to
m
20 hasIndicationHospitalDepartment
21 hasIndicationPathway
22 hasIndicationSite
23 hasLink
24 hasFunctions
hasMainFunction
hasSpecificFunction
25 hasMedicalInsuranced
26 hasOrganisationName
27 hasOriginalID
28 hasProductAdverseReaction
29 hasProductCompany
hasProductApplicantHolde
r
hasProductDistributor
hasProductLabeller
hasProductManufacturer
hasProductPacka
g
e
r
30 hasProductBrand
31 hasProductCompany
32 hasProductInfo
hasProductDosage
hasProductRoute
hasProductStrength
33 hasProductInstruction hasProductSpecification
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
174

Table 4: Data properties and subproperties defining the basic attributes of pharmaceutical entities (cont.).
No. Data property Subproperty
34 hasTitle
hasPublicTitle
hasScientificTitle
35 hasReferenceInfo
hasReferenceDoi
hasReferenceExternalDatabaseID
hasReferenceFile
hasReferenceLink
hasReferencePatentID
hasReferencePmi
d
36 hasRegistered
37 hasSource
38 hasDates
hasStartDate
hasU
p
dateTime
39 hasType
REFERENCES
Wouters, O. J., McKee, M., & Luyten, J. (2020). Estimated
Research and Development Investment Needed to
Bring a New Medicine to Market, 2009-
2018. JAMA, 323(9), 844–853.
Hanna, J., Joseph, E., Brochhausen, M., & Hogan, W. R.
(2013). Building a drug ontology based on RxNorm and
other sources. Journal of biomedical semantics, 4(1),
44.
Hur, J., Özgür, A., & He, Y. (2018). Ontology-based
literature mining and class effect analysis of adverse
drug reactions associated with neuropathy-inducing
drugs. Journal of biomedical semantics, 9(1), 17.
Cai, M. C., Xu, Q., Pan, Y. J., Pan, W., Ji, N., Li, Y. B., Jin,
H. J., Liu, K., & Ji, Z. L. (2015). ADReCS: an ontology
database for aiding standardization and hierarchical
classification of adverse drug reaction terms. Nucleic
acids research, 43(Database issue), D907–D913.
Podchiyska, T., Hernandez, P., Ferris, T., Weber, S., &
Lowe, H. J. (2010). Managing Medical Vocabulary
Updates in a Clinical Data Warehouse: An RxNorm
Case Study. AMIA ... Annual Symposium proceedings.
AMIA Symposium, 2010, 477–481.
Nelson, S. J., Zeng, K., Kilbourne, J., Powell, T., & Moore,
R. (2011). Normalized names for clinical drugs:
RxNorm at 6 years. Journal of the American Medical
Informatics Association: JAMIA, 18(4), 441–448.
Bodenreider, O., Cornet, R., & Vreeman, D. J. (2018).
Recent Developments in Clinical Terminologies -
SNOMED CT, LOINC, and RxNorm. Yearbook of
medical informatics, 27(1), 129–139.
Bona, J. P., Brochhausen, M., & Hogan, W. R. (2019).
Enhancing the drug ontology with semantically-rich
representations of National Drug Codes and RxNorm
unique concept identifiers. BMC bioinformatics, 20
(Suppl 21), 708.
Nikiema, J. N., Liang, M. Q., Després, P., & Motulsky, A.
(2021). OCRx: Canadian Drug Ontology. Studies in
health technology and informatics, 281, 367–371.
Sharp M. E. (2017). Toward a comprehensive drug
ontology: extraction of drug-indication relations from
diverse information sources. Journal of biomedical
semantics, 8(1), 2.
Musen, M. A., & Protégé Team (2015). The Protégé
Project: A Look Back and a Look Forward. AI
matters, 1(4), 4–12.
Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S.,
Hassanali, M., Stothard, P., Chang, Z., & Woolsey, J.
(2006). DrugBank: a comprehensive resource for in
silico drug discovery and exploration. Nucleic acids
research, 34(Database issue), D668–D672.
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J.,
Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li, C.,
Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu, Y.,
Maciejewski, A., Gale, N., Wilson, A., Chin, L.,
Cummings, R., Le, D., Pon, A., … Wilson, M. (2018).
DrugBank 5.0: a major update to the DrugBank
database for 2018. Nucleic acids research, 46(D1),
D1074–D1082.
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S.,
Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B.,
Zaslavsky, L., Zhang, J., & Bolton, E. E. (2019).
PubChem in 2021: new data content and improved web
interfaces. Nucleic Acids Res., 49(D1), D1388–D1395.
A Comprehensive and Scientifically Accurate Pharmaceutical Knowledge Ontology based on Multi-source Data
175
