
Survival Status Prediction for Non-small Cell Lung Cancer Patients
using Machine Learning
Aishwarya Mohan and Aleksandar Jeremic
Department of Electrical and Computer Engineering McMaster University, Hamilton, ON, Canada
Keywords:
Survival Prediction, Logistic Regression, Machine Learning.
Abstract:
Lung cancer is the leading cause among cancer-related deaths worldwide. Clinically, it could be divided
into several groups: 1) the non-small cell lung cancer (NSCLC, 83.4%), 2) the small cell lung cancer
(SCLC,13.3%), 3) not otherwise specified lung cancer (NOS,3.1%), 4) aarcoma lung carcinoma (0.2%), and
5) other specified carcinoma (0.1%). According to SEER Cancer Statistics Review, 5-year survival rate of
patients with advanced non-small cell lung cancer (NSCLC) who received chemotherapy was less than 5%.
Our ability to provide survival status at any time in future is important from at least two standpoints: a) from
the clinical standpoint it enables clinicians to provide optimal delivery of healthcare and b) from a personal
standpoint, by providing patient’s family with opportunities to plan their life ahead and potentially cope with
emotional aspect of loss of life. In this paper we propose to utilize machine learning techniques to achieve
this goal and evaluate several techniques in order to determine their prediction performance using publicly
available dataset.
1 INTRODUCTION
According to American Cancer Society, lung cancer
is the leading cause of cancer death among men and
women, for almost 25% of all cancer deaths. Since
the mortality rate of lung cancer is high, it belongs to a
group that has the worst survival prognosis (Matuzzi,
2019). Generally, after diagnosis the patient’s family
expects to know the patient’s chances of survival from
a clinician. An ability to predict life expectancy can
be beneficial from both emotional standpoint and clin-
ical standpoint, as it reduces stress on patient’s family
and enables them to cope with situation. It can also al-
low clinicians to evaluate patients’ risk, likelihood of
survival and postoperative treatment procedures. Due
to the very nature of the disease, lung cancer datasets
are generally imbalanced where majority of patient
population has low chances of survival. As a result,
predictive modelling on imbalanced datasets where
the majority of patients have low chances of survival
(Liang, 2017) makes it more challenging to accurately
predict survival status of patients with higher chances
of survival. Thus, for clinicians to accurately evalu-
ate patients’ risk and further design appropriate post
treatment procedures it is equally important to accu-
rately predict both true negatives and true positives.
Increasing the number diagnostic lab tests indi-
cates a potential of vast biomedical data assuming
there are plenty of electronic health records of pa-
tients. As a result, rapid increase in volume and com-
plexity of biomedical data can be utilized to draw pat-
terns and inferences. One of the promising techniques
that can be helpful in finding patterns from a large
patient cohort data is predictive modelling which uti-
lizes biomedical data to investigate relationships be-
tween the factors and the dependencies that further
help us predict survival. Ultimately, this can help pa-
tients with personalized medication and risk assess-
ment. Developing algorithms and mathematical mod-
els that can generate reliable predictions on an imbal-
anced dataset is a daunting task because of the under-
lying dependencies and bias which can be complex.
As a result, number of factors influencing the predic-
tions are huge. To implement this technique in medi-
cal practice we need rigorous training procedures for
complexities. Even in this case, the underlying as-
sumption of these techniques is that certain statisti-
cal/probabilistic models can describe these dependen-
cies which may not be true in certain cases (i.e., there
may exist certain number of outliers in every dataset).
In addition, we need to design vigorous testing, val-
idation, and verification procedures because of over-
whelming intricacies such as variability from patient-
to -patient that needs to be evaluated.
Mohan, A. and Jeremic, A.
Survival Status Prediction for Non-small Cell Lung Cancer Patients using Machine Learning.
DOI: 10.5220/0010916000003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 4: BIOSIGNALS, pages 273-277
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
273

Unequal distribution of data between majority
class i.e. patients that are less likely to survive and mi-
nority class i.e. patients that are likely to survive can
induce bias towards majority class, leaving minority
class samples to be often misclassified. Misclassifica-
tion of minority class can lead to hectic postoperative
treatment procedures, high dosage of recommended
drugs and accelerated health follow-ups and diagnos-
tic tests which can cause stress both physically and
psychologically. An ability to predict survival status
of patient at a given time by clinician can alleviate
this stress. Hence, to use machine learning models
in clinical practice they should be designed in such
a way that they are robust towards bias induced by
majority class. These models can also be used as a
risk assessment tool to help us determine which pa-
tients should be offered imaging. However, all these
tools suffer from aforementioned common challenge
of bias towards majority class. Furthermore, they are
also dynamic in nature and need to be updated con-
tinuously as the environment changes. Hence, model
should be constructed and designed in such a way that
it can adjust if there are changes in the subset of the
population.
In this paper, we investigate different approaches
for predicting survival status of patients suffering
from non-small cell lung cancer. In Section 2 we
present signal model, i.e. different classifiers on
which our analysis will be performed and later in the
paper we list evaluation metrics for measuring perfor-
mance. In addition we define a fusion algorithm that
can be used to combine decisions of different machine
learning algorithms. In Section 3, related dataset and
results from different tests performed on training data
will be discussed. In Section 4 we conclude our find-
ings for this study and present suggestions for future
work.
2 SIGNAL MODELS
2.1 Data Set
The dataset used for evaluation of the proposed model
is from MAASTRO Clinic, (Maastricht, The Nether-
lands). This dataset is open source and can be found
at TCIA (The cancer imaging archive) under NSCLC
(Aerts, 2019). Four hundred and twenty-two con-
secutive patients were included (132 women and 290
men), with inoperable, histologic or cytologic con-
ferred NSCLC, UICC stages I-IIIb, treated with rad-
ical radiotherapy alone (n = 196) or with chemora-
diation (n = 226). Mean age was 67.5 years (range:
33–91 years). The study has been approved by the
institutional review board. All research was carried
out in accordance with Dutch law. The Institutional
Review Board of the Maastricht University Medical
Centre (MUMC+) waved review due to the retrospec-
tive nature of this study. Out of 422 records, we have
only 365 patients with all the information. The sur-
vival time (in days) in the dataset is from the start of
the treatment and there is a possibility that the sta-
tus of patient recorded may not be accurate i.e. the
clinicians may not have received the information right
when the event outcome occurred.
2.2 Machine Learning Models
Training a model that predicts the survival status at
a given time, means forecasting the odds of outcome
instead of forecasting the point estimate of the occur-
rence. In our case there are two disease outcomes i.e.
alive and dead, defined so that if the result of odds are
greater than 50% then the predicted class is assigned
value 1 (alive) otherwise it is 0 (dead). We investi-
gate applicability of several models: gradient boost-
ing, XGboost and random forrest. The main difficulty
in this particular application are the unbalanced data
sets since the number of patients surviving the lung
cancer after certain period of time is relatively small.
To this purpose we propose to fuse the the proposed
machine learning algorithms using our information
fusion algorithm proposed in (Liu et al., 2007).
2.3 Gradient Boosting
Boosting is defined as a strategy that involves combi-
nation of multiple simple models resulting in an over-
all stronger model. The simple models are called as
weak learners. For example, the flow chart in Figure
1 below explains the gradient boosting method for N
trees. Tree 1 is trained using a feature matrix X and
target variable y. The predictions labelled ˆy
1
are used
to determine the training set loss function r
1
. Tree2
is then trained using the feature matrix X and the loss
function r
1
of Tree1 as labels. The predicted results
hatr
1
are then used to determine the loss function r
2
.
The process is repeated until all the N trees forming
the ensemble are trained.
In other words, instead of fitting a model on the
data at each iteration, it fits a new model to the resid-
ual errors made by the previous model. The details
of gradient boosting method are outlined in (Ke et al.,
2017).
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
274

Figure 1: Gradient Boosting algorithm scheme.
2.4 XGBoost
XGBoost stands for extreme gradient boosting as it
uses second-order Taylor expansion of the loss func-
tion to iterate and calculate weights at leaf nodes of
the new tree K (Zhao, 2020). Additionally, a regular-
ization term is added to the loss function to control the
complexity of the model and prevent it from overfit-
ting. Therefore, XGBoost performs better in training
efficiency, massive parallelism, and quadratic conver-
gence (Zhao, 2020).
It can perform well on imbalanced datasets as it
calculates the second order gradients i.e., second par-
tial derivativesof loss function ultimately giving more
information about the direction of gradients and min-
imizes loss function.
2.5 Random Forrest
In addition to the aforementioned models, we investi-
gate applicability of the ensemble methods that utilize
machine learning methods using different learning al-
gorithms. To this purpose we select decision tree ap-
proach and utilize commonly used Random Forrest
(Dai et al., 2018) technique which uses bagging and
feature randomness when building each tree creating
an uncorrelated forest of trees which makes decision
by aggregating the votes from different trees. To illus-
trate the performance of this algorithm in Fig. 2-4 we
illustrate the tree growth for our dataset. Due to ran-
dom feature selection, the trees are more independent
of each other as compared to regular bagging, which
often results in better predictive performance.
2.6 Fusion Algorithm
Each of the aforementioned classifiers can be treated
as a single channel detector making a decision in a bi-
nary classification problem. In order to improve their
overall performance we propose to combine their
classifications using the distributed system illustrated
in Figure 5.
Figure 2: First decision tree.
Figure 3: Fourth decision tree.
The global decision in the fusion centre is then
made by minimizing the overall probability of er-
ror/misclassification.
P
e
= P(H
0
)P(u
0
= 1|H
0
) + P(H
1
)P(u
0
= 0|H
1
)
The optimality criterion for N is given by (Varshney,
1986).
u
0
=
(
1, if w
0
+
∑
3
n=1
w
n
> 0
0, otherwise
(1)
where, w
0
= log
P
1
P
0
(2)
and w
n
=
(
log((1− P
m
n
)/P
f
n
), if u
n
= 1
log(P
m
n
/(1− P
f
n
)), if u
n
= 0
(3)
The probabilities of false alarm and missed detec-
tion of the nth local detector are denoted as P
f
n
and
P
m
n
, respectively. Note that in (Mirjalily, 2003) the
authors presented analytical solution for the above
problem in the case of binary classification. Note
that in a particular setting if the data size is limited
Figure 4: Fourth Decision Tree.
Survival Status Prediction for Non-small Cell Lung Cancer Patients using Machine Learning
275
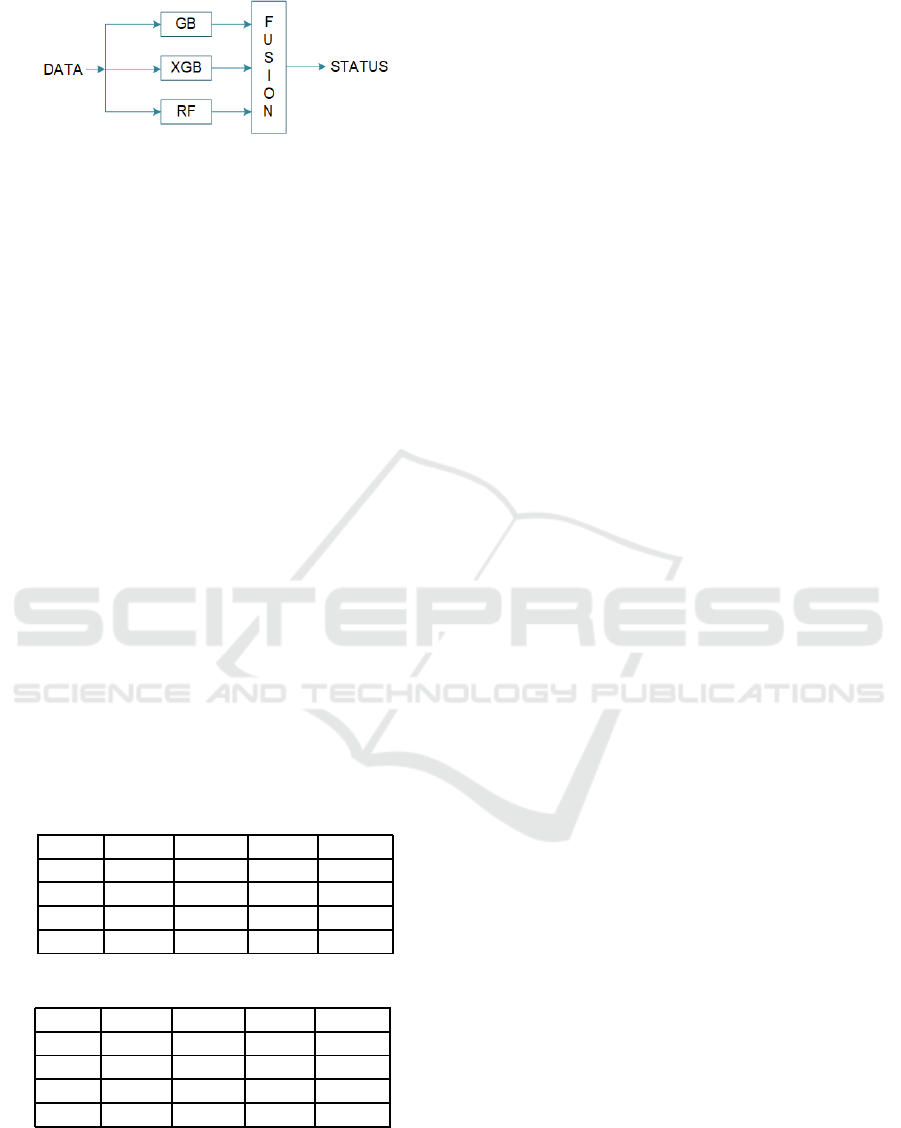
Figure 5: Classification Fusion System.
and/or the number of events needed for accurate cal-
culation of anomalies is not sufficient we developed
a maximum likelihood based algorithm that exploits
the multinomial probability mass function describing
the decision vector and utilized in order to estimate
the anomalies as well as prior probabilities (seizure
and no-seizure). We presented the details of these al-
gorithms in (Liu et al., 2014).
3 RESULTS
To evaluate the performance of the proposed algo-
rithm we plan to use several commonly used perfor-
mance metrics F1-score and recall as most of the lung
cancer datasets are imbalanced due to the nature of
the disease. Recall is defined as a ratio of true posi-
tives and summation of true positives and false neg-
atives and F1-score is defined as a harmonic mean
of the precision and recall.. In Table 1 we illustrate
the performance results for 50-50 split in which only
50% of the data was used for training. The results in-
clude both average value and variance since the per-
formance of machine learning algorithms is heavily
dependent on the training dataset. In Table 2 we illus-
trate similar results but for training ratio split 90-10.
Table 1.
av. R. av. F1 var R var F1
GB 79% 77% 0.7% 3%
XGB 73% 67% 0.6% 2.1%
RF 82% 63% 0.9% 0.8%
Fus, 86% 82% 0.8%. 0.6%
Table 2.
av. R. av. F1 var R var F1
GB 80% 83% 0.9% 1.9%
XGB 79% 80% 1.1% 3.1%
RF 88% 84% 1.2% 0.9%
Fus, 92% 89% 1.1%. 0.6%
4 CONCLUSIONS
In this paper we demonstrated applicability of sev-
eral machine learning models in order to determine
the life status of lung cancer patients after certain pe-
riod of time. Due to the limited nature of the dataset
available fully temporal model was not developed as
it would require larger data set in order to evaluate
performance dependence on the time passed. Our pre-
liminary results indicate that significant accuracy can
be achieved assuming that all the relevant parameters
are measured/monitored and available which further
emphasizes the need for standardized data manage-
ment. Due to the fact that the performanceof machine
learning models is heavily dependent on data set, an
effort should be made in order to investigate perfor-
mance of the proposed techniques, especially fusion,
on larger data sets. Given a sufficiently large data set,
we would be able to compare the performance of our
fusion model to an unsupervised model in which the
prediction results would be fused using another layer
of machine learning models.
Furthermore, the proposed techniques can be ex-
tended to create soft decision algorithms in which out-
comes would be given with certain probabilisticconfi-
dence. However to achieve this goal, which would in-
clude temporal dependence, an effort should be made
to obtain a database in which sufficient status infor-
mation exists for variety of patients and sufficiently
large temporal points. The main advantage of this ap-
proach would be to provide life expectancy estimate
in addition to survival probability at a particular time.
REFERENCES
Dai, B., Chen, R.-C., Zhu, S.-Z., and Zhang, W.-W. (2018).
Using random forest algorithm for breast cancer diag-
nosis. In 2018 International Symposium on Computer,
Consumer and Control (IS3C), pages 449–452.
Ke, G., Meng, Q., Finley, T., Wang, T., Chen, W., Ma,
W., Ye, Q., and Liu, T.-Y. (2017). Lightgbm: A
highly efficient gradient boosting decision tree. In
Guyon, I., Luxburg, U. V., Bengio, S., Wallach, H.,
Fergus, R., Vishwanathan, S., and Garnett, R., editors,
Advances in Neural Information Processing Systems,
volume 30. Curran Associates, Inc.
Liang, H. (2017). Evaluation and accurate diagnoses of
pediatric diseases using artificial intelligence. Nature
Medicine, 9(4):433–438.
Liu, B., Jeremic, A., and Wong, K. (2007). Blind adaptive
algorithm for M-ary distributed detection. In IEEE In-
ternational Conference on Acoustics, Speech and Sig-
nal Processing, 2007. ICASSP 2007, volume 2.
Liu, B., Jeremic, A., and Wong, K. (2014). Optimal dis-
tributed detection of multiple hypotheses using blind
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
276

algorithm. IEEE Trand. on Aerospace and Electronic
Systems, 50:1190–1203.
Matuzzi, C. (2019). Current cancer epidemiology. Journal
of Epidemiology and Global Health, 9(4):217–222.
Mirjalily, G. e. (2003). Blind adaptive decision fusion
for distributed detection. IEEE Transactions on
Aerospace and Electronic Systems, 39(1):34–52.
Varshney, P. (1986). Optimal data fusion in multiple sen-
sor detection systems. IEEE Trans. on Aerospace and
Electronic Systems, 40:98–101.
Zhao, W. (2020). Fast intelligent cell phenotyping for high-
throughput optofluidic time-stretch microscopy based
on the xgboost algorithm. Journal of biomedical op-
tics, 25(6):1–12.
Survival Status Prediction for Non-small Cell Lung Cancer Patients using Machine Learning
277
