
A New Neural Network Model for Prediction Next Stage of Alzheimer’s
Disease
Nour Zawawi
1,3
, Heba Gamal Saber
2
, Mohamed Hashem
1
and Tarek F. Gharib
1
1
Ain Shams University, Faculty of Computer and Information Science, Cairo, Egypt
2
Ain Shams University, Faculty of Medicine, Geriatric Department, Cairo, Egypt
3
October University for Modern Sciences and Arts, Faculty of Computer Science, 6th October City, Egypt
Keywords:
Feature Selection, Time-series Forecasting, LSTM.
Abstract:
Alzheimer’s disease (AD) is a brain-related illness; The risk of development is minimized when diagnosed
early. The early detection and treatment of Alzheimer’s disease are crucial since they can decrease disease
progression, improve symptom management, allow patients to receive timely guidance and support, and save
money on healthcare. Regrettably, much current research focuses on characterizing illness states in their
current phases rather than forecasting disease development. Because Alzheimer’s disease generally progresses
in phases over time, we believe that analyzing time-sequential data can help with disease prediction. Long
short-term memory (LSTM) is a recurrent neural network that links previous input to the current task. A new
Alzheimer’s Disease Random Forest (RF) LSTM Prediction Model (RFLSTM-PM) is proposed to capture the
conditions between characteristics and the next stage of Alzheimer’s Disease after noticing that a patient’s data
could be beneficial in predicting disease progression. Experiments reveal that our approach beats most existing
models and can help with early-onset AD prediction. Furthermore, tests show that it can recognize disease-
related brain regions across multiple data modalities (Magnetic resonance imaging (MRI), Neurological Test).
Also, it showed decreased value in Mean Absolute Error and Root Mean Square Error for forecasting the
progression of the disease.
1 INTRODUCTION
Alzheimer’s Disease (AD) is a form of dementia
that impacts humans’ memory, daily activities’ per-
formance, and communication abilities(Association,
2019). Effective early diagnosis and treatment of AD
is of fundamental importance as it can reduce dis-
ease progression, and therefore, reduce the substan-
tial cost for health care. Recent research shows that
only 20–40% of individual cases will change to AD
within three years; This is a lower rate of exchange
reported in medical samples than in clinical cases (Ve-
muri et al., 2017). However, AD’s progression starts
several years before any symptoms become visible
and progressive (Association, 2019). Thus, identify-
ing high-risk patients who will convert to AD is sub-
stantial (Association, 2019; Edwards III et al., 2019).
As a result, early diagnosis is essential for making a
treatment strategy to slow down the progress. It is
where the disease altered from one symptom to an-
other. At the same time, current research mainly fo-
cuses on predicting the possibility that it converts into
another stage.
In recent years, the growth of neurodegenerative
disorders such as AD has gained much interest from
researchers worldwide to develop high-performing
methods for diagnosis, treatment, preventive thera-
pies, and target drug discovery. In the risk assessment
of conversion from MCI, these variables’ change rate
could represent an additional source of knowledge
(Perneczky, 2018). Researches made it possible to di-
agnose AD using advanced diagnostic tools and com-
bine markers from different clinical features (Davda
and Corkill, 2020; Tanveer et al., 2020). Combining
markers from different clinical features and investi-
gation modalities has been shown to maximize their
sensitivity and specificity in clinical use compared to
individual biomarkers. Unfortunately, current stud-
ies mainly focus on classifying disease states in their
current stage using MRI instead of combining multi-
ple features. As a result, these studies serve as proof
of concept without being tested in the real world.
Machine learning models can help to forecast the
disease progression(Yang et al., 2018; El-Sappagh
Zawawi, N., Saber, H., Hashem, M. and Gharib, T.
A New Neural Network Model for Prediction Next Stage of Alzheimer’s Disease.
DOI: 10.5220/0010892400003122
In Proceedings of the 11th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2022), pages 689-696
ISBN: 978-989-758-549-4; ISSN: 2184-4313
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
689

et al., 2020) accurately. Time-series forecasting mod-
els based on recurrent memory-based approaches are
used to examine extracting patterns from sequential
healthcare data and classifying data based on diag-
nostic categories. A variety of settings in healthcare
uses predictions(Piccialli et al., 2021). They ranged
from predicting future medical outcomes and diag-
nosis to univariate time-series predictions of monthly
expenditures of patients for medication. We can find
that it has lots of applications in the healthcare in-
dustry. Time series forecasting has been a grow-
ing science subject due to its utility in real-world
applications, but technique development has been a
concern. In medical applications, time series fore-
casting models predict sickness progression, estimate
death rates, and assess time-dependent risk(Bui et al.,
2018). However, the large number of various method-
ologies available, each of which thrives in different
scenarios, makes selecting an effective model more
complicated.
This paper addressed several of the mentioned
issues for generating time-series predictions of AD
data. On the other hand, studies show that devel-
opments linked with Alzheimer’s disease can begin
more than 20 years before symptoms arise. The pro-
posed model can identify the stage of transforma-
tion throughout time. In contrast to earlier research,
a model using LSTM (Hong et al., 2019; Basher
et al., 2021) presents to predict the progression of
Alzheimer’s disease. It employs the time step data
acquired by a data preprocessing pipeline because the
time series data may alter the prediction. It predict
time progression of AD based on these data. The pa-
per is organized as follows: Related work and previ-
ous work is discussed in section 2. Section 3 illustrate
the proposed data and methods. Section 4 discusses
the proposed prediction model (RFLSTM-PM). Fi-
nally, experiments where the models sensitivity was
tested to different features and test its stability in var-
ious data sizes shown in section 5
2 RELATED WORK
Most of the studies on Alzheimer’s disease have fo-
cused on using medical imaging as the only fac-
tor. (Mart
´
ı-Juan et al., 2020) is a survey concen-
trating on longitudinal imaging data. It focused on
papers that have been published between 2007 and
2019. (Hong et al., 2019) introduce Long short-
term memory (LSTM) to predict the development of
AD. It carries out the future state prediction for the
disease, rather than the state of a current diagnosis.
While (Janghel, 2020) develops and compares dif-
ferent methods to diagnose and predict AD by using
MRI scans only. It implements one model which is
the convolution neural network (CNN). At the same
time, it uses four different architectures of CNN.
An embedded feature selection method based on the
least-squares loss function and within-class scatter for
selecting the optimal feature subset are proposed by
(Cai et al., 2020). The optimal subsets of features
used for binary classification are based on a support
vector machine (SVM). Also, deep learning technol-
ogy was discussed by (Bi et al., 2019). It focused on
the problem of automatic prediction of AD based on
MRI images. It applies two main steps: 1-implement
the unsupervised CNN for feature extraction. 2- uti-
lizes the unsupervised predictor to achieve the final
diagnosis.
According to our knowledge, (Grassi et al., 2019)
and (Liu et al., 2020) are the only work that em-
ploys more realistic and affordable data for diagnosis.
First, (Grassi et al., 2019) use a weighted rank aver-
age grouped by different supervised machine learning
methods to predict 3 years conversion. Only a limited
set of diverse characteristics are used to make pre-
dictions. The employment of algorithmic decision-
making tools is the key benefit. While,(Liu et al.,
2020) provides a new method for detecting AD based
on spectrogram characteristics collected from voice
data. This can assist families in better understand-
ing the progression of a patient’s sickness at an early
stage.
The following studies serve as the foundation
for our study. They are listed in ascending order.
The first, (Qiu et al., 2018), explains how MRI data
can improve the accuracy of diagnoses for the Mini-
Mental State Examination and logical memory tests.
It accesses model correctness via Multilayer Precep-
tor. The second, (Grassi et al., 2019), shows how
clinically translatable strategies for conversion can
be predicted. It also detects high-risk people who
are converted. Then, (Haaksma et al., 2018) address
the link between Alzheimer’s disease and its pre-
dictors. It included some Alzheimer’s disease cases
that have had at least one examination following di-
agnosis. To determine whether there are any latent
classes of Mini Mental State Examination and Clini-
cal Dementia Rating sum of boxes routes across time.
To find baseline predictors of class membership, re-
searchers utilised bias-corrected multinomial logistic
regression. A multimodal data (Shikalgar and Sona-
vane, 2020) classifier that employs a hybrid deep neu-
ral network classifier. It is based on a set of MRI pic-
tures as well as EEG inputs. The goal is to improve
the learning process by incorporating the weight com-
ponent of DNN into CNN. Then it explains how the
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
690

accuracy of hybrid classifiers is determined.
To find correlations between brain areas and
genes, use the appropriate correlation analysis ap-
proach at the conclusion. (Bi et al., 2020) was
proposed via a cluster evolutionary random forest
(CERF). It adds the concept of clustering evolution
to increase the random forest’s generalisation perfor-
mance. (Farouk and Rady, 2020) investigate the use
of unsupervised clustering methods for the early iden-
tification of Alzheimer’s disease. This research de-
veloped a two-stage technique for effectively predict-
ing Alzheimer’s Disease (AD)(Soliman et al., 2021).
Using an upgraded sparse autoencoder (SAE), an un-
supervised neural network, the initial stage includes
finding the optimal representation of the training data.
Based on the learned records and brain MRI scan, the
second stage involves employing a 3D-Convolutional
Neural Network (3D-CNN) to discern between health
and ill status.
3 METHODS AND
PREPROCESSING
3.1 Data Preparation
Data used in this article’s preparation is obtained
from the Alzheimer’s Disease Neuroimaging Initia-
tive (ADNI) database (adni.loni.usc.edu). The ADNI
was launched in 2003 as a public-private partner-
ship. ADNI’s primary goal is developing clinical,
imaging, genetic, and biochemical biomarkers for
the early detection and tracking of Alzheimer’s dis-
ease. It contains nine classes cognitive normal nor-
mal aging (NL), Mild Cognitive Impairment (MCI),
Alzheimer’s disease (AD), early mild cognitive im-
pairment (EMCI), and late mild cognitive impairment
(LMCI) to dementia or AD. The subjects recruited in
over 50 different US and Canada centers, with follow
up assessments performed every six months. The pro-
posed work uses only two types of data: 1)Magnetic
resonance imaging (MRI), 2)Assessment data. Due to
the numbers of records in each classes the only ones
that contains multiple data, only three classes is se-
lected(AD, MCI, and NL).
3.2 Subjects
Our prediction model was trained and tested on data
extracted from the ADNI; 18-month longitudinal tra-
jectories of 900 cases on each class (MCI, NL, AD).
It covers a total of 2700 instances and 90 attributes.
Each patient profile consisted of multiple data sources
(24 tests and 7 image files with personal records).
Data is classified as ordinal, continuous, or image.
Patient trajectories described the time evolution of all
variables in 3-month intervals. The following subsec-
tion describes data processing steps in more detail.
The subject inclusion criteria employed in this study
are: 1- Age ranges from 55 to 90; 2-Education lev-
els range from primary to graduate; 3-All colors and
ethnicities included. The proposed work uses differ-
ent types of data: 1-Neurological test (neuropsycholo-
gist), and 2-Brain image technology (MRI only). Per-
sonal Information is excluded from the study, where
the main objective of this work is to predict the next
stage of ad over 3-month interval values.
3.3 Data Preprocessing
ADNI data (per subject) was captured multiple times
over a maximum period of 120 months. The chal-
lenge for the prediction is that it encounters many
real-world data problems, such as the following three:
• Incomplete data: Some subjects lack physical ex-
amination data at specific time points. The num-
ber of values in a set is known, but the values
themselves are unknown, resulting in incomplete
data from missing data.
• Missing data: Some subjects lack several values
of data. As a result, they are deleting the missing
values exceeding 60%. Table 1 views the rest of
the data it shows the percentage of missing data.
Other missing data are replaced by mean or vari-
ance depending on the data type.
• Time-frequency data: The different aspects of the
subjects over specific time value. However, not all
the subjects had the same time-frequency. Some
of the subjects may have only one type of data
model.
3.4 Methods
Alzheimer’s disease is a progressive neurological ill-
ness that begins gradually and worsens with time.
A primary diagnostic system, comprised of a ran-
dom forest selection and multiple illness prediction
models, is constructed to prevent disease progression.
Different methods exhibit different changes as the
disease progresses and identify relevant components.
The methodology is applied to different inputs sepa-
rately to uncover relevant biomarkers for each type.
It is critical to identify the most significant disease-
related risk factors (Remeseiro and Bolon-Canedo,
2019; Feng et al., 2021). In recent years, most au-
thors have concentrated on hybrid approaches to fea-
ture selection (Hancer et al., 2020). Variable selection
A New Neural Network Model for Prediction Next Stage of Alzheimer’s Disease
691
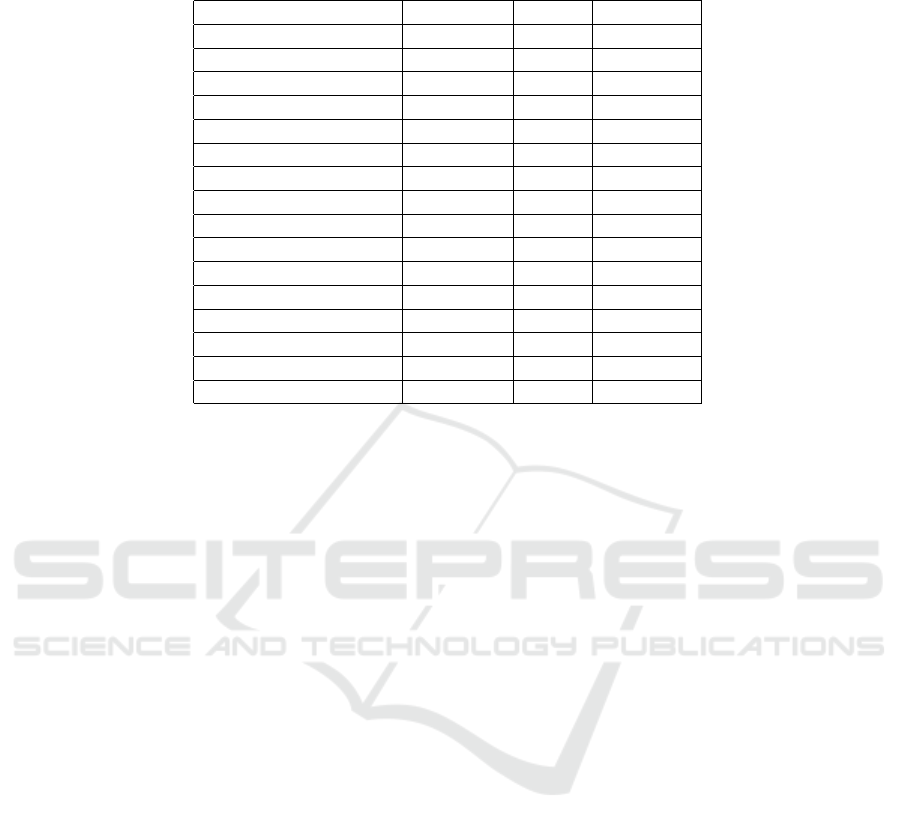
Table 1: The model contains cognitive function variables, as well as MRI images. The percentage of missing data for each
feature is indicated in the missing percentage column.
Name Type Mean Missing %
CDRSB continuous 2.425 1
MMSE continuous 25.862 0
ADAS11 continuous 13.009 0
ADAS13 continuous 19.939 2
RAVLT immediate continuous 31.223 1
RAVLT learning continuous 3.547 1
RAVLT forgetting continuous 4.067 1
RAVLT perc forgetting continuous 63.86 3
FAQ continuous 6.769 0
Ventricles image – 2
Hippocampus image – 2
WholeBrain image – 1
Entorhinal image – 2
Fusiform image – 2
MidTemp image – 2
ICV image – 1
approaches include filter methods, wrapper methods,
ensemble methods, and embedding methods.
Feature selection becomes more critical in data
sets with many variables and features. Random For-
est (RF) has proven to be a practical feature selection
approach, even when dealing with many variables.
It will eliminate insignificant variables and increase
classification accuracy and performance. The fact that
it is simple to calculate the relevance of each variable
on the tree decision contributes to its interpretabil-
ity(Hameed et al., 2015; Khaire and Dhanalakshmi,
2019). It falls under the area of embedded techniques.
Embedded methods combine filter and wrapper meth-
ods to implement algorithms with built-in feature se-
lection methods used. It indicates critical advantages
over other methodologies regarding handling highly
non-linearly correlated data, robustness to noise, tun-
ing simplicity, and opportunity for efficient parallel
processing (Dimitriadis et al., 2018). Moreover, It
presents another essential characteristic: an intrinsic
feature selection step, applied before the classifica-
tion task, to reduce the variables’ space by giving an
importance value to each feature (Zhong et al., 2021;
Helal et al., 2015).
LSTM networks are recurrent neural networks
that may learn order dependence in sequence predic-
tion challenges. They are a problematic area of deep
learning to master. It is required in various compli-
cated issue domains, including machine translation,
speech recognition, and others (Alom et al., 2019).
Developing and selecting accurate time series models
is a difficult task because it requires training several
different models and selecting the best among them.
At the same time, it needs extensive feature engineer-
ing to derive informative features and find optimal
time lags, which are commonly used input features
for time series models. For the processing of tempo-
ral data, LSTM models are prominent. The majority
of articles that utilize LSTM models do so with mod-
est variations. The following section describe it in
more details. Almost all of the gates in this model
contain the concept of peepholes.
Figure 1 shows the model preprocessing pipeline;
It uses two different data types. One is an MRI image,
while the second one is neurological test results. It
contains 24 data attributes (9 neurological tests) and 8
MRI that views the participant’s brain. The first stage
in the proposed model got the patients data as input
values. In this stage the missing values and outliers
data is dealt with. Second come the feature impor-
tance where the best feature is selected. To include all
this type of data costs a high amount of money and
processing power. As a result, the proposed model
chooses the best features that will increase perfor-
mance. The output of this is the attributes that best
descripe patients next progression time. Prediction
phase organize the data with specific time and each
feature with corresponding disease stage. Finally, the
future prediction of next test value appears in sequen-
tial data preprocess.
4 AD PREDICTION MODEL
The proposed model is a combination between LSTM
and RF. This paper proposed a new hybrid model
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
692
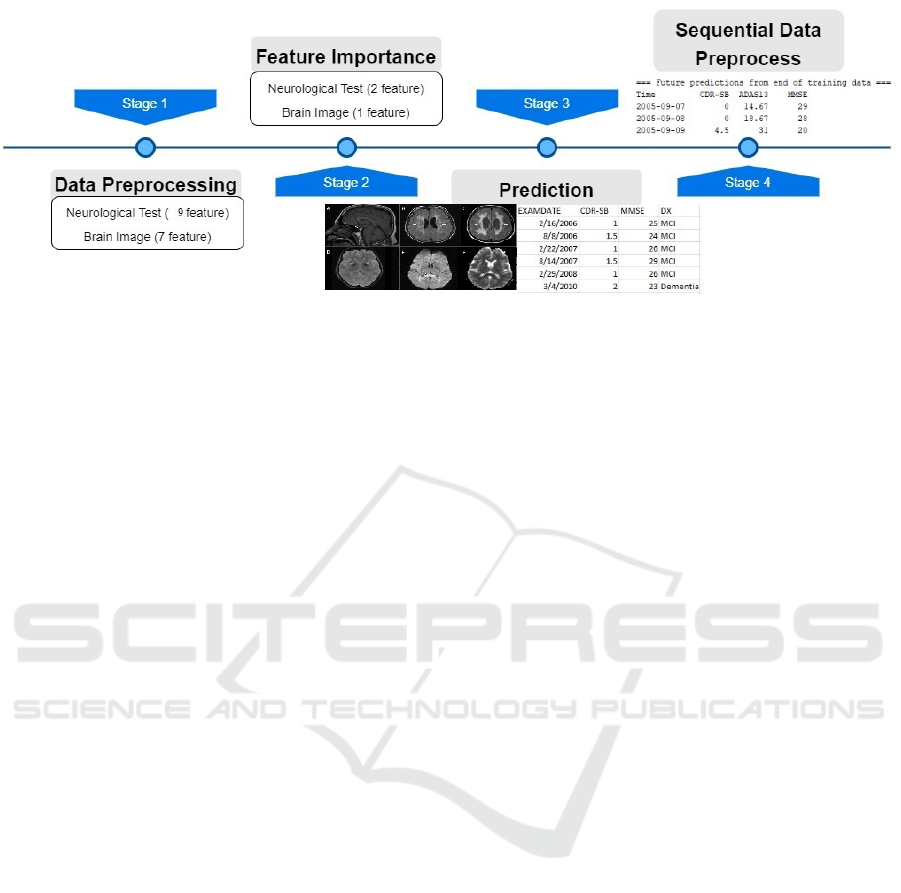
Figure 1: Propsed Model Pipeline.
called Random Forest Least Short Term Memory Pre-
diction Model(RFLSTM-PM). It is a recurrent neu-
ral network that links the previous state to the current
one. A network with fully connected and activation
layers is created to encode the temporal relationship
between features and the next stage of AD (Hochreiter
and Schmidhuber, 1997). It is supposed to avoid the
long-term dependency problem by using a sequence
of repeating neural network modules.
Figure 2 illustrates the proposed architecture. It
involves two layers: Fully Connected Layer and Out-
Fully Connected Layer. A fully connected layer is
used to find the correlation between these selected
features and Alzheimer’s Disease. It is connected
with the exponential linear unit (ELU) as the activa-
tion function predicts AD by feeding these features to
the layer. This model uses two types of data (MRI and
Neurological test), as stated earlier. The Out-Fully
Connected layer consists of a sigmoid layer fully con-
nected. It consist of four layers that communicate
with one another, such as:
• forget gate: The decision of whether the informa-
tion is thrown away from the cell state is made by
the forget gate, shown in Equation1
• input gate: Equation 2 and 3 show the input gate
that decides which values to update with sigmoid
and tanh layers.
• update gate: The update gate in Equation4. up-
dates the old cell state with the value from the in-
put gate.
• output gate: The output gate in Equation5 and 6
decides which value is to be output from the layer.
f
t
= σ(W
f
[h
t−1
, x
t
] + b
f
) (1)
where W
f
is the weight matrix, b
f
is the bias vector,
and f
t
is a number between 0 and 1, where 0 repre-
sents the forget and 1 represents the keep.
i
t
= σ(W
i
[h
t−1
, x
t
] + b
i
) (2)
ˇ
C
t
= tanh(W
c
[h
t−1
, x
t
] + +b
C
) (3)
where W
i
and W
c
are the weight matrices; b
i
and b
C
are the bias vectors; and i
t
,
ˇ
C
t
are the outputs of these
two equations.
C
t
= f
t
∗ C
t−1
+ i
t
∗
ˇ
C
t
(4)
where f
t
decides which information is to be forgotten,
and i
t
∗
ˇ
C
t
chooses the updated values for the cell.
i
o
= σ(W
o
[h
t−1
, x
t
] + b
o
) (5)
h
t
= o
t
∗ tanh(C
t
) (6)
where the value of i
o
in Equation 5 decides which part
of the cell state will be the output. The new cell state
C
t
multiplied by o
t
, and function tanh have selected,
to obtain h
t
in Equation 6, which is the output of the
parts t
o
The proposed work based on one assumption is
that each time-dependent variable in a patient’s clini-
cal record is stochastic. It is effective at differentiat-
ing between different aspects of data. RFLSTM-PM
represents the underlying time-dependent probability
distribution of value. It sampled from a range of val-
ues rather than taking on a single deterministic value.
During model training, preprocess sequential data
with time steps feds into the model, and the model
predicts the status of the following year. The model
predicts the stage of the 18-th month, one year fol-
lowing the final month, as AD, MCI, or NI. During
model testing patient’s 18th and 24-th month features
data are entered into the model, the output is a forecast
of his state in the next year. The model selects MMSE
and CDR-SB from the test, the Ventricles view from
the MRI image. Mean Absolute Error , Absolute Per-
centage Error, and Root Mean Square Error are three
of the most critical evaluation metrics for evaluating
the forecasting model’s performance. The following
section describes the experiments and test results in
more details.
A New Neural Network Model for Prediction Next Stage of Alzheimer’s Disease
693
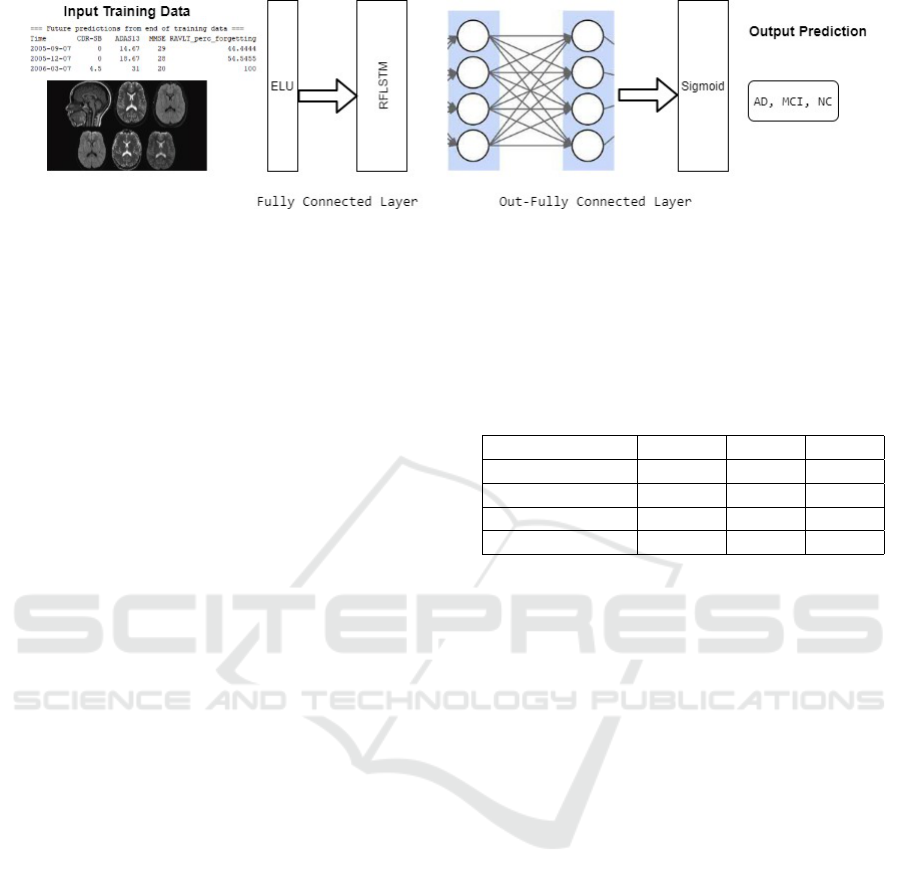
Figure 2: Proposed Model architecture.
5 EXPERIMENTS
Studies show that symptoms associated with AD may
begin several years before it already appears. In this
work, the prediction stated of AD after two years for
MRI image and neurological tests. As a result, the
states of disease are labeled as prediction status. The
proposed model can predict the next state with an er-
ror near 0.04. Also, the development of AD is pre-
dicted by putting into consideration multiple factors.
In order to evaluate model performance over time,
they are computing three distinct measurements and
comparing the anticipated value to the actual value.
First, Mean Absolute Error (MAE) is the most basic
measure of forecast performance. Second, compare
forecasts of different series in different scales using
the Mean Absolute Percentage Error (MAPE). Be-
cause both of these strategies are dependent on the
mean error, the impact of significant errors is underes-
timated. The uncommon error will catch us off guard
if it focuses too much on the mean. That is the reason
why the Root Mean Square Error (RMSE) adjusts for
huge, infrequent faults.
A recurrent neural network (RNN) is a type of
Artificial Neural Network used to execute prediction
operations on sequential or time-series-based data.
These Deep learning layers are widely employed for
ordinal or temporal problems like Natural Language
Processing, Neural Machine Translation, and auto-
mated picture captioning. The LSTM is a type of
RNN that can learn long-term sequences. It is created
to avoid long-term dependency issues. Its method
of operation is to remember large sequences for an
extended period. A Gated Recurrent Unit (GRU)
workflow is similar to that of an RNN, except for
the operation and gates associated with each GRU
unit. GRU integrates two gate operating techniques
named Update gate and Reset gate to solve the prob-
lem presented by ordinary RNN. Table 2 shows the
model performance against the previously described
models. It shows good prediction responses over a
specific time sequence; Lower RMSE values find a
better match. If the model objective is prediction,
then RMSE is a good indicator of how accurately the
model predicts the response, and it is the most impor-
tant criterion for fit.
Table 2: Performance Metrices.
Model MAE MAPE RMSE
Proposed Model 0.0298 2.9795 0.0358
RNN 0.0363 3.8431 0.0448
GRU 0.03889 4.0859 0.0471
LSTM 0.0394 4.1171 0.0470
Furthermore, To validate the resulting model and
analyze the summary results, cross-validation based
on prediction is used. It is a statistical tool for deter-
mining how well machine learning models perform. It
is used to safeguard our predictive model from over-
fitting, especially when the amount of data available
is minimal. Following the discovery of the best lag
length and the number of layers, the following are the
tested meta-parameters for the RFLSTM-PM:
1. batch size as 50, the learning rate as 0.001
2. the number of pre-fully connected cells as 128
3. the number of post-fully connected cells as 3 and
keep probability as 0.8
4. the number of fully connected cells is 128, and the
number of layers is 2
5. Sigmoid, hyperbolic tangent (tanh), exponential
linear unit (ELU) activation functions in hidden
layers. The exponential linear unit helps reach the
best values.
At the same time, the proposed model tuned with
the following parameters to discover the optimal pa-
rameters: the number of fully connected cells, the
number of cells, and the number of layers. Experi-
ments on AD vs. MCI vs. NC predict that the model
obtains the best MAE, MAPE, and RMSE. Our model
achieves the best prediction for the MMSE exam,
CDR-SB neurological test and Ventricles. Figure 3
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
694
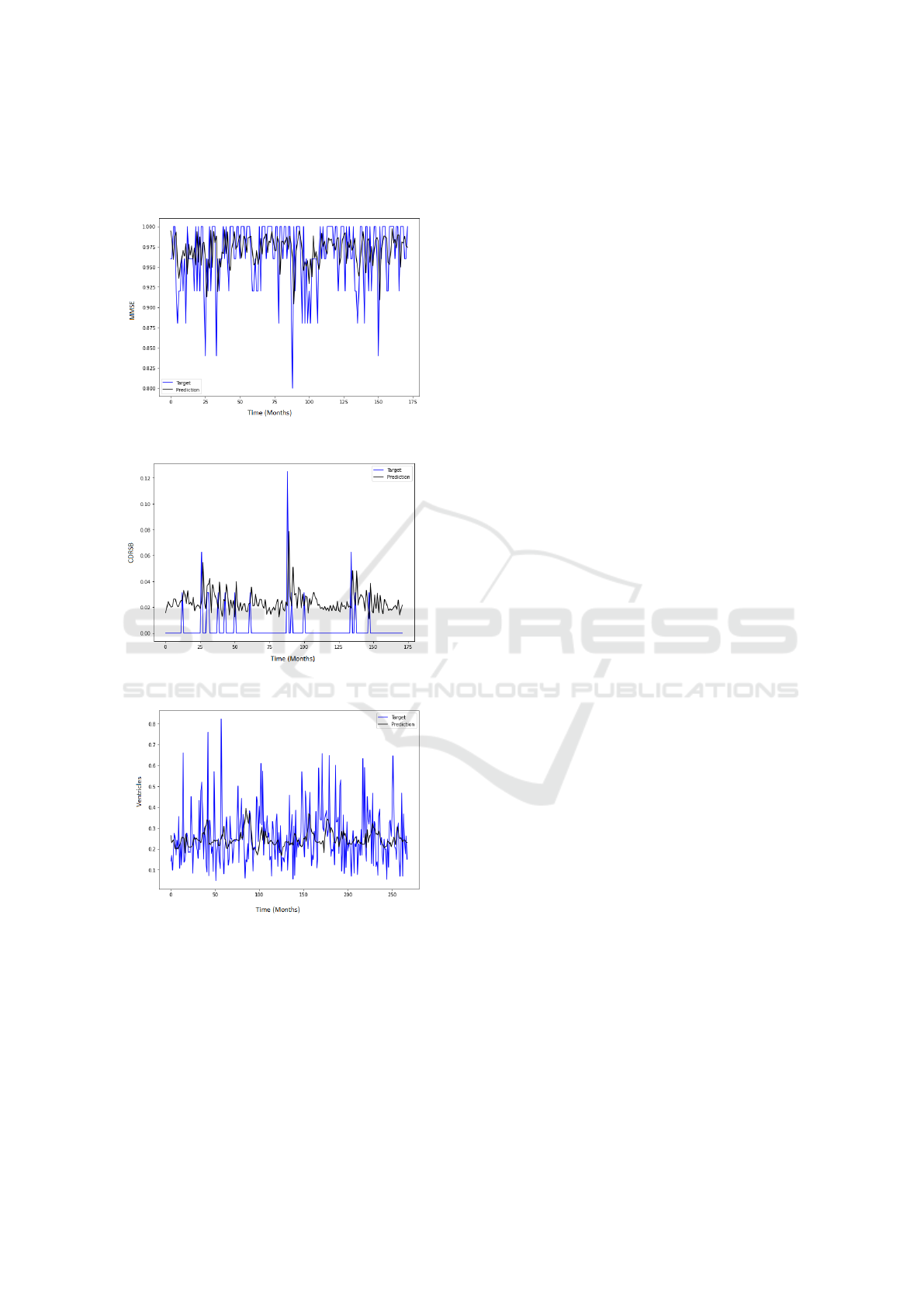
,figure 4 and figure 5 shows a good fit and stable pre-
diction for the medium term horizon of next stages in
symmetric order.
Figure 3: MMSE Prediction Results.
Figure 4: CDRSB Prediction Results.
Figure 5: Ventricles Prediction Results.
6 CONCLUSION
This research introduces a deep learning model for
predicting the next stage of Alzheimer’s disease. Be-
cause the condition is essentially progressive, the
model considers the timing data gathered from the
cases. In contrast to previous methodologies, our
model can predict the disease’s future condition rather
than classify the state of a current diagnosis. Exper-
iments have shown that our model outperforms the
vast majority of existing techniques. Improving the
model’s performance will require further research in
future studies. Ventricles’ prediction also needs to
be improved. Personal data could improve the ac-
curacy and efficiency of AD prediction at an earlier
stage. Furthermore, the proposed methodology will
be tested using actual data.
REFERENCES
Alom, M. Z., Taha, T. M., Yakopcic, C., Westberg, S.,
Sidike, P., , Nasrin, M. S., Hasan, M., Essen, B. C. V.,
Awwal, A. A. S., and Asari, V. K. (2019). A state-of-
the-art survey on deep learning theory and architec-
tures. Electronics, 8(3).
Association, A. (2019). 2019 alzheimer’s disease facts and
figures. Alzheimer’s & Dementia, 15(3):321–387.
Basher, A., Kim, B. C., Lee, K. H., and Jung, H. Y.
(2021). Volumetric feature-based alzheimer’s disease
diagnosis from smri data using a convolutional neu-
ral network and a deep neural network. IEEE Access,
9:29870–29882.
Bi, X., Hu, X., Wu, H., and Wang, Y. (2020). Multimodal
data analysis of alzheimer’s disease based on clus-
tering evolutionary random forest. IEEE Journal of
Biomedical and Health Informatics.
Bi, X., Li, S., Xiao, B., Li, Y., Wang, G., and Ma, X. (2019).
Computer aided alzheimer’s disease diagnosis by an
unsupervised deep learning technology. Neurocom-
puting.
Bui, C., Pham, N., Vo, A., Tran, A., Nguyen, A., and Le,
T. (2018). Time series forecasting for healthcare di-
agnosis and prognostics with the focus on cardiovas-
cular diseases. In Vo Van, T., Nguyen Le, T. A.,
and Nguyen Duc, T., editors, 6th International Con-
ference on the Development of Biomedical Engineer-
ing in Vietnam (BME6), pages 809–818, Singapore.
Springer Singapore.
Cai, J., Hu, L., Liu, Z., Zhou, K., and Zhang, H. (2020).
An embedded feature selection and multi-class clas-
sification method for detection of the progression
from mild cognitive impairment to alzheimer’s dis-
ease. Journal of Medical Imaging and Health Infor-
matics, 10(2):370–379.
Davda, N. and Corkill, R. (2020). Biomarkers in the diag-
nosis and prognosis of alzheimer’s disease. Journal of
Neurology, 267:2475–2477.
Dimitriadis, S. I., Liparas, D., Tsolaki, M. N., Initiative, A.
D. N., et al. (2018). Random forest feature selection,
fusion and ensemble strategy: Combining multiple
morphological mri measures to discriminate among
healhy elderly, mci, cmci and alzheimer’s disease pa-
tients: From the alzheimer’s disease neuroimaging
initiative (adni) database. Journal of neuroscience
methods, 15(302):14–23.
A New Neural Network Model for Prediction Next Stage of Alzheimer’s Disease
695

Edwards III, G. A., Gamez, N., Escobedo Jr, G., Calderon,
O., and Moreno-Gonzalez, I. (2019). Modifiable risk
factors for alzheimer’s disease. Frontiers in aging neu-
roscience, 11(164).
El-Sappagh, S., Abuhmed, T., Islam, S. R., and Kwak, K. S.
(2020). Multimodal multitask deep learning model
for alzheimer’s disease progression detection based on
time series data. Neurocomputing, 412:197–215.
Farouk, Y. and Rady, S. (2020). Early diagnosis of
alzheimer’s disease using unsupervised clustering. In-
ternational Journal of Intelligent Computing and In-
formation Sciences, 20(2):112–124.
Feng, J., Zhang, S.-W., Chen, L., and Xia, J.
(2021). Alzheimer’s disease classification using
features extracted from nonsubsampled contourlet
subband-based individual networks. Neurocomputing,
421:260–272.
Grassi, M., Rouleaux, N., Caldirola, D., Loewenstein, D.,
Schruers, K., Perna, G., Dumontier, M., Initiative,
A. D. N., et al. (2019). A novel ensemble-based
machine learning algorithm to predict the conversion
from mild cognitive impairment to alzheimer’s disease
using socio-demographic characteristics, clinical in-
formation, and neuropsychological measures. Front
Neurology, 10(756).
Haaksma, M. L., Calder
´
on-Larra
˜
naga, A., Rikkert, M.
G. O., Melis, R. J., and Leoutsakos, J. S. (2018). Cog-
nitive and functional progression in alzheimer disease:
A prediction model of latent classes. International
journal of geriatric psychiatry, 33(8).
Hameed, B., Elfetouh, A., and Elkheir, M. A. (2015).
Data cleaningtool: Usageoffuzzyroughsettheory as
machine learningpre-processing. International Jour-
nal of Intelligent Computing and Information Sci-
ences, 15:41–54.
Hancer, E., Xue, B., and Zhang, M. (2020). A survey on
feature selection approaches for clustering. Artificial
Intelligence Review volume, 53:4519–4545.
Helal, M., Elmogy, M., and Al-Awady, R. (2015). Using
rough set and boosting ensemble techniques to en-
hance classification performance of hepatitis c virus.
International Journal of Intelligent Computing and
Information Sciences, 15:45–59.
Hochreiter, S. and Schmidhuber, J. (1997). Long Short-
Term Memory. Neural Computation, 9(8):1735–1780.
Hong, X., Lin, R., Yang, C., Zeng, N., Cai, C., Gou, J., and
Yang, J. (2019). Predicting alzheimer’s disease using
lstm. IEEE Access, 7:80893–80901.
Janghel, R. R. (2020). Deep-Learning-Based Classification
and Diagnosis of Alzheimer’s Disease, chapter 76,
page 25. IGI Global.
Khaire, U. M. and Dhanalakshmi, R. (2019). Stability of
feature selection algorithm: A review. Journal of
King Saud University – Computer and Information
Sciences.
Liu, L., Zhao, S., Chen, H., and Wang, A. (2020). A new
machine learning method for identifying alzheimer’s
disease. Simulation Modelling Practice and Theory,
99.
Mart
´
ı-Juan, G., Sanroma-Guell, G., and Piellaa, G. (2020).
A survey on machine and statistical learning for lon-
gitudinal analysis of neuroimaging data in alzheimer’s
disease. Computer Methods and Programs in
Biomedicine.
Perneczky, R., editor (2018). Biomarkers for Preclinical
Alzheimer’s Disease, volume 137 of Neuromethods.
Humana Press, New York, NY.
Piccialli, F., Giampaolo, F., Prezioso, E., Camacho, D.,
and Acampora, G. (2021). Artificial intelligence and
healthcare: Forecasting of medical bookings through
multi-source time-series fusion. Information Fusion,
74:1–16.
Qiu, S., Chang, G. H., Panagia, M., Gopal, D. M., Au,
R., and Kolachalama, V. B. (2018). Fusion of deep
learning models of mri scans, mini-mental state ex-
amination, and logical memory test enhances diagno-
sis of mild cognitive impairment. Alzheimers Dement
(Amst)., 28(10):737–749.
Remeseiro, B. and Bolon-Canedo, V. (2019). A review
of feature selection methods in medical applications.
Computers in Biology and Medicine, 112.
Shikalgar, A. and Sonavane, S. (2020). Hybrid deep learn-
ing approach for classifying alzheimer disease based
on multimodal data. In Iyer, B., Deshpande, P. S.,
Sharma, S. C., and Shiurkar, U., editors, Computing
in Engineering and Technology, pages 511–520, Sin-
gapore. Springer Singapore.
Soliman, S. A., El-Dahshan, E.-S. A., and Salem, A.-B. M.
(2021). Diagnosis of alzheimer’s disease by three
dimensional convolutional neural network using un-
supervised feature learning method. International
Journal of Intelligent Computing and Information Sci-
ences.
Tanveer, M., Richhariya, B., Khan, R. U., Rashid, A. H.,
Khanna, P., Prasad, M., and Lin, C. T. (2020).
Machine learning techniques for the diagnosis of
alzheimer’s disease: A review. ACM Transactions on
Multimedia Computing, Communications, and Appli-
cations, 16(15).
Vemuri, P., Lesnick, T. G., Przybelski, S. A., Knopman,
D. S., Lowe, V. J., Graff-Radford, J., Roberts, R. O.,
Mielke, M. M., Machulda, M. M., Petersen, R. C.,
et al. (2017). Age, vascular health, and alzheimerdis-
ease biomarkers in an elderly sample. Annals of neu-
rology, 82:706–718.
Yang, C., Delcher, C., Shenkman, E., and Ranka, S. (2018).
Machine learning approaches for predicting high cost
high need patient expenditures in health care. BioMed-
ical Engineering OnLine, 17(113).
Zhong, Y., Yang, H., Zhang, Y., Li, P., and Ren, C. (2021).
Long short-term memory self-adapting online random
forests for evolving data stream regression. Neuro-
computing, 457:265–276.
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
696
