
Bone Segmentation of the Human Body in Computerized
Tomographies using Deep Learning
Angelo Antonio Manzatto
a
, Edson José Rodrigues Justino
b
and Edson Emílio Scalabrin
c
Programa de Pós Graduação em Informática, PPGIa - PUCPR, Curitiba, Paraná, Brazil
Keywords: Deep Learning, Medical Segmentation, Computed Tomography, Bones.
Abstract: The segmentation of human body organs in medical imaging is a widely used process to detect and diagnose
diseases in medicine and to help students learn human anatomy in education. Despite its significance,
segmentation is time consuming and costly because it requires experts in the field, time, and the requisite
tools. Following the advances in artificial intelligence, deep learning networks were employed in this study
to segment computerized tomography images of the full human body, made available by the Visible Human
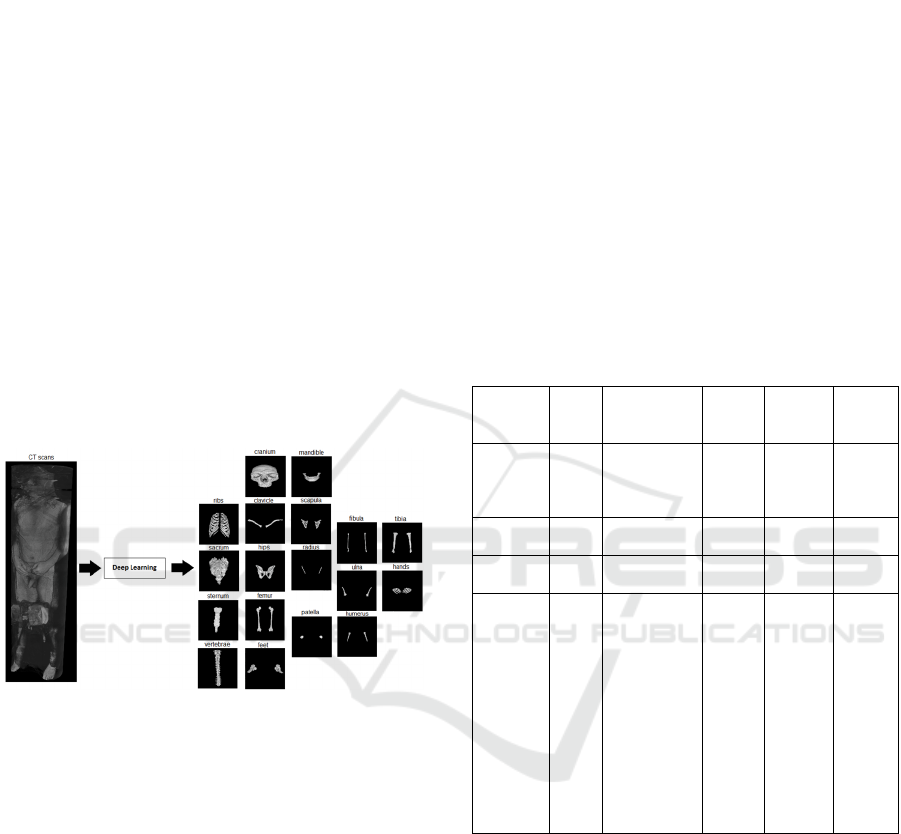
Project (VHP), which included among 19 classes (18 types of bones and background): cranium, mandible,
clavicle, scapula, humerus, radius, ulna, hands, ribs, sternum, vertebrae, sacrum, hips, femur, patella, tibia,
fibula, and feet. For the proposed methodology, a VHP male body tomographic base containing 1865 images
in addition to the 20 IRCAD tomographic bases containing 2823 samples were used to train deep learning
networks of various architectures. Segmentation was tested on the VHP female body base containing 1730
images. Our quantitative evaluation of the results with respect to the overall average Dice coefficient was
0.5673 among the selected network topologies. Subsequent statistical tests demonstrated the superiority of
the U-Net network over the other architectures, with an average Dice of 0.6854.
1 INTRODUCTION
Segmentation in medical imaging is a crucial area
both within medicine, assisting in the identification
and treatment of diseases, and in education, teaching
students about the anatomy of the human body using,
for example, a digital anatomy table with such
capability while examining the tomography set from
a patient (Brongel et al, 2019). Recent studies, such
as those of Hesamian et al., (2019), have shown that
the performance achieved by deep learning networks
in image segmentation is superior to that of other
existing methods, leading to an impressive increase in
research efforts aimed at developing new and more
promising architectures in this area.
In medicine, studies have primarily focused on the
detection, prevention, and combat of serious diseases.
Research highlights with regard to deep learning
networks include segmentation of the computerized
tomography (CT) scans of the lungs ((Chunran and
Yuanyuan, 2018), (Shaziya et al, 2018), (Huang et al,
a
https://orcid.org/0000-0002-6263-1399
b
https://orcid.org/0000-0001-9145-6879
c
https://orcid.org/0000-0002-3918-1799
2018), (Kumar et at, 2019), (Alves et al, 2018), (Jin
et al, 2017), (Gerard and Reinhardt, 2019)), with the
purpose of detecting pulmonary nodules to fight lung
cancer as well as segmentation of the CT images of
the liver ((Jiang et al, 2019), (Wang et al, 2019),
(Shrestha and Salari, 2018), (Ahmad et al, 2019),
(Wang et al, 2019), (Chen et al, 2019), (Li et al,
2018), (Rafiei et al, 2018), (Xia et al, 2019). (Truong
et al, 2018), (Zhou et al, 2019)), both to track tumors
and lesions, and to aid in preoperative activities.
Other studies directly applicable to the medical field
and linked to the use of deep learning networks
include segmentation of the brain surface in post-
surgical activities of epilepsy patients ((Shell and
Adam, 2020)), segmentation of the esophagus for
cancer treatment ((Chen et al, 2019), (Trullo et al,
2017), (Trullo et al, 2017)), and segmentation of the
spine ((Fang et al, 2018), (Tang et al, 2019), (Kuok et
al, 2018)) to assist in pre-surgical activities.
In the course of the evolution of deep learning
networks, a few studies have thoroughly tested the
Manzatto, A., Justino, E. and Scalabrin, E.
Bone Segmentation of the Human Body in Computerized Tomographies using Deep Learning.
DOI: 10.5220/0010891200003182
In Proceedings of the 14th International Conference on Computer Supported Education (CSEDU 2022) - Volume 2, pages 17-26
ISBN: 978-989-758-562-3; ISSN: 2184-5026
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
17
limits of a single network segmenting all body organs
for the possible creation of a medical atlas. A 3D deep
learning network model/architecture, trained in a
semi-supervised manner combining supervised
learning using a small amount of labelled data with
unsupervised learning on massive unlabelled data,
was proposed to segment 16 abdominal organs (Zhou
et al, 2019). The use of a U-Net type deep learning
network was also proposed for the computerized
segmentation of six different types of bones (La Rosa,
2017). The lack of complete body tomographic image
bases and the lack of available specialists to create
ground truth masks of segmented images for each
organ have been a major impediment to advances
toward the creation of a medical atlas.
To advance along this direction, in this paper, we
present the results of segmenting one of the most
complete tomographic bases in the world, the Visible
Human Project (VHP), into 19 classes using deep
learning networks, with 18 of the classes denoting 18
different bones, and one class denoting the
background, as illustrated in Figure 1.
Figure 1: Segmentation of a CT image base into 18 bone
classes. The images were extracted from the VHP male
database and compiled by the author.
Five network topologies were selected to perform
the segmentation task: U-Net, DenseNet, ResNet,
DeepLab, and FCN; all of which were trained in the
supervised mode. The Dice coefficient was used as a
metric to evaluate the results. Statistical tests such as
ANOVA and the non-parametric post-hoc Games-
Howell test were used to compare the performances
of the five different architectures of deep learning
networks in question.
The remainder of this paper is organized as
follows. Section 2 presents studies related to bone
segmentation using deep learning networks. Section
3 describes the method used both in the creation of
the ground truth bases and in the training, testing, and
analysis of the segmentation results. Section 4
presents an analysis of the results and discussion.
Section 5 concludes by highlighting the main results
and proposals for future research.
2 RELATED WORK
Three studies ((Fang et al, 2018), (Kuok et al, 2018),
(La Rosa, 2017)) with focus on the application of
deep learning networks to bone segmentation were
chosen to compare against. Some characteristics of
the items common to all these research studies were
compared against those of the current study. Table 1
shows the size of the databases used for training,
number of bone classes to be segmented, and metrics
for measuring the performance of the deep learning
networks pertaining to these studies.
Table 1: Deep learning networks specifically for bone
segmentation.
Reference
Qty.
classes
Bone classes
Qty
Training
Base
Network Metric
La Rosa, F.
(2017)
6
vertebrae,
sacrum, hips,
ribs, femur,
and sternum
15653 U-Net Dice
Kuok et al.
(2018)
1 vertebrae 200 DenseNet Dice
Fang et al.
(2018)
1 vertebrae 4500 FCN Accuracy
This article 18
cranium,
mandible,
clavicle,
scapula,
humerus,
radius, ulna,
hands, ribs,
sternum,
vertebrae,
sacrum, hips,
femur, patella,
tibia, fibula,
and feet
4688
(Male
VHP +
20
IRCAD
bases)
U-Net,
DenseNet,
ResNet,
DeepLab
and FCN
Dice
Although La Rosa uses a base containing 15653
samples to train a single U-Net for the segmentation
of six classes of bones of the abdominal region, in this
study, we used multiple bases for a total of 4688
(1865 male VHP + 2823 IRCAD) training samples,
which were segmented into 18 classes. An additional
test base (1730 female VHP), which represented a
significant challenge in terms of volume of
experiments, was used to evaluate five distinct
network topologies. The only class common to the
aforementioned studies was the vertebra, and as noted
earlier, the Dice metric was the predominant metric
for performance measurement. Although the three
studies sought to achieve the best segmentation by
CSEDU 2022 - 14th International Conference on Computer Supported Education
18

training a deep learning network topology, here in
addition to segmentation, we investigate whether
there is a significant difference in the segmentation
results among the five different deep learning
network topologies, using statistical analysis.
3 METHOD
The experimental method is structured as follows:
database selection, creation of ground truth base,
definition of deep learning network topologies,
supervised training of deep learning networks, cross-
validation, and collection and statistical analyses of
the results.
3.1 Databases
Two different tomographic image databases were
selected for experimentation: the first database
provided by the National Library of Medicine, is
referred to as the Visible Human Project (VHP)
1
, and
the second database provided by the Institute for
Research Against Digestive Cancer (IRCAD)
2
.
The selection of the VHP database was because of
its uniqueness, in that it consisted of full-body CT
scans, allowing us to explore the challenges of
segmenting all types of different bones, a challenge
not put into practice in other works ((Fang et al,
2018), (Kuok et al, 2018), (La Rosa, 2017)). These
studies focused on one type of bone or a single area
of the body, such as the abdominal region. The
IRCAD database was selected to complement the
model training database because it has segmentation
of the bones in the abdominal region.
The VHP Project image base includes two
tomographic sets: male and female. The male body
base contains 1865 512 × 512 pixel copies, whereas
the female human body base contains 1730 512 × 512
pixel copies. The database provided by IRCAD
consists of 2823 512 × 512 pixel CT images from 20
clinical studies, involving lesions and tumors of
various patients, anonymized in DICOM format. In
conducting the experiments, we segregated the
datasets into two groups: S1 and S2. Set S1,
exclusively used for training, contains the VHP male
database plus the 20 IRCAD databases, totaling 4688
images. In contrast, group S2 contains only the
female VHP database intended for testing, data
collection, and results analysis to verify the
1
https://www.nlm.nih.gov/research/visible/visible_human.
html
2
https://www.ircad.fr/research/3d-ircadb-01/
generalization power of the segmentation on a base
other than the one used during training. In the pre-
processing step, all images from the VHP and IRCAD
bases were converted to 8-bit monochrome color
scale with grayscales ranging from 0 to 255 and saved
in PNG format.
The procedure adopted in the preparation of each
ground truth mask for the 18 bone classes consisted
of applying: (a) thresholding layers to each
tomographic image to eliminate a significant part of
the background and other organs, and (b) using
different combinations of selection tools to segment
each bone class and subsequently create a binary
image in PNG format for the generation of the mask.
This procedure was applied to all 6418 CT images,
generating a total of 14 602 masks, as shown in Table
2 and took approximately eight months to complete.
Table 2: Number of ground truth masks of bone class in the
Visible Human Project male and female human body bases
along with the 20 IRCAD bases.
The entire manual segmentation process for the
creation of the ground truth was performed only by
the first author of this article, using a CT scan
specialist at CETAC (Centro de Tomografia
Computadorizada LTDA) in the city of Curitiba
(Brazil) and a medical atlas (Fleckenstein and
Tranum-Jensen, 2018) in case of doubts.
Our dataset is publicly available for download at the
following link: https://www.ppgia.pucpr.br/datasets/SA/
Classes Male VHP VHP female IRCAD
clavicle 94 86 17
craniu
m
174 160 0
feet 152 162 0
femu
r
486 423 42
fibula 401 345 0
hands 143 167 0
hips 219 213 255
humerus 240 280 12
mandible 106 82 0
p
atella 52 52 0
radio 129 216 0
ribs 390 366 1929
sacru
m
157 127 65
scapula 176 164 100
sternu
m
210 188 407
tibia 410 351 0
ulna 146 198 0
vertebrae 619 598 2707
Total 4304 4178 5534
Bone Segmentation of the Human Body in Computerized Tomographies using Deep Learning
19
3.2 Network Topologies
Five topologies of deep learning networks were
chosen for the study: U-Net, DenseNet, ResNet,
DeepLab, and FCN. As each network can be
assembled in countless ways and encompass an
infinite number of variations, we selected
architectures already discussed in image
segmentation related manuscripts. Some changes
were introduced to adapt the topologies to the
proposal of this work, such as accepting a 512 × 512
× 1 tensor (height, width, and depth) monochrome
computed tomography image as input and producing
a 512 × 512 × 19 tensor (height, width, and number
of classes, 18 bones and the background) for output,
referred to as the resulting segmentation.
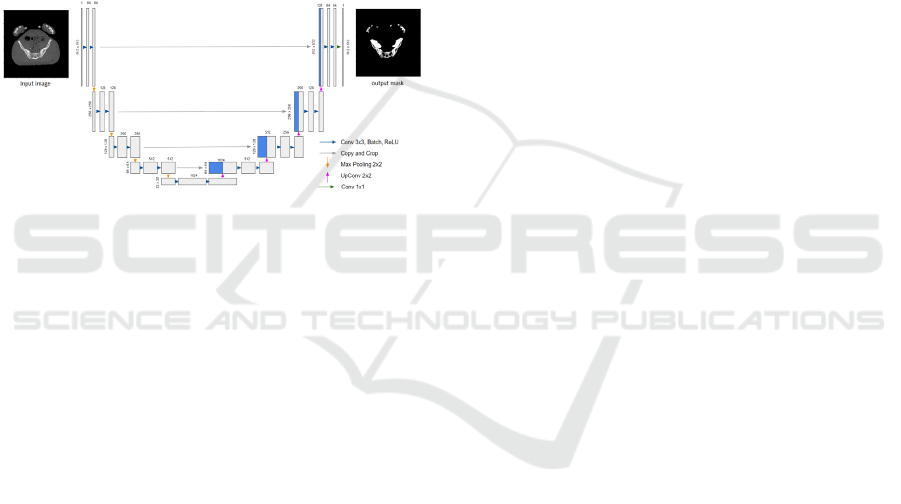
Figure 2: Topology of the U-Net network.
The U-Net network architecture (Figure 2), of
Ronneberger et al., (2015), consists of a contraction
part and an expansion part. Each path of the
contraction contains two blocks formed by a 3 × 3
convolution layer without padding, followed by an
activation layer resembling a rectifier linear unit
(ReLU), and a 2 × 2 max pooling contraction layer.
The number of filters is doubled after each
contraction block following the sequence of 64, 128,
256, 512, and 1024 filters to the deepest level while
halving the number of filters at each expansion block.
Expansion blocks are created by upsampling or by
concatenating 2 × 2 transposed convolution layers to
the output along with the features of the appropriate
contraction layer. A 1 × 1 convolution layer is applied
at the end to go from a multidimensional space of 64
to a monochrome image.
To conserve space, only a very brief description
of the other topologies is presented. The DenseNet
topology was implemented using the FC-
DenseNet103 model (Jegou et al, 2017), which deals
with semantic segmentation issues. The ResNet
architecture was implemented as described in
(Liciotti et al, 2018), where the original version was
applied to the task of people tracking crowded
environments through aerial view images. ResNet
deals with the vanishing/exploding gradient problem
in which the network weights are practically zero or
tend to infinite values when the network has multiple
layers, introducing the concept of shortcut
connection.
The DeepLab topology was implemented using
the DeepLab V3 Plus model (Chen et al, 2018). In
addition to depthwise separable convolution layers
(Chollet, 2017), the model includes a refinement in
the decoder part of the architecture, allowing the use
of convolutional layers with upsampling, referred to
as atrous convolution, which solves the problem of
loss of resolution in the multiple layers resulting from
downsampling during convolution or pooling with a
stride greater than one. The model also employs a
process called atrous spatial pyramid pooling
(ASPP), in which the fusion of multiple atrous
convolution layers with different sampling rates is
applied to the input image, allowing the capture of
features at different scales of the objects to be
segmented.
A fully convolutional network (FCN) topology
has also been implemented as described in
(Shelhamer et al, 2017). Here the VGG16 topology
(Simonyan and Zisserman, 2015) constitutes the body
of the encoder part and the output segmentation is
refined by concatenating the deconvolution layers
with the max pooling layers.
All the aforementioned networks had their last
convolutional layer adapted to generate an output
tensor of size 512 × 512 × 19 pixels before passing
through a Softmax-type activation layer.
3.3 Training and Collection of Results
Figure 3 presents the general scheme of the
experiments, which consists of four steps. Each of the
five networks were subjected to a training process to
obtain the segmentation model (step 1). As the next
step, each model was used to segment the
tomographic image base of test S2 (step 2). A
statistical analysis was carried out on the results to
evaluate the quality of the segmentation. The number
of true positive (TP), true negative (TN), false
positive (FP), and false negative (FN) were
determined for each channel of the network output
segmented image in relation to the corresponding
class in the ground truth image. The statistics were
subsequently used to calculate the Dice coefficient as
an evaluation metric (step 3). The experiments were
concluded with a statistical analysis on the
performances of the five deep learning network
topologies in question vis-à-vis the VHP and IRCAD
image bases (step 4).
CSEDU 2022 - 14th International Conference on Computer Supported Education
20

Figure 3: Basic scheme of evaluation of deep learning
networks.
The training of the networks was performed using
a GPU with variable memory from 12 to 16 GB. The
software algorithms were programmed in Python
using the OpenCV and Matplotlib libraries for image
pre-processing and Keras for training and testing of
the deep learning networks. The number of network
parameters and layers as well as the average time
required to complete one training along with the batch
size for each network topology are listed in Table 3.
Table 3: Configuration parameters of the networks during
post-training of the experiments.
Network
Total
p
arameters
Layer Training time
Batch
Size
U-Net 31
053
965 73 27 h 43 min 20 s 4
DenseNet 9
426
355 494 35 h 13 min 20 s 2
ResNet 2,754
771 276 28 h 31 min 40 s 4
DeepLab 41
257
123 412 29 h 20 min 00 s 4
FCN 134
455
833 35 28 h 35 min 00 s 2
To avoid biased results, we used the cross-
validation method with five folds for each deep
learning network topology, using the following
protocol in each training session (Table 4):
Table 4: Training protocol of the experiments.
Parameters Value
Number of e
p
ochs 100
Trainin
g
base S1
(
male VHP + IRCAD
)
Trainin
g
/ Validation 80 % / 20 %
Weight optimization
al
g
orithm
Adam
Learnin
g
rate 0.0001
Batch size 2 or 4
Cost function Weighted Cross Entrop
y
Chance to apply data
augmentation per technique
for each sample
15%
Data augmentation
techniques
Rotation, translation, scaling,
horizontal mirroring, elastic
deformation, saturation,
contrast
The cost function for calculating the gradients, in
the process of updating the weights of each network,
is the weighted cross entropy (Equation 1):
𝐿
𝑙,𝑞
1
1
𝑀
𝑤
𝑥
𝑝
log
𝑝
𝑥
(1)
where M represents the number of pixels of the
segmented image; N is the number of classes; w
n
(x)
is the value of the class weight n applied to pixel x;
p
gt
represents the value of the ground truth pixel; and
p
pr
is the probability predicted by the Softmax layer
for the output of the deep learning network for pixel
x.
To reduce the overfitting generated by the
unbalanced samples, different weights were applied
to the cost function (Equation 1) for each bone class.
Each weight was calculated using Equation 2, where
freq(c) is the pixel frequency of a given bone class
divided by the total number of pixels in all images in
which the class appears, and freq
m
is the median pixel
frequency of all classes, as displayed in Table 5.
𝛼
𝑓
𝑟𝑒𝑞
𝑓
𝑟𝑒𝑞
𝑐
(2)
Table 5: Weights assigned to each bone class for the
Weighted Cross Entropy cost function.
Class Weight
background 0.00033
clavicle 3.851657
cranium 0.189668
feet 0.681439
femur 0.236127
fibula 2.563772
hands 1.884704
hips 0.194719
humerus 1
mandible 1.740797
patella 6.942333
radio 6.094191
ribs 0.09294
sacrum 0.653063
scapula 1.145436
sternum 2.256164
tibia 0.431686
ulna 4.130787
vertebrae 0.041536
Bone Segmentation of the Human Body in Computerized Tomographies using Deep Learning
21
All weights were calculated prior to training
globally.
After each training, the data collection process
was performed in the S2 base by following the
sequential steps outlined below:
1-The trained network performs the segmentation of
the CT scan, obtaining an image consisting of 19
channels, 18 for the bone classes, and one for the
background.
2-With the ground truth mask for each of the 19
classes, the number of true positive (TP), true
negative (TN), false positive (FP), and false negative
(FN) were calculated.
3-The data calculated for each class were saved in a
line of the "csv" file with the first column having the
tomography identifier name, and the rest of the
information following the previously outlined steps in
Figure 3, for each class.
Twenty-five training runs were performed, that is,
five experiments for each of the following networks:
U-Net, DenseNet, ResNet, DeepLab, and FCN.
4 EXPERIMENTAL RESULTS
This section presents the results of the segmentation
of the CT scan images, as well as a three-dimensional
visualization of the segmentation of some of the bone
classes.
4.1 Bone Segmentation of the Human
Body
Segmentation on set S2 followed the training step
conducted on set S1 with 5-fold cross-validation. The
training and validation workflow were executed five
times for each network topology. The Dice
coefficient, or Sørensen-Dice, is given by Equation 3,
where TP was chosen as an evaluation metric to
compare the degree of similarity between the
segmentation output of the network and the ground
truth mask.
𝐷𝑖𝑐𝑒
2𝑇𝑃
2𝑇𝑃
𝐹𝑃
𝐹𝑁
(3)
There are several situations in which the presence
of the evaluated class simply does not exist in the
tomography, for example, the class "feet" or the class
"femur" when the head region is being evaluated. In
these cases, only true negative (TN) values are
acceptable as correct; however, the Dice coefficient
does not make an exception for these situations,
generating a division by zero. In such cases, one is not
sure whether the model predicted false correctly, by
way of learning or whether there is an overfitting
problem of the dominant class “background”.
Therefore, we decided to disregard the coefficient
measurements for each class outside the range of
images in which they do not appear. This implies that,
for the segmentation of the S2 base, of the generated
images with 19 channels totaling 32 870 masks, only
4178 were considered in the evaluation.
Table 6 presents the Dice coefficients calculated
from the S2 set, for the five network topologies: U-
Net, DenseNet, ResNet, DeepLab, and FCN. For each
topology, the average Dice coefficient was calculated
across the five individual trainings for each network
in the k-fold process, with k equal to five.
Table 6: Overall average of the Dice coefficients for each
class. The green color highlights the coefficients that
exceeded the value of 0.700.
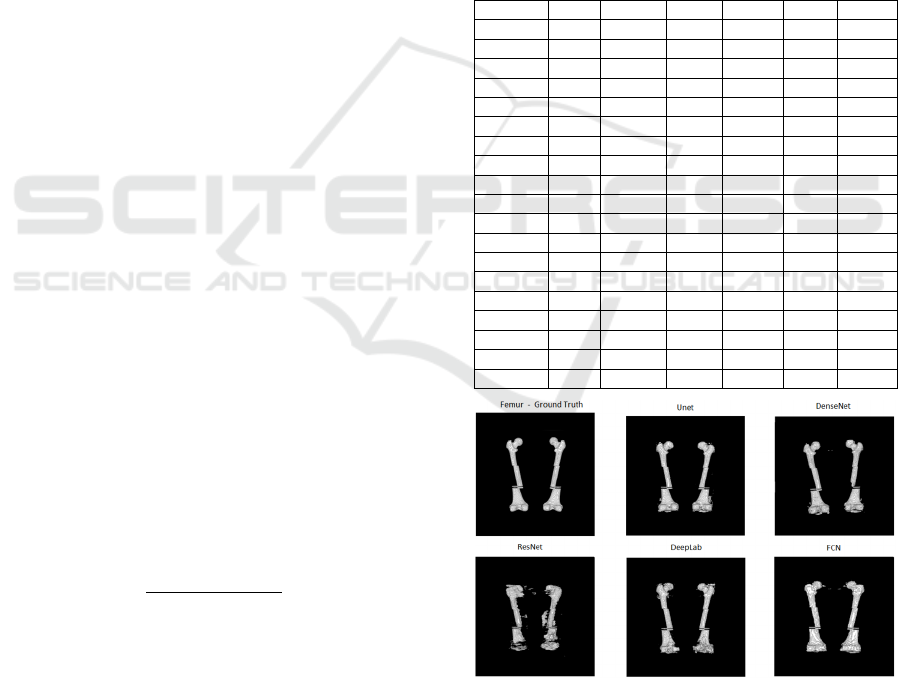
Figure 4: Segmentation of femur bones from the base of the
VHP female body for each network topology compared to
ground truth.
Dice U-Net DenseNet ResNe
t
DeepLab FCN Average
clavicle 0.7130 0.6744 0.4494 0.6468 0.5280 0.6023
cranium 0.8092 0.8170 0.7296 0.8239 0.6779 0.7715
feet 0.6475 0.3444 0.3062 0.3403 0.5878 0.4452
femu
r
0.8761 0.7748 0.6718 0.8298 0.7435 0.7792
fibula 0.7368 0.5357 0.4482 0.6728 0.5547 0.5896
hands 0.5411 0.4810 0.2125 0.2404 0.2676 0.3485
hips 0.7940 0.7433 0.6638 0.7609 0.7150 0.7354
humerus 0.6453 0.6575 0.4426 0.7285 0.5912 0.6130
mandible 0.7991 0.7559 0.6845 0.7961 0.6695 0.7410
patella 0.6951 0.7623 0.1798 0.2106 0.4704 0.4636
radio 0.3620 0.3700 0.3149 0.3926 0.3058 0.3491
ribs 0.5687 0.5701 0.4128 0.4984 0.4304 0.4961
sacrum 0.4957 0.4069 0.3294 0.3889 0.4931 0.4228
scapula 0.6489 0.5623 0.4200 0.5637 0.4774 0.5345
sternum 0.5355 0.5394 0.3072 0.5152 0.3405 0.4476
tibia 0.8051 0.7683 0.5648 0.7517 0.7195 0.7219
ulna 0.5122 0.5663 0.2978 0.5708 0.4133 0.4721
vertebrae 0.7710 0.6746 0.5727 0.7166 0.6563 0.6782
Global 0.6642 0.6113 0.4449 0.5804 0.5357 0.5673
CSEDU 2022 - 14th International Conference on Computer Supported Education
22
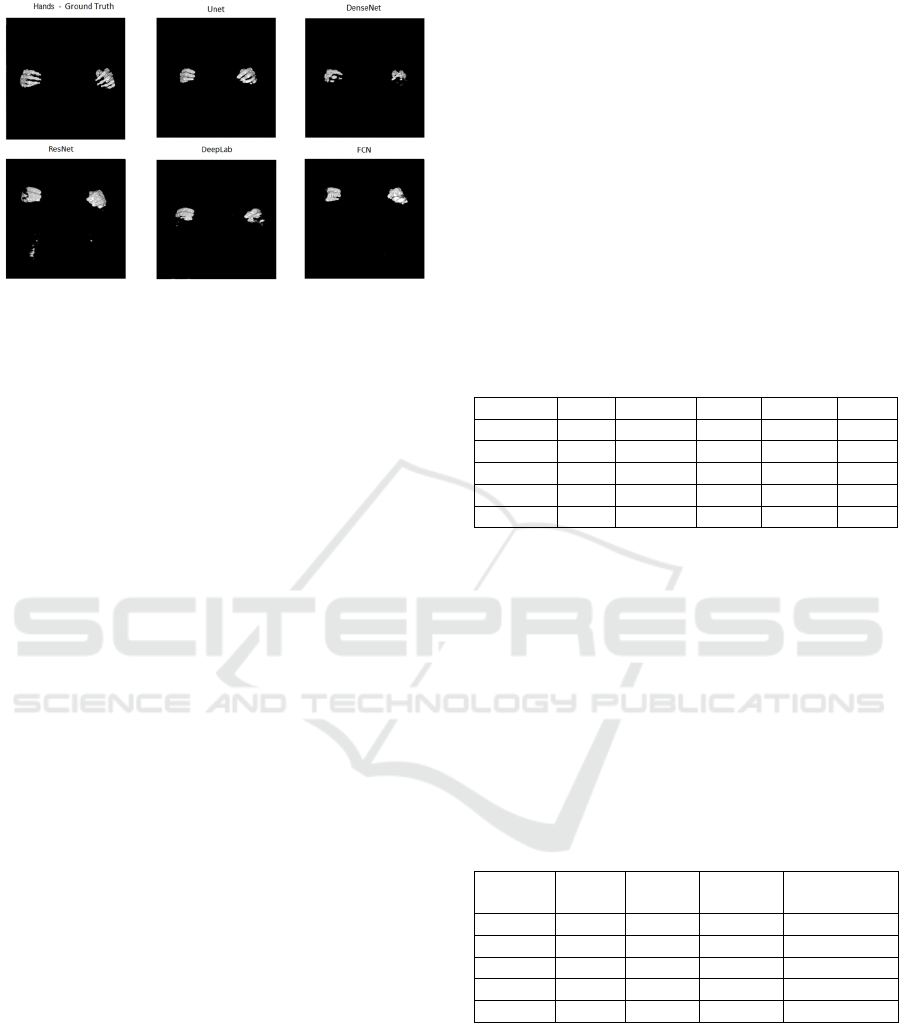
Figure 5: Segmentation of the bones of the hands from the
base of the female body of the VHP for each network
topology in comparison with the ground truth. It is observed
that there was practically no segmentation of the hand
bones by the networks.
In the images of Figures 4 and 5, it can be
observed that of among all bone classes, the
segmentation of the femur was the most successful,
as the femur Dice coefficient of 0.7729 was the
highest and the worst Dice coefficient of all bone
classes was the hand with a Dice coefficient of
0.3485. This finding can be observed in the
tridimensional images generated by each network.
Starting with the average Dice coefficients for
each deep learning network, calculated for the 4178
valid samples in set S2 and displayed in Table 6, we
performed several statistical tests using the IBM's
SPSS™ software to validate or refute the following
research hypotheses: Null Hypothesis H0: The
average Dice coefficient of all deep learning networks
are equal. Alternative Hypothesis H1: One of the
deep learning networks has at least one average Dice
coefficient that is different.
One-way analysis of variance (ANOVA) was
performed. Data normality was assessed using the
Kolmogorov-Smirnov and Shapiro-Wilk tests and
homogeneity of variance using Levene’s test.
Bootstrapping was also used with 1000 samples with
a reliability index (CI) of 95%, which increases the
confidence in the results obtained (Haukoos & Lewis,
2005). We also applied Welch correction on the
results and calculated the Dice coefficients to predict
the possibility of heterogeneity of variance in the
samples.
The preliminary results indicated that the samples
did not have a normal distribution (Kolmogorov-
Smirnov = 0.10, p-value < 0.001; Shapiro-Wilk =
0.95, p-value < 0.001), nor did they have
homogeneity of variance (Levene F(4, 20885), p-
value < 0.001). The results of ANOVA analysis of
variance, in contrast, showed that there is a
statistically significant difference between the means
of the Dice coefficients for the deep learning
networks [Welch's F(4, 10434.86) = 614.84, p-value
< 0.001], which refutes the null hypothesis H0 and
corroborates with the alternative hypothesis H1.
To find out which pairs of deep learning network
topologies show statistical differences, the non-
parametric post-hoc Games-Howell test was
performed, whereby it was found that only the deep
learning networks DenseNet and DeepLab were
similar (p-value = 0.8890 > 0.05), whereas all other
pairwise combinations were statistically different (p-
value < 0.01), as shown in Table 7.
Table 7: Games-Howell test shows that only the deep
learning networks DenseNet and DeepLab have statistical
similarity.
Finally, by analyzing the averages of the Dice
coefficients given in Table 8, it is observed that the
deep learning network U-Net has average Dice
coefficients significantly higher (0.6854) than those
of the other deep learning topologies analyzed. The
U-Net deep learning network performed the best in
the automatic segmentation of the VHP female body
as indicated by the Dice coefficients. It should also be
highlighted that U-Net has the second lowest variance
and standard deviation, and is the median with respect
to the number of parameters.
Table 8: Dice coefficient statistics on 4178 samples given
the number of training parameters.
N
etwor
k
Average Variance
Standard
Deviation
Parameters
U-Net 0.6854 0.0408 ±0.2021 31
053
965
DenseNet 0.6224 0.0475 ±0.2179 9
426
355
ResNet 0.4780 0.0417 ±0.2042 2
754
771
DeepLab 0.6270 0.0533 ±0.2308 41
257
123
FCN 0.5630 0.0394 ±0.1984 134
455
833
The box-and-whisker plot displayed in Figure 6
shows the distributions of the Dice coefficients for all
the female body CT images of the VHP or S2 set.
Such a graph allows visualization of the similarity
between the average Dice coefficient values of the
deep learning networks such as DenseNet and
DeepLab and the superior performance of the deep
learning network U-Net vis-à-vis the others alongside
P-value U-Net DenseNet ResNet Dee
p
Lab FCN
U-Net - 0.0000 0.0000 0.0000 0.0000
DenseNet 0.0000 - 0.0000 0.8890 0.0000
ResNet 0.0000 0.0000 - 0.0000 0.0000
DeepLab 0.0000 0.8890 0.0000 - 0.0000
FCN 0.0000 0.0000 0.0000 0.0000 -
Bone Segmentation of the Human Body in Computerized Tomographies using Deep Learning
23
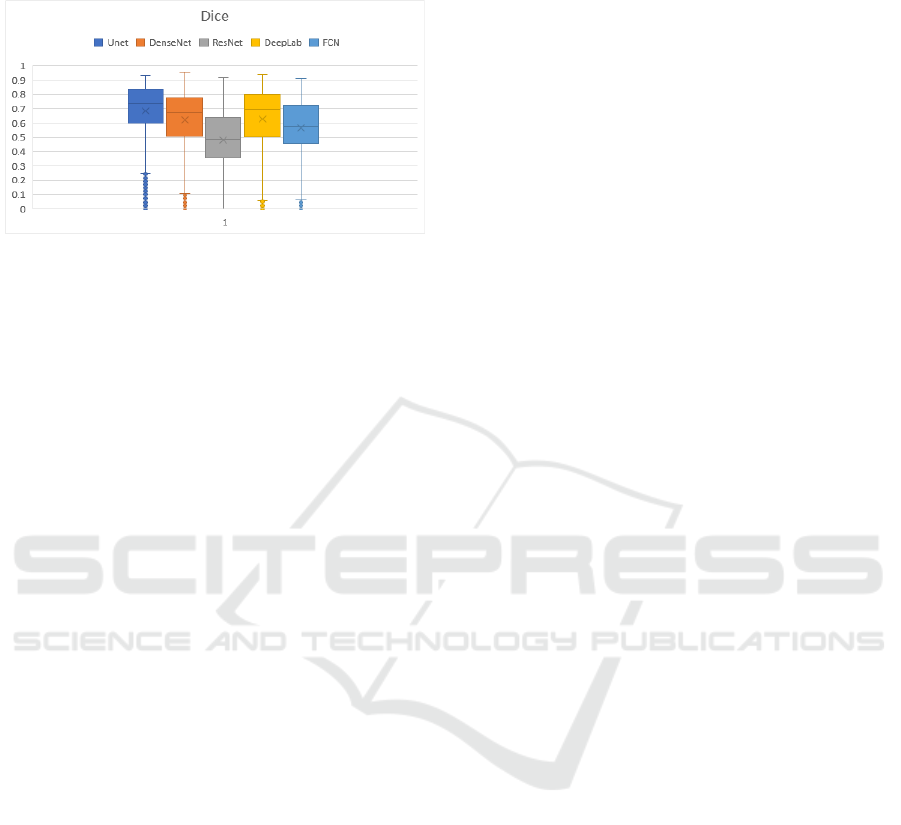
the low performance of the deep learning networks
ResNet and FCN.
Figure 6: Box-and-whisker plot for the Dice coefficient
associated with the performance of each deep learning
network used in the segmentation of tomographic images.
It is important to emphasize that the results
obtained depend on the topologies of the deep
learning networks assembled. The addition of extra
layers in the composition of each network can change
the results obtained. Therefore, generic conclusions
on each tested topology should be avoided.
5 CONCLUSIONS
In this study, we proposed the segmentation of a set
of male human body CT images from the Visible
Human Project (VHP) into 18 distinct bone classes
using deep learning networks by applying a workflow
consisting of the classical steps of: training, testing,
data collection, and pairwise statistical analysis. Until
the present moment of this paper we created the
largest ground truth bone dataset with respect to the
number of classes used in supervised training on a
medical segmentation task.
The results obtained by our tomographic image
segmentation experiments provided an overall
average Dice coefficient of 0.5673 for all classes of
segmented bones, considering the U-Net, DenseNet,
ResNet, DeepLab, and FCN networks together. The
main challenges were the limited number of
tomographic samples available for training, as well as
the great irregularity in the shape of the bones,
excessive presence of the "background" class, and
very close edges between bones of different classes.
Among the five U-Net, DenseNet, ResNet,
DeepLab, and FCN network topologies compared
using statistical tests, the U-Net topology
outperformed the others with a global average Dice
coefficient of 0.6854. DenseNet and DeepLab
networks exhibited slightly lower performance than
U-Net, but were statistically similar to each other,
with Dice coefficients of 0.6224 and 0.6270,
respectively. The FCN and ResNet topologies had the
worst performance with Dice coefficients of 0.5630
and 0.4780, respectively.
In future work, we will study more efficient
techniques that mitigate the influence of the
"background" class. We will also explore three-
dimensional topologies with 3D convolutional layers
and generative adversarial network networks. We will
examine techniques that allow the addition of new
classes from different databases without explicitly
changing the ground truth already produced. This
represents a stride towards the incremental
construction of an atlas of the human body containing
the segmentation of all organs using a single network
model.
REFERENCES
Brongel, A.; Brobouski, W.; Pierin, L.; Gomes, C.;
Almeida, M. and Justino, E. (2019). An Ultra-high
Definition and Interactive Simulator for Human
Dissection in Anatomic Learning.In Proceedings of the
11th International Conference on Computer Supported
Education - Volume 2: CSEDU, ISBN 978-989-758-
367-4, ISSN 2184-5026, pages 284-291. DOI:
10.5220/0007707102840291
Hesamian, M. H., Jia, W., He, X., & Kennedy, P. (2019).
Deep Learning Techniques for Medical Image
Segmentation: Achievements and Challenges. Journal
of Digital Imaging, 32(4), 582–596.
https://doi.org/10.1007/s10278-019-00227-x
Chunran, Y., & Yuanyuan, W. (2018). Nodule on CT
Images. 2–6.
Shaziya, H., Shyamala, K., & Zaheer, R. (2018). Automatic
Lung Segmentation on Thoracic CT Scans Using U-Net
Convolutional Network. Proceedings of the 2018 IEEE
International Conference on Communication and
Signal Processing, ICCSP 2018, 643–647.
https://doi.org/10.1109/ICCSP.2018.8524484.
Huang, C. H., Xiao, W. T., Chang, L. J., Tsai, W. T., & Liu,
W. M. (2018). Automatic tissue segmentation by deep
learning: From colorectal polyps in colonoscopy to
abdominal organs in CT exam. VCIP 2018 - IEEE
International Conference on Visual Communications
and Image Processing, 1–4. https://doi.org/10.1109/
VCIP.2018.8698645.
Kumar, A., Fulham, M., Feng, D., & Kim, J. (2019). Co-
Learning Feature Fusion Maps from PET-CT Images of
Lung Cancer. IEEE Transactions on Medical Imaging,
1–1. https://doi.org/10.1109/tmi.2019.2923601.
Alves, J. H., Neto, P. M. M., & Oliveira, L. F. (2018).
Extracting Lungs from CT Images Using Fully
Convolutional Networks. Proceedings of the
International Joint Conference on Neural Networks,
2018-July. https://doi.org/10.1109/IJCNN.2018.8489223.
CSEDU 2022 - 14th International Conference on Computer Supported Education
24

Jin, T., Cui, H., Zeng, S., & Wang, X. (2017). Learning
Deep Spatial Lung Features by 3D Convolutional
Neural Network for Early Cancer Detection. DICTA
2017 - 2017 International Conference on Digital Image
Computing: Techniques and Applications, 2017-
December, 1–6. https://doi.org/10.1109/DICTA.2017.
8227454.
Gerard, S. E., & Reinhardt, J. M. (2019). Pulmonary lobe
segmentation using a sequence of convolutional neural
networks for marginal learning. Proceedings -
International Symposium on Biomedical Imaging,
2019-April(Isbi), 1207–1211. https://doi.org/10.1109/
ISBI.2019.8759212.
Jiang, H., Shi, T., Bai, Z., & Huang, L. (2019). AHCNet:
An Application of Attention Mechanism and Hybrid
Connection for Liver Tumor Segmentation in CT
Volumes. IEEE Access, 7, 24898–24909.
https://doi.org/10.1109/ACCESS.2019.2899608
Wang, C., Song, H., Chen, L., Li, Q., Yang, J., Hu, X. T.,
& Zhang, L. (2019). Automatic Liver Segmentation
Using Multi-plane Integrated Fully Convolutional
Neural Networks. Proceedings - 2018 IEEE
International Conference on Bioinformatics and
Biomedicine, BIBM 2018, 518–523.
https://doi.org/10.1109/BIBM.2018.8621257
Shrestha, U., & Salari, E. (2018). Automatic Tumor
Segmentation Using Machine Learning Classifiers.
IEEE International Conference on Electro Information
Technology, 2018-May, 153–158. https://doi.org/10.
1109/EIT.2018.8500205
Ahmad, M., Ai, D., Xie, G., Qadri, S. F., Song, H., Huang,
Y., … Yang, J. (2019). Deep Belief Network Modeling
for Automatic Liver Segmentation. IEEE Access, 7,
20585–20595.
https://doi.org/10.1109/ACCESS.2019.2896961
Wang, Z. H., Liu, Z., Song, Y. Q., & Zhu, Y. (2019).
Densely connected deep U-Net for abdominal multi-
organ segmentation. Proceedings - International
Conference on Image Processing, ICIP, 2019-
September, 1415–1419. https://doi.org/10.1109/ICIP.
2019.8803103
Chen, X., Zhang, R., & Yan, P. (2019). Feature fusion
encoder decoder network for automatic liver lesion
segmentation. Proceedings - International Symposium
on Biomedical Imaging, 2019-April(Isbi), 430–433.
https://doi.org/10.1109/ISBI.2019.8759555
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C. W., & Heng, P. A.
(2018). H-DenseU-Net: Hybrid Densely Connected U-
Net for Liver and Tumor Segmentation from CT
Volumes. IEEE Transactions on Medical Imaging,
37(12), 2663–2674. https://doi.org/10.1109/TMI.2018.
2845918.
Rafiei, S., Nasr-Esfahani, E., Soroushmehr, S. M. R.,
Karimi, N., Samavi, S., & Najarian, K. (2018). Liver
segmentation in ct images using three dimensional to
two dimensional fully convolutional network. ArXiv,
2067–2071.
Xia, K., Yin, H., Qian, P., Jiang, Y., & Wang, S. (2019).
Liver semantic segmentation algorithm based on
improved deep adversarial networks in combination of
weighted loss function on abdominal CT images. IEEE
Access, 7, 96349–96358. https://doi.org/10.1109/
ACCESS.2019.2929270
Truong, T. N., Dam, V. D., & Le, T. S. (2018). Medical
Images Sequence Normalization and Augmentation:
Improve Liver Tumor Segmentation from Small Data
Set. Proceedings - 2018 3rd International Conference
on Control, Robotics and Cybernetics, CRC 2018, 1–5.
https://doi.org/10.1109/CRC.2018.00010
Zhou, Y., Wang, Y., Tang, P., Bai, S., Shen, W., Fishman,
E. K., & Yuille, A. (2019). Semi-supervised 3D
abdominal multi-organ segmentation via deep multi-
planar co-training. Proceedings - 2019 IEEE Winter
Conference on Applications of Computer Vision,
WACV 2019, 121–140. https://doi.org/10.1109/
WACV.2019.00020.
Shell, Adam (2020), How to invest in artificial intelligence,
https://www.usatoday.com/story/money/2020/01/27/ar
tificial-intelligence-how-invest/4542467002/.
Chen, S., Yang, H., Fu, J., Mei, W., Ren, S., Liu, Y., …
Chen, H. (2019). U-Net Plus: Deep Semantic
Segmentation for Esophagus and Esophageal Cancer in
Computed Tomography Images. IEEE Access, 7,
82867–82877.
https://doi.org/10.1109/ACCESS.2019.2923760
Trullo, R., Petitjean, C., Ruan, S., Dubray, B., Nie, D., &
Shen, D. (2017). Segmentation of Organs at Risk in
thoracic CT images using a SharpMask architecture and
Conditional Random Fields. Proceedings -
International Symposium on Biomedical Imaging,
1003–1006.
https://doi.org/10.1109/ISBI.2017.7950685
Trullo, R., Petitjean, C., Nie, D., Shen, D., & Ruan, S.
(2017). Fully automated esophagus segmentation with
a hierarchical deep learning approach. Proceedings of
the 2017 IEEE International Conference on Signal and
Image Processing Applications, ICSIPA 2017, 503–
506. https://doi.org/10.1109/ICSIPA.2017.8120664
Fang, L., Liu, J., Liu, J., & Mao, R. (2018). Automatic
Segmentation and 3D Reconstruction of Spine Based
on FCN and Marching Cubes in CT Volumes. (2018)
10th International Conference on Modelling,
Identification and Control (ICMIC), (Icmic), 1–5.
Tang, Z., Chen, K., Pan, M., Wang, M., & Song, Z. (2019).
An Augmentation Strategy for Medical Image
Processing Based on Statistical Shape Model and 3D
Thin Plate Spline for Deep Learning. IEEE Access, 7,
133111–133121. https://doi.org/10.1109/ACCESS.
2019.2941154
Kuok, Chan-Pang and Hsue, Jin-Yuan and Shen, Ting-Li
and Huang, Bing-Feng and Chen, Chi-Yeh and Sun, Y.-
N. (2018). Segmentation from 3D CT Images. Pacific
Neighborhood Consortium Annual Conference and
Joint Meetings (PNC), (c), 1–6.
Zhou, Y., Wang, Y., Tang, P., Bai, S., Shen, W., Fishman,
E. K., & Yuille, A. (2019). Semi-supervised 3D
abdominal multi-organ segmentation via deep multi-
planar co-training. Proceedings - 2019 IEEE Winter
Conference on Applications of Computer Vision,
Bone Segmentation of the Human Body in Computerized Tomographies using Deep Learning
25

WACV 2019, 121–140. https://doi.org/10.1109/
WACV.2019.00020.
La Rosa, F. (2017). A deep learning approach to bone
segmentation in CT scans. 66. Retrieved from AMS
Laurea Institutional Thesis Repository.
P. Fleckenstein, J. Tranum-Jensen (2004), Anatomia em
Diagnóstico Por Imagens: 2. ed.,São Paulo: Manole,.
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net:
Convolutional networks for biomedical image
segmentation. Lecture Notes in Computer Science
(Including Subseries Lecture Notes in Artificial
Intelligence and Lecture Notes in Bioinformatics),
9351, 234–241. https://doi.org/10.1007/978-3-319-
24574-4_28
Jegou, S., Drozdzal, M., Vazquez, D., Romero, A., &
Bengio, Y. (2017). The One Hundred Layers Tiramisu:
Fully Convolutional DenseNets for Semantic
Segmentation. IEEE Computer Society Conference on
Computer Vision and Pattern Recognition Workshops,
2017-July, 1175–1183. https://doi.org/10.1109/
CVPRW.2017.156
Liciotti, D., Paolanti, M., Pietrini, R., Frontoni, E., &
Zingaretti, P. (2018). Convolutional Networks for
Semantic Heads Segmentation using Top-View Depth
Data in Crowded Environment. Proceedings -
International Conference on Pattern Recognition,
2018-August, 1384–1389. https://doi.org/10.1109/
ICPR.2018.8545397
Chen, L. C., Zhu, Y., Papandreou, G., Schroff, F., & Adam,
H. (2018). Encoder-decoder with atrous separable
convolution for semantic image segmentation. Lecture
Notes in Computer Science (Including Subseries
Lecture Notes in Artificial Intelligence and Lecture
Notes in Bioinformatics), 11211 LNCS, 833–851.
https://doi.org/10.1007/978-3-030-01234-2_49
Chollet, F. (2017). Xception: Deep learning with depthwise
separable convolutions. Proceedings - 30th IEEE
Conference on Computer Vision and Pattern
Recognition, CVPR 2017, 2017-January, 1800–1807.
https://doi.org/10.1109/CVPR.2017.195
Shelhamer, E., Long, J., & Darrell, T. (2017). Fully
Convolutional Networks for Semantic Segmentation.
IEEE Transactions on Pattern Analysis and Machine
Intelligence, 39(4), 640–651. https://doi.org/10.1109/
TPAMI.2016.2572683
Simonyan, K., & Zisserman, A. (2015). Very deep
convolutional networks for large-scale image
recognition. 3rd International Conference on Learning
Representations, ICLR 2015 - Conference Track
Proceedings, 1–14.
Haukoos JS, Lewis RJ. Advanced statistics: bootstrapping
confidence intervals for statistics with "difficult"
distributions. Acad Emerg Med. 2005 Apr;12(4):360-5.
doi: 10.1197/j.aem.2004.11.018. PMID: 15805329.
CSEDU 2022 - 14th International Conference on Computer Supported Education
26
