
Bias Assessment in Medical Imaging Analysis:
A Case Study on Retinal OCT Image Classification
Gabriel Oliveira, Lucas David, Rafael Padilha, Ana Paula da Silva,
Francine de Paula, Lucas Infante, Lucio Jorge, Patricia Xavier and Zanoni Dias
Institute of Computing, University of Campinas, Campinas, SP, Brazil
Keywords:
Deep Learning, Dataset Bias, Model Interpretability, Medical image diagnosis, Retinal OCT Analysis.
Abstract:
Deep learning classifiers can achieve high accuracy in many medical imaging analysis problems. However,
when evaluating images from outside the training distribution — e.g., from new patients or generated by
different medical equipment — their performance is often hindered, highlighting that they might have learned
specific characteristics and biases of the training set and can not generalize to real-world scenarios. In this
work, we discuss how Transfer Learning, the standard training technique employed in most visual medical
tasks in the literature, coupled with small and poorly collected datasets, can induce the model to capture such
biases and data collection artifacts. We use the classification of eye diseases from retinal OCT images as the
backdrop for our discussion, evaluating several well-established convolutional neural network architectures
for this problem. Our experiments showed that models can achieve high accuracy in this problem, yet when
we interpret their decisions and learned features, they often pay attention to regions of the images unrelated to
diseases.
1 INTRODUCTION
With data-driven approaches achieving promising
results in many inference tasks, numerous deep
learning-based methods were proposed in the past
decade for the medical domain (Litjens et al., 2017).
Many works focus on obtaining highly accurate mod-
els for tasks such as diagnostics, medical imaging
analysis, referral assessment, drug discovery — all of
which could significantly improve healthcare in our
society. Although important, accuracy alone is not
enough for an automatic approach to be applied in a
real scenario in which its decision might affect peo-
ple’s lives. In such cases, it is essential to guarantee
the transparency and interpretability of the model, as-
sessing the factors that influence each automatic an-
swer. Doing so improves the trustworthiness of the
algorithm, which is pivotal for its acceptance in prac-
tical scenarios.
When transparency is overlooked, black-box
models might output correct answers for the wrong
reasons. A recent example of this occurred during
the COVID-19 outbreak. Moved by the urgency of
the pandemic, a large body of works proposed image-
based diagnostic methods that reported high accuracy
in diverse scenarios (Shi et al., 2020). However, in
a recent analysis (Roberts et al., 2021), the authors
evaluated a pool of 415 works proposing machine
learning-based approaches for COVID-19 diagnostic
through chest X-ray and computational tomography
scans. According to their assessment, none of the
works could be used in clinical practice, primarily due
to biases and inference flaws learned by the model
during training. Such undesired effects may originate
from methodological errors during dataset collection
and sanitization that are mistakenly leveraged during
model optimization. For example, if images from a
patient present a frequent acquisition artifact that dis-
tinguishes them from other patients’, the model might
learn to identify that artifact to classify a disease in-
stead of learning the correct features for a diagnosis.
Deep learning models often consist of complex
and parameter-heavy neural networks able to directly
learn the most discriminative characteristics for the
target problem from available data. To properly learn
them, most models require vast amounts of annotated
data, which are not always available, especially in the
medical domain. To overcome such limitations, re-
searchers employ Transfer Learning, pre-training the
model on a different domain with plenty of data and
further fine-tuning the acquired knowledge to the tar-
get task. However, when trained with datasets whose
collection and sanitization were not rigorously per-
formed to mitigate possible biases (e.g., lack of pa-
tient and sensor representativity, data leakage during
the organization of training and validation splits), the
574
Oliveira, G., David, L., Padilha, R., Paula da Silva, A., de Paula, F., Infante, L., Jorge, L., Xavier, P. and Dias, Z.
Bias Assessment in Medical Imaging Analysis: A Case Study on Retinal OCT Image Classification.
DOI: 10.5220/0010867400003116
In Proceedings of the 14th International Conference on Agents and Artificial Intelligence (ICAART 2022) - Volume 3, pages 574-580
ISBN: 978-989-758-547-0; ISSN: 2184-433X
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

optimization may lead the model to learn whichever
features better solve the task, regardless of their qual-
ity. Consequently, this behavior might reflect in an
artificially high accuracy that does not generalize to
real application scenarios. Once trained, it becomes
hard to identify and fix such issues due to their black-
box nature and the insufficient reproducibility details
presented by most works.
In this work, we extend the discussion about
dataset biases and how modern data-driven tech-
niques are prone to capture them instead of focusing
on the task at hand. We examine the training proce-
dure frequently resorted by deep learning-based ap-
proaches and discuss the characteristics of medical
datasets employed in their training that might lead to
low generalization at inference time. To better exem-
plify these issues and what type of artifacts and bi-
ases are captured in a real scenario, we analyze convo-
lutional neural networks (CNN) to diagnose eye dis-
eases on retinal optical coherence tomography (OCT)
scans. Finally, we discuss interpretability techniques
that can aid us in identifying and mitigating biases
captured during training.
The remainder of the work is organized as follows.
In Section 2, we discuss how researchers leverage
datasets from other domains with the Transfer Learn-
ing technique, as well as the issues and biases that
originated during dataset collection and sanitization
that might affect generalization. Whereas, in Sec-
tion 3, we present the case analysis of retinal OCT
classification, evaluating several deep learning mod-
els under an experimental scenario and interpreting
their decisions. Finally, in Section 4, we discuss our
final thoughts, highlighting the importance of explain-
ability approaches for bias mitigation.
2 RELATED CONCEPTS
Convolutional Neural Networks have achieved
promising results in different visual domains, includ-
ing medical problems (Deepak and Ameer, 2019;
Oliveira et al., 2020). Due to their data requirement
and the lack of annotated data in medical tasks, most
methods in the literature employ Transfer Learning as
the standard approach to deal with smaller datasets.
The technique originated from educational psychol-
ogy, which states that experiences in one domain
can be generalized to another (Zhuang et al., 2021).
This approach is widely used by the deep learning
community as it alleviates the data requirements for
complex models whose training from scratch would
be unfeasible (Tan et al., 2018).
The rationale behind Transfer Learning is that,
once optimized, the initial and intermediate layers of
CNNs tend to capture low-level information in im-
ages, such as edges, corners, and color blobs. These
superficial representations are often shared between
tasks of distinct visual domains (Yosinski et al., 2014;
Hussain et al., 2018) — from object classification
to medical imaging analysis — allowing them to be
transferred for new tasks. On the other hand, deeper
layers build on top of the low-level concepts, learning
domain-specific characteristics specialized in the task
at hand that cannot be applied to other problems.
The pre-trained models are optimized in large an-
notated datasets before having their features trans-
ferred. Most works pre-train their approaches on
ImageNet (Deng et al., 2009), an object classifica-
tion dataset with 14 million images and 1000 classes.
The adaptation and adjustment to the new domain are
made by fine-tuning the weights of the CNN with the
target dataset, using the pre-trained weights as the
starting point for the optimization. This is usually
done with a lower learning rate, as the objective is
solely to tweak (and not to re-train) the network to
generalize for the new domain.
Even though the training procedure plays a crucial
part in the performance of the final model, it is only
one of its critical components. The quality of data
has a significant impact on the generalization of deep
learning models. Due to the sensitive nature of med-
ical tasks, biases and flaws introduced in the dataset
during its collection and sanitization could have se-
vere repercussions during inference in real-world sce-
narios. As discussed by previous works (Roberts
et al., 2021), dataset bias has many possible origins
that should be considered, but most of them tend to
fall under the lack of data representativity, flaws in the
collection process, and data leakage or contamination
during training and evaluation.
Medical images are captured with expensive
equipment (e.g., computed tomography, ultrasound,
and optical coherence tomography) that can differ in
characteristics and parameters from one machine to
another even when considering similar models (Wu
et al., 2018). Additionally, images are captured from
several patients, usually following a protocol oriented
by a technician. Issues with the equipment (e.g., sen-
sor noises from a particular machine), in the collec-
tion procedure (e.g., the person moving during the
exam), and patient profile (e.g., imagery captured
from a particular age and gender), if not accounted,
can all introduce artifacts that might be exploited by
the model afterward.
Besides those, several issues can be introduced
when organizing the dataset into training and testing
Bias Assessment in Medical Imaging Analysis: A Case Study on Retinal OCT Image Classification
575
splits, as well as posteriorly during the model evalu-
ation. Data leakage is a methodological mistake that
can have subtle consequences. For example, having
shared patients between splits might induce the model
to recognize the patient’s identity (through body char-
acteristics or how that person posed for capture) in-
stead of focusing on the disease features. A similar
issue can happen when the data from one class of
our task is all captured by the same equipment that
was not used for patients of other classes, allowing
the model to correlate the sensor noise of the machine
with the diagnostic.
3 CASE ANALYSIS:
CLASSIFICATION OF
RETINAL OCT IMAGES
Vision impairment is a growing medical concern in
our society,
1
in which early diagnosis plays an im-
portant role in prevention and treatment assessment.
Approximately 30 million OCT scans are performed
each year worldwide(Swanson and Fujimoto, 2017),
requiring extensive human supervision to filter and
analyze potential patients. Considering this, auto-
matic medical imaging analysis methods are becom-
ing important tools to process the high volume of
scans in a timely and effective manner.
In this section, we evaluate several CNN architec-
tures over a scenario of classification of eye diseases
through the analysis of retinal OCT scans. Firstly, we
describe the employed dataset and its properties. We
then discuss the methodology and the experimental
evaluation setup. Finally, we present the results and
interpret them using explainability techniques.
3.1 Dataset
The dataset used in this work was collected from pa-
tients from several hospitals and ophthalmology in-
stitutes in the USA and China between 2013 and
2017 (Kermany et al., 2018b; Kermany et al., 2018a).
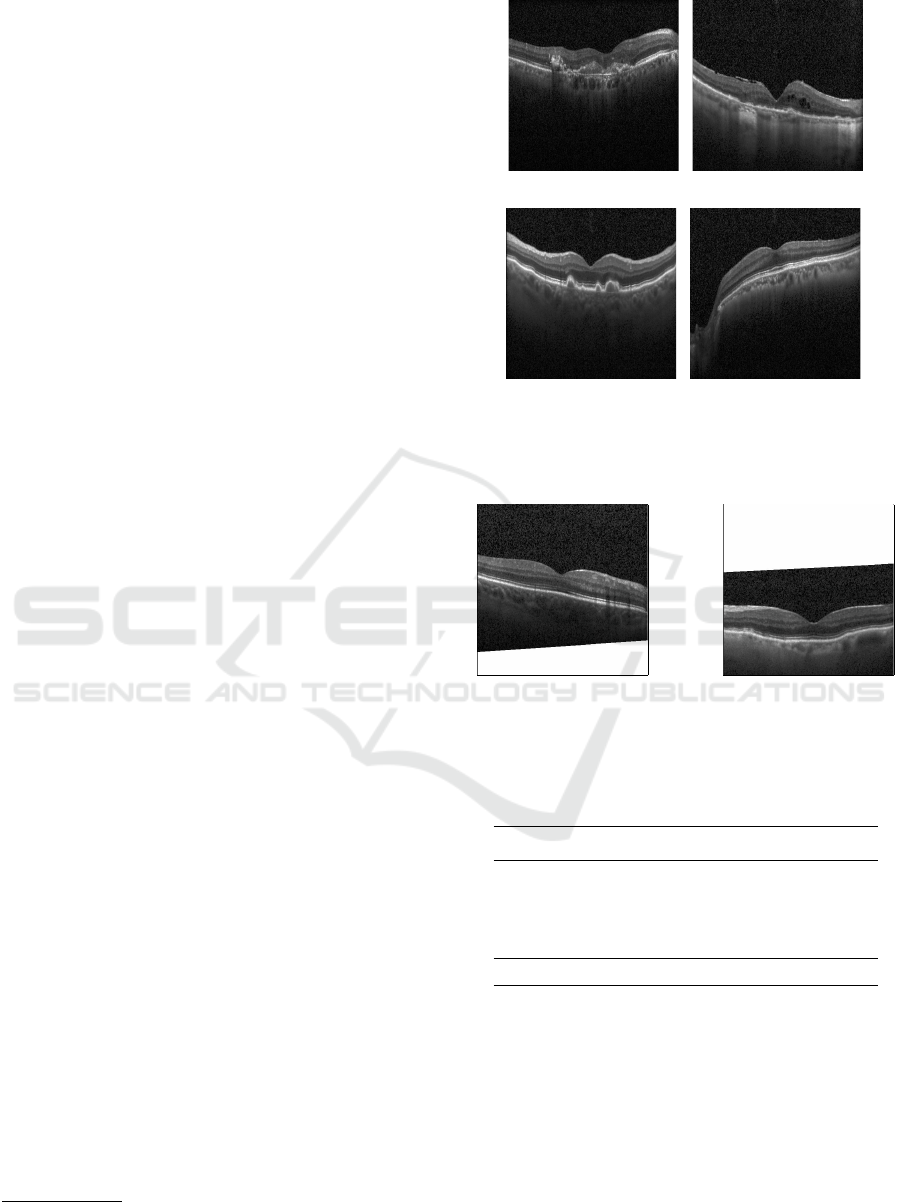
Each OCT scan belongs to one of four classes:
Choroidal Neovascularization (CNV), Diabetic Mac-
ular Edema (DME), Drusen, and Normal. Images as-
signed as Normal indicate that they belong to healthy
patients without any sign of diseases, such as fluids
or edemas. Figure 1 presents examples of each avail-
able class. Several OCT images in the dataset present
some type of noise, such as in- and out-of-plane ro-
tations and image shearing, as presented in Figure 2.
1
https://www.who.int/en/news-room/fact-sheets/detail/
blindness-and-visual-impairment
(a) (b)
(c) (d)
Figure 1: Examples of OCT scans from (a) Choroidal
Neovascularization (CNV), (b) Diabetic Macular Edema
(DME), (c) Drusen, and (d) Normal classes.
Figure 2: Examples of noisy OCT images, with different
degrees of rotation, crop and shearing.
Table 1: OCT images distribution on training, validation,
and testing sets. Class distribution is unbalanced on the
training and validation sets, but balanced on the testing set.
Class Train Validation Test
CNV 19,115 3,245 242
DME 5,484 1,414 242
Drusen 3,269 590 242
Normal 18,214 4,466 242
Total 46,082 9,715 968
Besides that, images vary in resolution, ranging from
512 × 512 up to 1536 × 496.
The dataset has already been organized into train-
ing, validation and testing splits. However, when ana-
lyzing the split composition, we noticed that some pa-
tients had images in multiple sets. By sharing patients
between the sets, the models might be encouraged
to learn patient-specific characteristics instead of dis-
criminative features of the diseases. Even though this
might lead to a higher classification accuracy on the
testing set, it is an undesired effect as it hinders their
ICAART 2022 - 14th International Conference on Agents and Artificial Intelligence
576
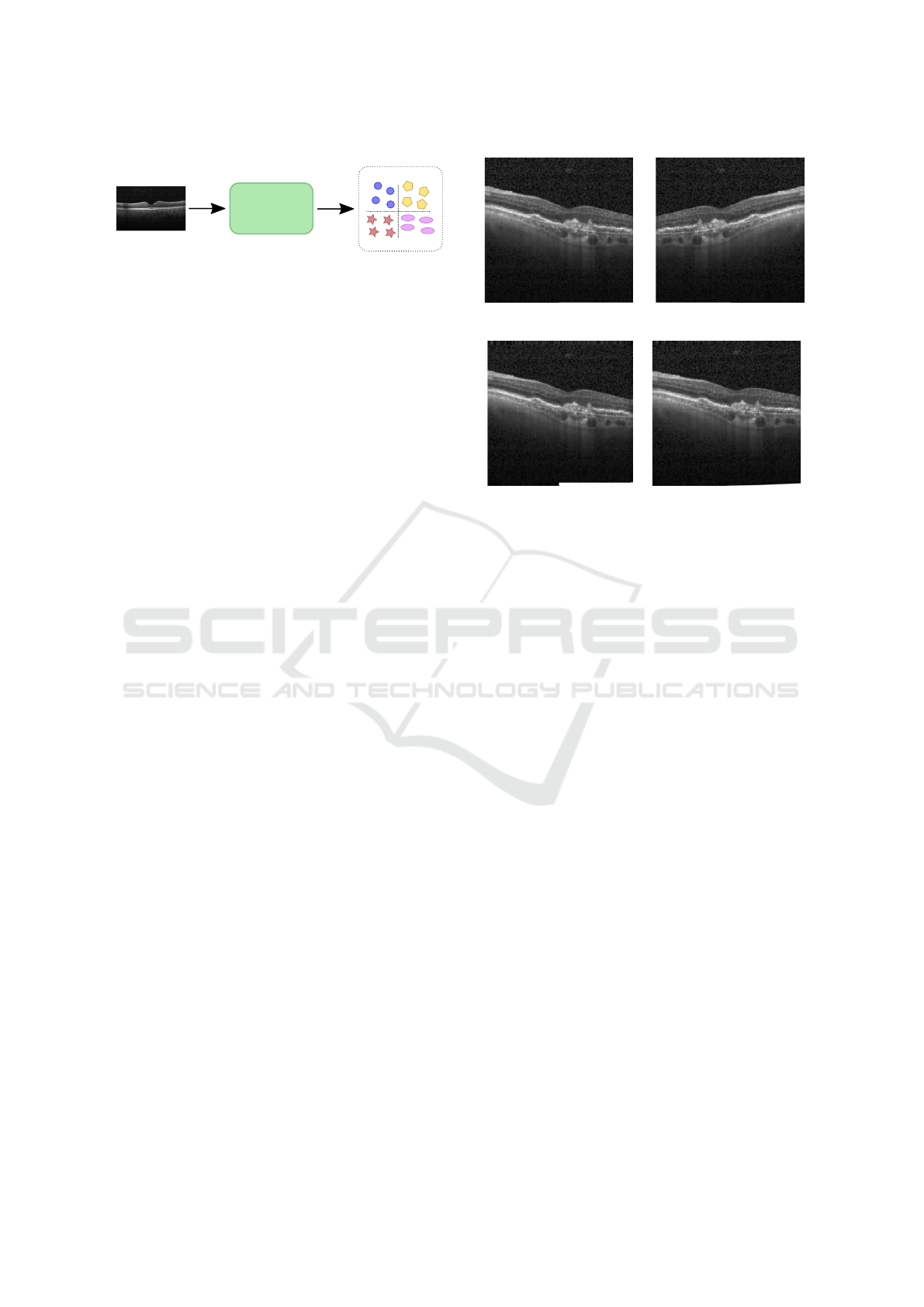
CNN
CNV
DME
Drusen
Normal
Classification
Figure 3: Pipeline of our method.
generalization to scans from new patients. To avoid
data leakage and not harm the learning capability of
the models, we redefined the training and validation,
removing the images from patients that are also pre-
sented on the testing set. We gathered all the images
from the training and validation sets and divided them
into 80% for training and 20% for validation, ensur-
ing that the same patient was present in only one of
the sets. Table 1 shows the number of OTC scans on
training, validation, and testing sets with the new con-
figuration.
3.2 Methodology
In this subsection, we present our approach for the
problem of classification of retina-related diseases.
The pipeline of our method is presented in Figure 3.
Firstly, we employ well-established CNN archi-
tectures, pre-trained over the ImageNet dataset (Deng
et al., 2009), to perform feature extraction. The
output features are summarized with a Global Aver-
age Pooling layer (GAP) and used to train a dense
softmax classifier. We experiment with the fol-
lowing architectures: ResNet-50 (He et al., 2016),
MobileNet (Howard et al., 2017), VGG-16 and
VGG-19 (Simonyan and Zisserman, 2014), Efficient-
NetB0 (Tan and Le, 2019), and InceptionV3 (Szegedy
et al., 2016). For each feature extracting network, the
softmax classifier is trained for ten epochs using Nes-
terov Momentum SGD optimizer for the categorical
cross-entropy loss function. We consider a learning
rate of 0.001 and a momentum factor of 0.9. Also, we
use the early stopping technique to halt the training
after four epochs without decreasing the loss function
on the validation set.
In a second phase, we select the topmost scoring
architecture from the previous step and fine-tune it
over the Retinal OCT dataset. Unlike the previous
step, in which the networks’ weights were fixed, we
allow the CNN to update its parameters to match the
training OCT images to their respective labels. The
same training procedure, as well as hyperparameters,
are employed in this phase.
In both experiments, we augment the training data
using these three operations: random horizontal flip,
random zoom in the range of [0.8, 1.2], and random
(a) (b)
(c) (d)
Figure 4: Examples of data augmentation: (a) original im-
age, (b) horizontal flip, (c) zoom, and (d) shear.
shear in the range of [0.8, 1.2]. Figure 4 shows these
three operations of data augmentation.
3.3 Experimental Evaluation
In this subsection, we present the experimental eval-
uation of different CNN architectures considering
transfer learning and fine-tuning techniques. Firstly,
we report the feature extraction results using CNNs
trained over the ImageNet dataset. Then we report
and discuss the results of the fine-tuning experiment
and, finally, we evaluate the highest performing net-
work against the test set. Finally, we consider the in-
terpretability technique Grad-CAM (Selvaraju et al.,
2017) to evaluate the features being utilized by the
model’s decision process.
3.3.1 Transfer Learning with ImageNet Weights
The result of each architecture is presented in Ta-
ble 2. ResNet50 reached the highest balanced accu-
racy on the validation set between the selected archi-
tectures, with a score of 79.75%. The remaining net-
works achieved similar results, with InceptionV3 out-
performed by all of them.
3.3.2 Fine-tuned Network
With our results considering the convolutional neural
network architectures on the validation set with trans-
Bias Assessment in Medical Imaging Analysis: A Case Study on Retinal OCT Image Classification
577

Table 2: Results of transfer learning and fine-tuning on the validation set. ResNet50 (He et al., 2016) with transfer learning
achieved the highest balanced accuracy on the validation set. With that, we fine-tuned this architecture, outperforming the
previous results with transfer learning.
Network Balanced Accuracy (%)
Transfer Learning
ResNet50 (He et al., 2016) 79.75
MobileNet (Howard et al., 2017) 79.68
VGG-16 (Simonyan and Zisserman, 2014) 79.21
VGG-19 (Simonyan and Zisserman, 2014) 78.96
EfficientNetB0 (Tan and Le, 2019) 78.32
InceptionV3 (Szegedy et al., 2016) 75.98
Fine-tuning
ResNet50 (He et al., 2016) 89.95
Figure 5: Progress of the loss function for the training and
validation sets throughout the optimization of the fine-tuned
ResNet50.
fer learning technique, we performed further investi-
gations with the best CNN.
The best result was reached by ResNet50, which
achieved 79.75% of balanced accuracy on the valida-
tion set. With that, we employed the fine-tuning tech-
nique, i.e., we retrained ResNet50, initialized with
ImageNet weights, but allowing them to be freely up-
dated during optimization.
Added to the fine-tuning process, we analyzed the
loss function during the training step, as shown in
Figure 5. The loss curve of training and validation
phases indicates that the training converged quickly
to a global minimum in the training set. However, the
validation loss stayed close to the same point, stop-
ping the training process due to the early-stopping
technique. Considering the curve, we can highlight
the impact of the hyperparameters chosen for the
SGD optimizer, and further investigations can be done
with different values for the hyperparameters.
As a result, the fine-tuned ResNet50 obtained
89.95% of balanced accuracy on the validation set, as
shown in Table 2. We concluded that the fine-tuning
Figure 6: Confusion matrix of the predictions on the test
set.
technique is paramount to adapt the network trained
over a general problem domain to the Retinal OCT
images domain.
3.3.3 Test Set Evaluation
Driven by the results on the validation set, we evaluate
the fine-tuned ResNet50 model on the test set, achiev-
ing a balanced accuracy of 98.04%. We conclude that
our method can generalize the knowledge learned on
the training set to new and unseen images.
Additionally, we generated the confusion matrix
of the predictions on the test set and present it in Fig-
ure 6, which indicates that our method achieves more
than 90% of accuracy in each class. Also, our method
had 0% of false positive and false negative classifi-
cations for the Normal class, i.e., none of the patients
with some disease (CNV, DME, or Drusen) were clas-
sified as Normal, nor healthy patients were misdiag-
nosed with CNV, DMR or Drusen. This is particularly
important considering a triage scenario, in which an
automatic model would decide if a patient needs to
be analyzed by expert clinicians or not. However, the
classification of the Drusen class had 5.8% of mis-
ICAART 2022 - 14th International Conference on Agents and Artificial Intelligence
578
takes for CNV class, showing that further investiga-
tions can be done to mitigate these wrong predictions.
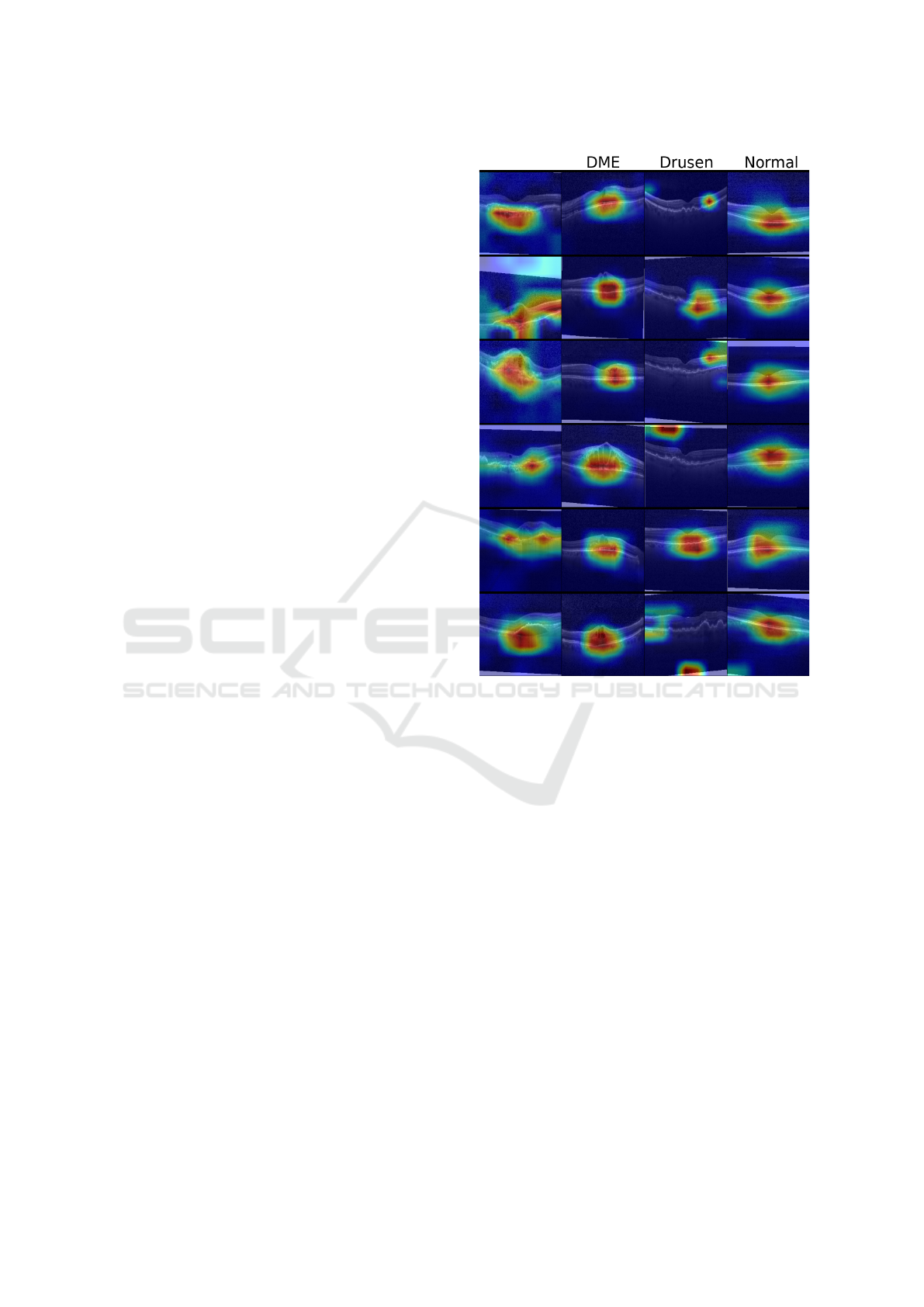
3.3.4 Interpreting Model Decisions
While our goal is to correctly distinguish among the
existing diseases present in OCT scans, developing
models with resilient and transparent decision rules is
also paramount. In this vein, we use Grad-CAM (Sel-
varaju et al., 2017), an AI explaining technique based
on Class Activation Maps, to highlight the most con-
tributing regions considered by the softmax classifier
in its decision process. Figure 7 illustrates the class-
based salient regions for multiple images in our test-
ing set.
We observe that the model focuses on reasonable
regions of interest when classifying samples belong-
ing to the classes CNV, DME, and Normal. On the
other hand, it has incorrectly focused on artificial cap-
turing properties (e.g., the presence of image borders
created by incorrect placement of the OCT sample
during capturing) on two degenerated cases of the
class Drusen. This indicates that the model tends to
use characteristics unrelated to the disease itself to
classify a scan. This goes in accordance with the re-
sults presented in Figure 6, whose errors might be a
reflex of the model incorrectly identifying unrelated
artifacts in the image. Besides that, these cases show
that additional preprocessing and regularization steps
that remove such capturing artifacts might lead to a
more robust model that correctly focuses on the dis-
criminative features of Drusen.
4 CONCLUSION
The use of deep learning to classify medical images
proved to be effective in multiple medical problems,
achieving high accuracy and potentially alleviating
the need for manual inspection. In practice, however,
automatic approaches are still far from being success-
fully deployed in most real-world scenarios. Deep
neural networks are complex non-linear models prone
to capture biases and flaws in the dataset, exploiting
them during training but failing to generalize to un-
seen data.
This work discusses potential biases that may
be introduced during dataset collection, organization,
and model training. To exemplify them, we evaluated
convolutional neural networks for retinal OCT image
classification of eye diseases — Choroidal Neovascu-
larization, Diabetic Macular Edema, and Drusen. We
employed transfer learning for different CNN archi-
tectures, training a fully-connected layer on top of the
CNV
Figure 7: Class-based saliency maps for samples in the test
set. Samples are labeled as, from the left-most to the right-
most columns: CVN, DME, Drusen and Normal.
extracted features. The best architecture, a ResNet50,
achieved a balanced accuracy of 79.75% on the vali-
dation set. In an additional experiment, we fine-tuned
it, allowing all network weights to be freely updated
for this task, which considerably improved the re-
sults to 89.95% on the same set. When evaluating
the test set, our method obtained a balanced accuracy
of 98.04%.
Interpretability experiments highlighted that the
model correctly considers relevant retinal regions for
most classes. However, for Drusen samples, the
model exploits artifacts on the image border instead
of focusing on discriminative portions of the retina.
These are common telltales that the network has in-
correctly captured a potential bias (in this case, due
to the noise in training images) that would probably
affect its performance in unseen Drusen imagery. Be-
sides that, such behavior indicates that a more rigor-
ous capturing procedure and preprocessing step could
improve model robustness and confidence for the im-
plementation in real-world scenarios.
Bias Assessment in Medical Imaging Analysis: A Case Study on Retinal OCT Image Classification
579

In future work, we will investigate further the pos-
sible biases present in medical imaging analysis prob-
lems. We will extend the evaluation to other datasets,
employing interpretability techniques to aid us in cat-
egorizing existing biases in data. Additionally, we
will investigate which preprocessing techniques are
viable to reduce the impact of noise and acquisition
artifacts of images on model performance.
ACKNOWLEDGMENTS
This research was supported by S
˜
ao Paulo Research
Foundation (FAPESP) [grant numbers 2015/11937-
9, 2017/12646-3 and 2017/21957-2], and the Na-
tional Council for Scientific and Technological De-
velopment (CNPq) [grant numbers 140929/2021-5,
161015/2021-2 and 304380/2018-0].
REFERENCES
Deepak, S. and Ameer, P. (2019). Brain tumor classification
using deep cnn features via transfer learning. Comput-
ers in Biology and Medicine, 111:103345.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei,
L. (2009). ImageNet: A large-scale hierarchical image
database. In IEEE International Conference on Com-
puter Vision and Pattern Recognition (CVPR), pages
248–255.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In IEEE Inter-
national Conference on Computer Vision and Pattern
Recognition (CVPR), pages 770–778.
Howard, A. G., Zhu, M., Chen, B., Kalenichenko,
D., Wang, W., Weyand, T., Andreetto, M., and
Adam, H. (2017). MobileNets: Efficient convolu-
tional neural networks for mobile vision applications.
arXiv:1704.04861.
Hussain, M., Bird, J. J., and Faria, D. R. (2018). A Study on
CNN Transfer Learning for Image Classification. In
UK Workshop on Computational Intelligence (UKCI),
pages 191–202. Springer.
Kermany, D., Zhang, K., and Goldbaum, M. (2018a).
Labeled Optical Coherence Tomography (OCT) and
Chest X-Ray Images for Classification. Mendeley
Data, 2(2).
Kermany, D. S., Goldbaum, M., Cai, W., Valentim, C. C.,
Liang, H., Baxter, S. L., McKeown, A., Yang, G., Wu,
X., Yan, F., Dong, J. D., Prasadha, M. K., Pei, J., Ting,
M. Y., Zhu, J., Li, C., Hewett, S., Dong, J., Ziyar, I.,
Shi, A., Zhang, R., Zheng, L., Hou, R., Shi, W., Fu,
X., Duan, Y., Huu, V. A., Wen, C., Zhang, E. D. Z.,
Zhang, C. L., Li, O., Wang, X., Singer, M. A., Sun, X.,
Xu, J., Tafreshi, A., Lewis, M. A., Xia, H., and Zhang,
K. (2018b). Identifying Medical Diagnoses and Treat-
able Diseases by Image-Based Deep Learning. Cell,
172(5):1122–1131.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., Van Der Laak, J. A.,
Van Ginneken, B., and S
´
anchez, C. I. (2017). A survey
on deep learning in medical image analysis. Medical
Image Analysis, 42:60–88.
Oliveira, G., Padilha, R., Dorte, A., Cereda, L., Miyazaki,
L., Lopes, M., and Dias, Z. (2020). COVID-19 X-
ray Image Diagnostic with Deep Neural Networks. In
2020 Brazilian Symposium on Bioinformatics (BSB),
pages 57–68. Springer.
Roberts, M., Driggs, D., Thorpe, M., Gilbey, J., Yeung,
M., Ursprung, S., Aviles-Rivero, A. I., Etmann, C.,
McCague, C., Beer, L., et al. (2021). Common pitfalls
and recommendations for using machine learning to
detect and prognosticate for covid-19 using chest ra-
diographs and ct scans. Nature Machine Intelligence,
3(3):199–217.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2017). Grad-cam: Visual
explanations from deep networks via gradient-based
localization. In IEEE International Conference on
Computer Vision (ICCV), pages 618–626.
Shi, F., Wang, J., Shi, J., Wu, Z., Wang, Q., Tang, Z., He, K.,
Shi, Y., and Shen, D. (2020). Review of artificial in-
telligence techniques in imaging data acquisition, seg-
mentation, and diagnosis for COVID-19. IEEE Re-
views in Biomedical Engineering, 14:4–15.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv:1409.1556, pages 1–14.
Swanson, E. A. and Fujimoto, J. G. (2017). The ecosystem
that powered the translation of oct from fundamental
research to clinical and commercial impact. Biomedi-
cal Optics Express, 8(3):1638–1664.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wojna,
Z. (2016). Rethinking the inception architecture for
computer vision. In IEEE International Conference
on Computer Vision and Pattern Recognition (CVPR),
pages 2818–2826.
Tan, C., Sun, F., Kong, T., Zhang, W., Yang, C., and Liu, C.
(2018). A Survey on Deep Transfer Learning. In In-
ternational Conference on Artificial Neural Networks
(ICANN), pages 270–279. Springer.
Tan, M. and Le, Q. (2019). Efficientnet: Rethinking model
scaling for convolutional neural networks. In IEEE In-
ternational Conference on Machine Learning (ICML),
pages 6105–6114.
Wu, J., Ruan, S., Lian, C., Mutic, S., Anastasio, M. A.,
and Li, H. (2018). Active learning with noise mod-
eling for medical image annotation. In 15th Inter-
national Symposium on Biomedical Imaging (ISBI),
pages 298–301. IEEE.
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014).
How transferable are features in deep neural net-
works? In Advances in Neural Information Process-
ing Systems (NIPS), pages 3320–3328.
Zhuang, F., Qi, Z., Duan, K., Xi, D., Zhu, Y., Zhu, H.,
Xiong, H., and He, Q. (2021). A Comprehensive Sur-
vey on Transfer Learning. Proceedings of the IEEE,
109(1):43–76.
ICAART 2022 - 14th International Conference on Agents and Artificial Intelligence
580
